Phylogenetic and Expression Analysis of Fos Transcription Factors in Zebrafish
Abstract
1. Introduction
2. Results
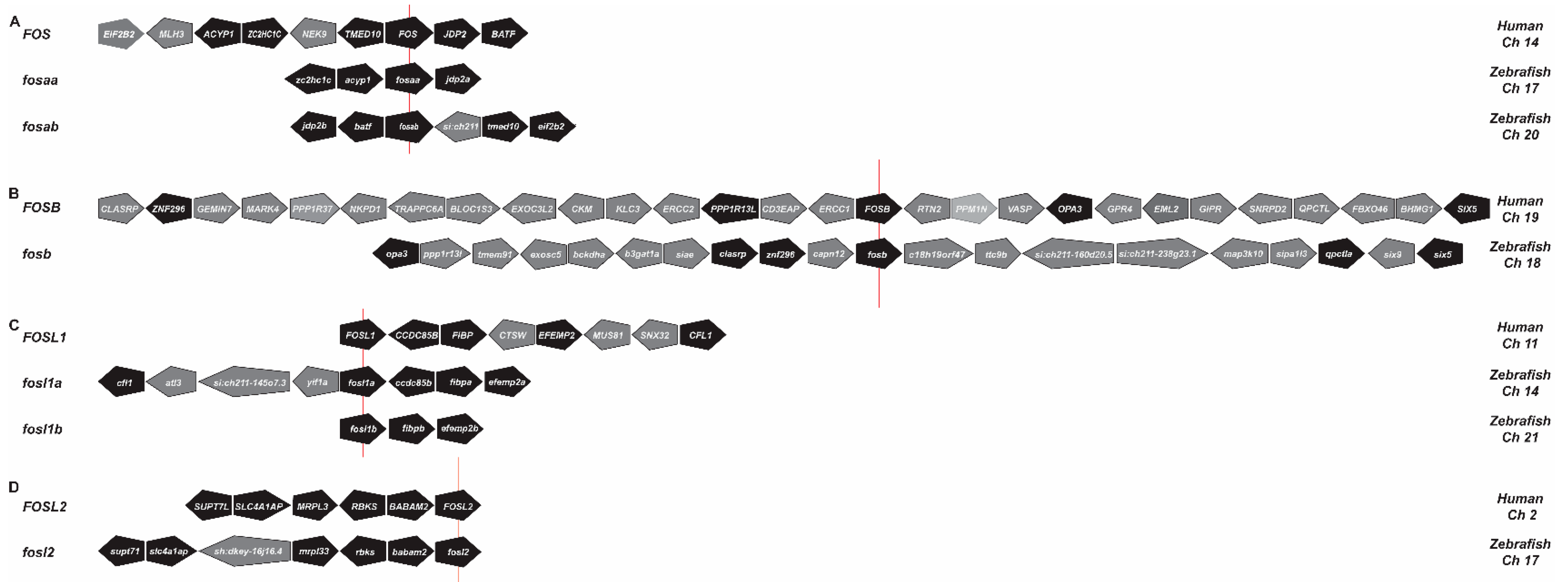
2.1. Identification of Zebrafish Fos Proteins
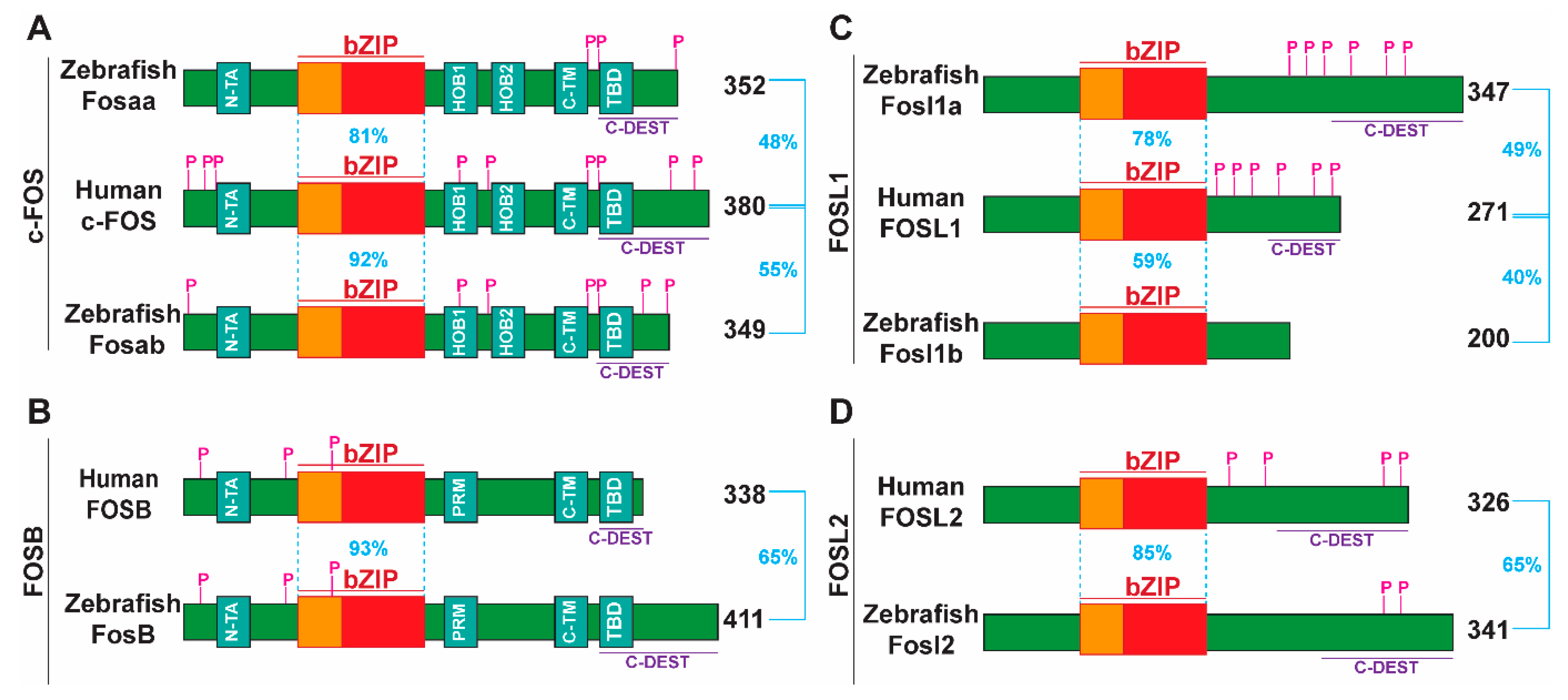
2.2. Major Functional Domains and Post-Translationally Modified Residues in Human FOS Proteins Are Conserved in Their Zebrafish Counterparts
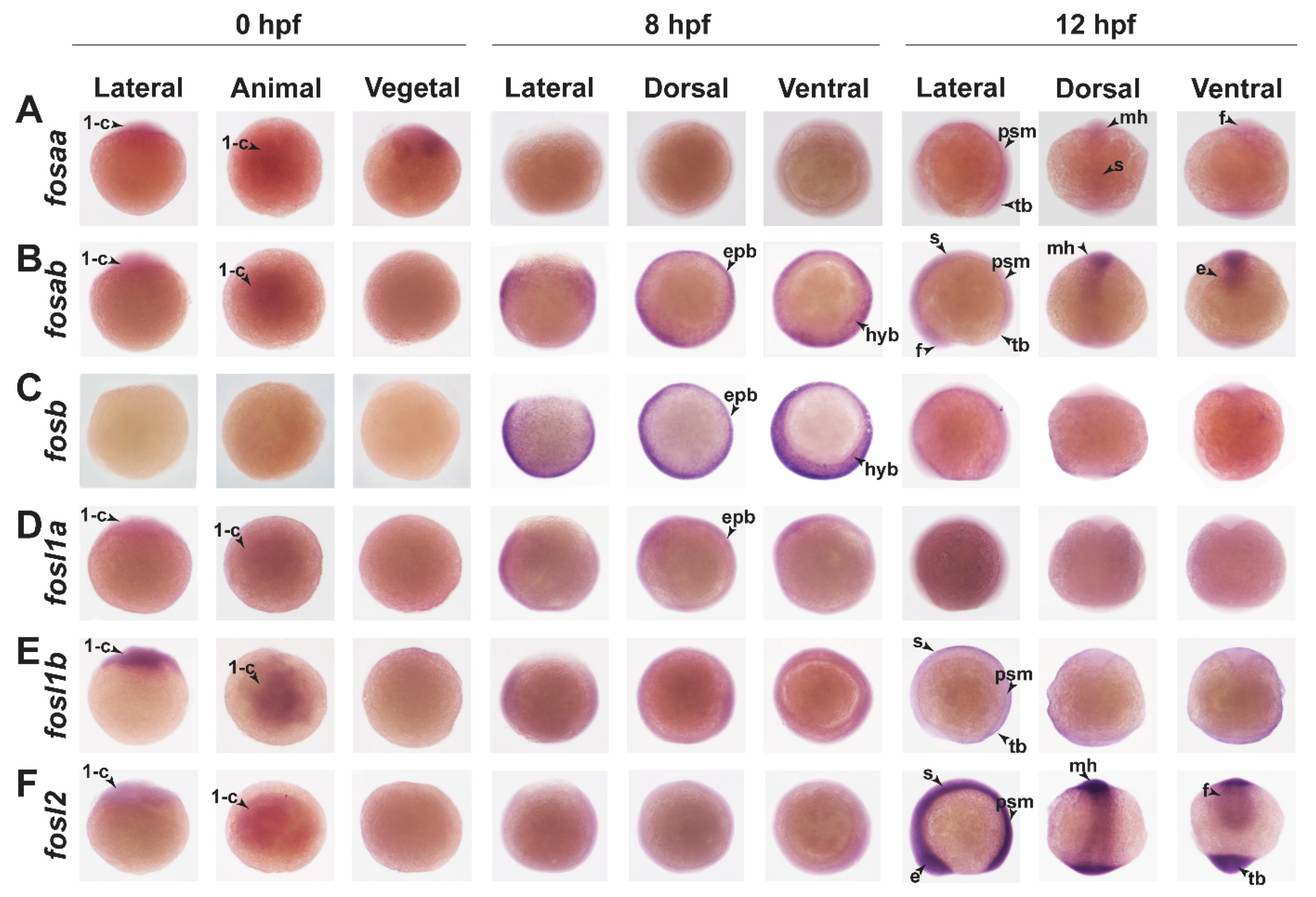
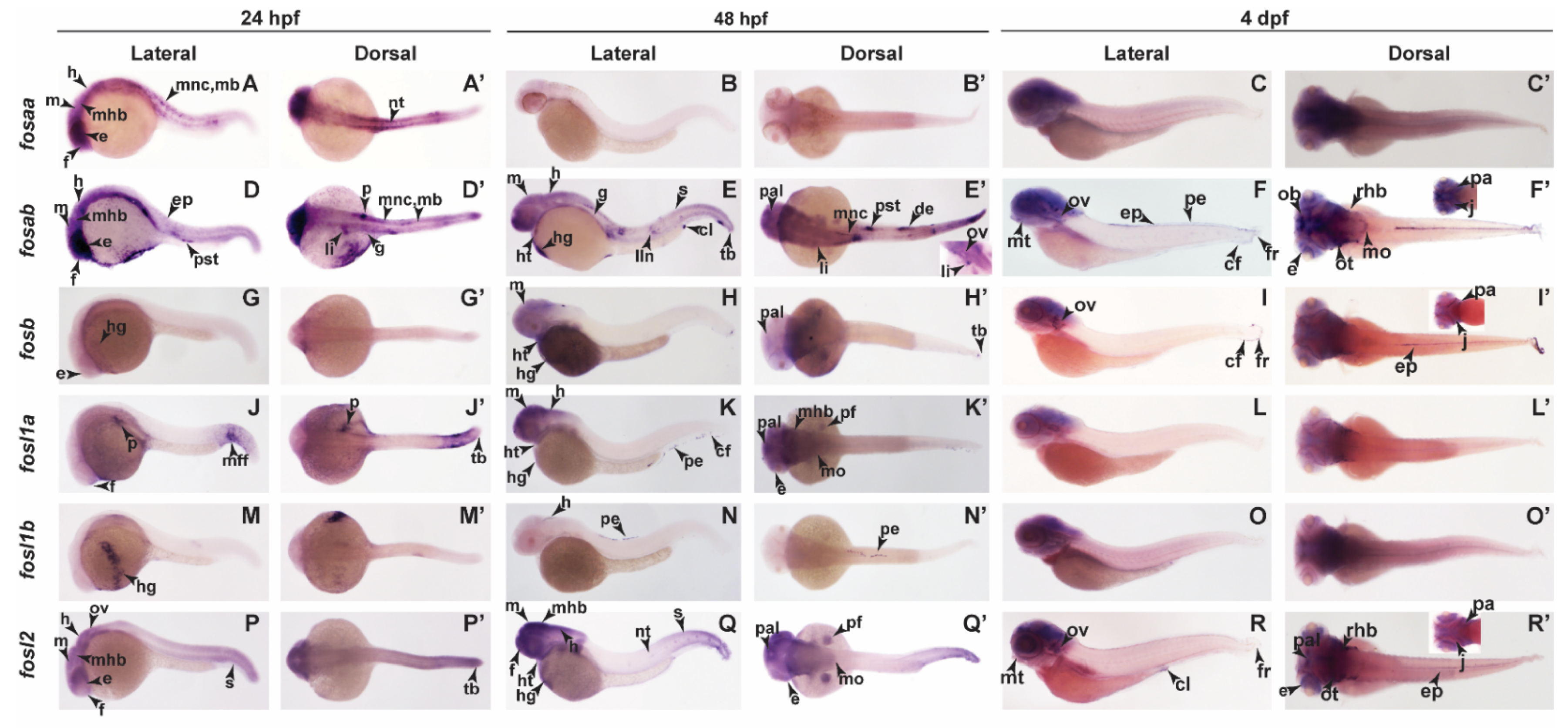
2.3. Zebrafish Fos Genes Show Distinct Expression Patterns during Embryonic Development
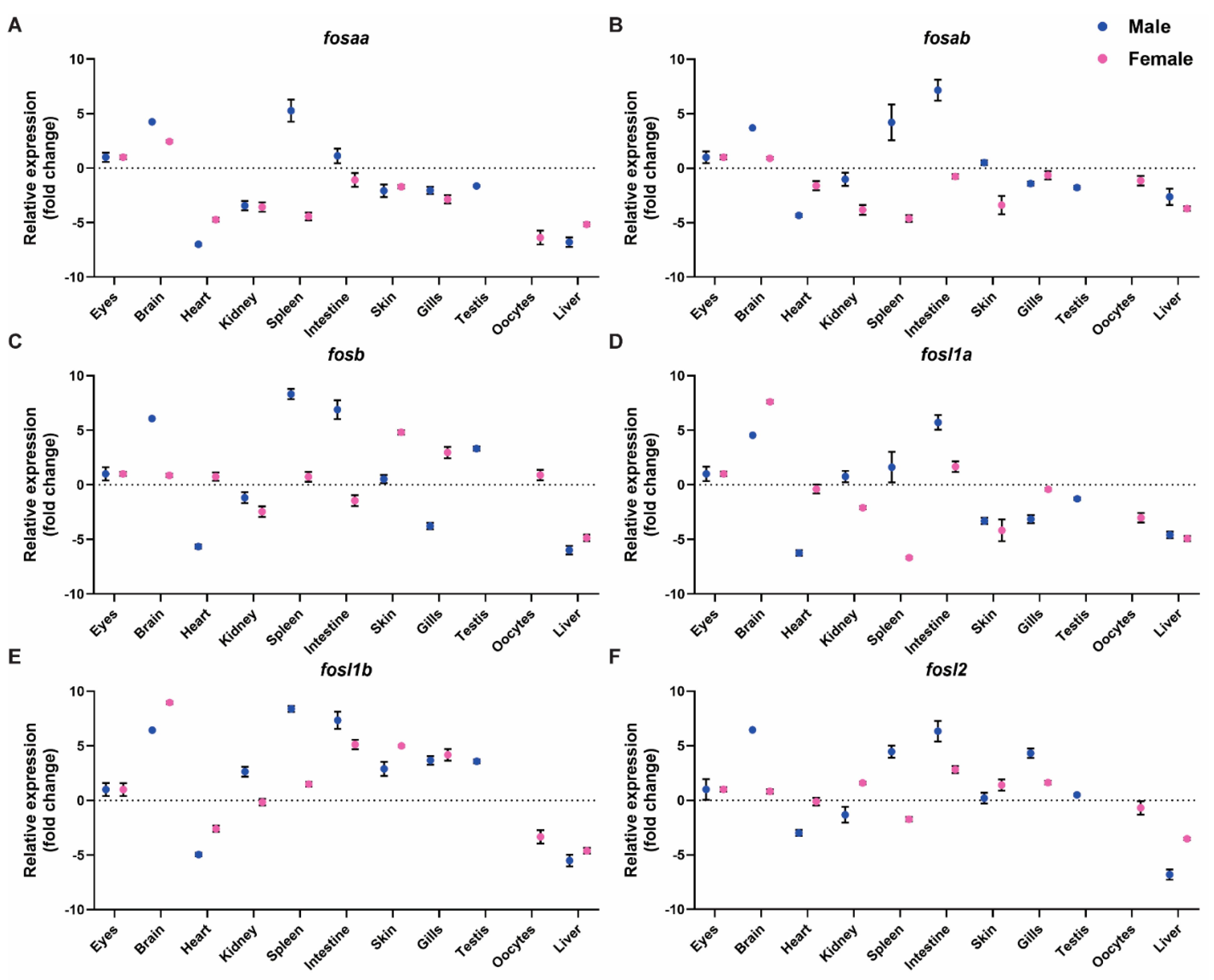
2.4. Expression of FOS Genes in Adult Zebrafish Tissues
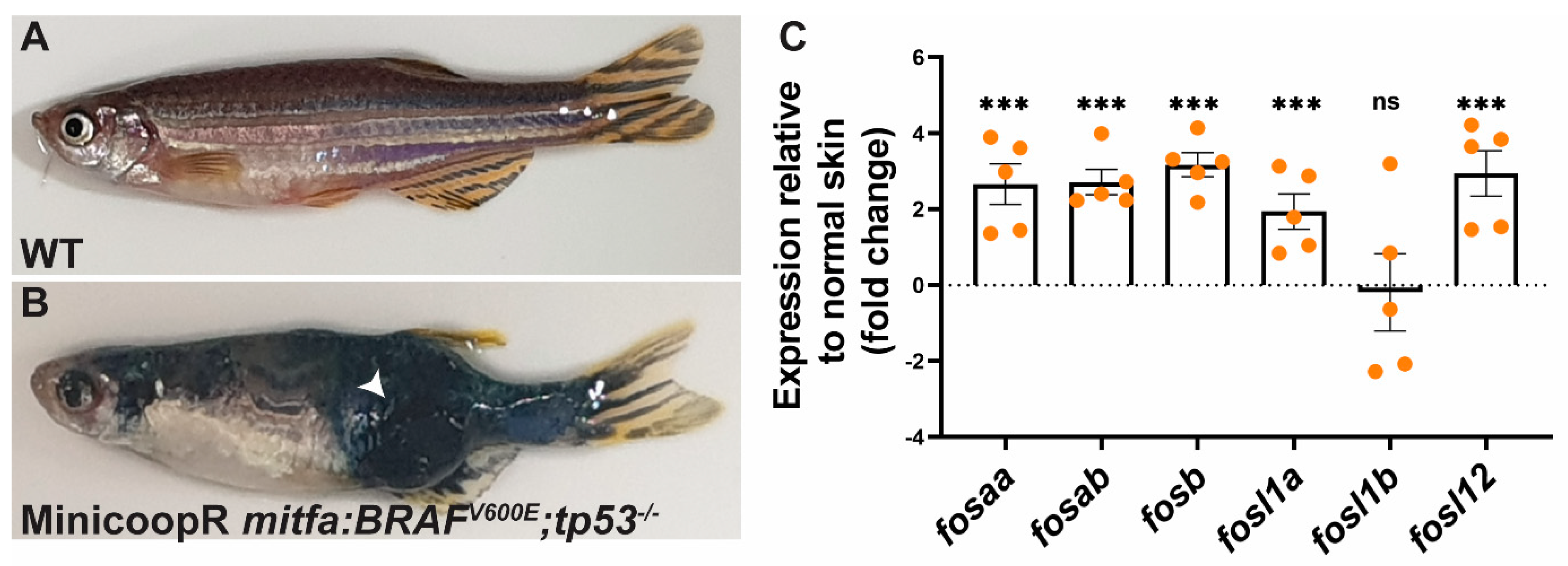
2.5. Fos Genes Are Induced in BRAF-Driven Melanoma in Zebrafish
3. Discussion
4. Materials and Methods
4.1. Identification of Zebrafish FOS Family Orthologues
4.2. Multiple Sequence Alignment (MSA) and Phylogenetic Comparison of FOS Family Orthologues in Human and Zebrafish
4.3. Zebrafish Maintenance and Embryo Collection
4.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.5. Whole-Mount In Situ Hybridization (WISH)
4.6. Melanoma Model
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tulchinsky, E. Fos family members: Regulation, structure and role in oncogenic transformation. Histol. Histopathol. 2000, 15, 921–928. [Google Scholar] [PubMed]
- Bejjani, F.; Evanno, E.; Zibara, K.; Piechaczyk, M.; Jariel-Encontre, I. The AP-1 transcriptional complex: Local switch or remote command? Biochim. Biophys. Acta (BBA)-Rev. Cancer 2019, 1872, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Jochum, W.; Passegué, E.; Wagner, E.F. AP-1 in mouse development and tumorigenesis. Oncogene 2001, 20, 2401–2412. [Google Scholar] [CrossRef] [PubMed]
- Shaulian, E.; Karin, M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002, 4, E131–E136. [Google Scholar] [CrossRef] [PubMed]
- Shaulian, E.; Karin, M. AP-1 in cell proliferation and survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef]
- Angel, P.; Szabowski, A.; Schorpp-Kistner, M. Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene 2001, 20, 2413–2423. [Google Scholar] [CrossRef]
- Chinenov, Y.; Kerppola, T.K. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 2001, 20, 2438–2452. [Google Scholar] [CrossRef]
- Macian, F.; López-Rodríguez, C.; Rao, A. Partners in transcription: NFAT and AP-1. Oncogene 2001, 20, 2476–2489. [Google Scholar] [CrossRef]
- Van Dam, H.; Castellazzi, M. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene 2001, 20, 2453–2464. [Google Scholar] [CrossRef]
- Trop-Steinberg, S.; Azar, Y. AP-1 expression and its clinical relevance in immune disorders and cancer. Am. J. Med. Sci. 2017, 353, 474–483. [Google Scholar] [CrossRef]
- Eferl, R.; Wagner, E.F. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 2003, 3, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Wisdon, R.; Verma, I.M. Transformation by Fos proteins requires a C-terminal transactivation domain. Mol. Cell. Biol. 1993, 13, 7429–7438. [Google Scholar] [PubMed]
- Sutherland, J.A.; Cook, A.; Bannister, A.J.; Kouzarides, T. Conserved motifs in Fos and Jun define a new class of activation domain. Genes Dev. 1992, 6, 1810–1819. [Google Scholar] [CrossRef][Green Version]
- Wisdom, R.; Yen, J.; Rashid, D.; Verma, I.M. Transformation by FosB requires a trans-activation domain missing in FosB2 that can be substituted by heterologous activation domains. Genes Dev. 1992, 6, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Gomard, T.; Jariel-Encontre, I.; Basbous, J.; Bossis, G.; Mocquet-Torcy, G.; Piechaczyk, M. Fos family protein degradation by the proteasome. Biochem. Soc. Trans. 2008, 36 Pt 5, 858–863. [Google Scholar] [CrossRef]
- Pakay, J.L.; Diesch, J.; Gilan, O.; Yip, Y.Y.; Sayan, E.; Kolch, W.; Mariadason, J.M.; Hannan, R.D.; Tulchinsky, E.; Dhillon, A.S. A 19S proteasomal subunit cooperates with an ERK MAPK-regulated degron to regulate accumulation of Fra-1 in tumour cells. Oncogene 2012, 31, 1817–1824. [Google Scholar] [CrossRef]
- Sayan, A.E.; Stanford, R.; Vickery, R.; Grigorenko, E.; Diesch, J.; Kulbicki, K.; Edwards, R.; Pal, R.; Greaves, P.; Jariel-Encontre, I.; et al. Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL. Oncogene 2012, 31, 1493–1503. [Google Scholar] [CrossRef]
- Hess, J.; Angel, P.; Schorpp-Kistner, M. AP-1 subunits: Quarrel and harmony among siblings. J. Cell Sci. 2004, 117, 5965–5973. [Google Scholar] [CrossRef]
- Angel, P.; Karin, M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta (BBA)-Rev. Cancer 1991, 1072, 129–157. [Google Scholar] [CrossRef]
- Reichmann, E.; Schwarz, H.; Deiner, E.M.; Leitner, I.; Eilers, M.; Berger, J.; Busslinger, M.; Beug, H. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial-fibroblastoid cell conversion. Cell 1992, 71, 1103–1116. [Google Scholar] [CrossRef]
- Saez, E.; Rutberg, S.E.; Mueller, E.; Oppenheim, H.; Smoluk, J.; Yuspa, S.H.; Spiegelman, B.M. c-Fos is required for malignant progression of skin tumors. Cell 1995, 82, 721–732. [Google Scholar] [CrossRef]
- Mahner, S.; Baasch, C.; Schwarz, J.; Hein, S.; Wölber, L.; Jänicke, F.; Milde-Langosch, K. C-Fos expression is a molecular predictor of progression and survival in epithelial ovarian carcinoma. Br. J. Cancer 2008, 99, 1269–1275. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-C.; Li, J.; Zhao, Y.-P.; Zhou, L.; Cui, Q.-C.; Zhou, W.-X.; Zhang, T.P.; You, L. Expression of c-fos was associated with clinicopathologic characteristics and prognosis in pancreatic cancer. PLoS ONE 2015, 10, e0120332. [Google Scholar] [CrossRef]
- Funk, M.; Poensgen, B.; Graulich, W.; Jérôme, V.; Müller, R. A novel, transformation-relevant activation domain in Fos proteins. Mol. Cell. Biol. 1997, 17, 537–544. [Google Scholar] [CrossRef]
- Tkach, V.; Tulchinsky, E.; Lukanidin, E.; Vinson, C.; Bock, E.; Berezin, V. Role of the Fos family members, c-Fos, Fra-1 and Fra-2, in the regulation of cell motility. Oncogene 2003, 22, 5045–5054. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, A.S.; Tulchinsky, E. FRA-1 as a driver of tumour heterogeneity: A nexus between oncogenes and embryonic signalling pathways in cancer. Oncogene 2015, 34, 4421–4428. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E. Functions of AP1 (Fos/Jun) in bone development. Ann. Rheum. Dis. 2002, 61 (Suppl. S2), ii40–ii42. [Google Scholar] [CrossRef]
- Wang, Z.-Q.; Ovitt, C.; Grigoriadis, A.E.; Möhle-Steinlein, U.; Rüther, U.; Wagner, E.F. Bone and haematopoietic defects in mice lacking c-fos. Nature 1992, 360, 741–745. [Google Scholar] [CrossRef]
- Grigoriadis, A.E.; Schellander, K.; Wang, Z.-Q.; Wagner, E.F. Osteoblasts are target cells for transformation in c-fos transgenic mice. J. Cell Biol. 1993, 122, 685–701. [Google Scholar] [CrossRef]
- Brown, J.R.; Ye, H.; Bronson, R.T.; Dikkes, P.; Greenberg, M.E. A defect in nurturing in mice lacking the immediate early gene fosB. Cell 1996, 86, 297–309. [Google Scholar] [CrossRef]
- Kent, L.N.; Rumi, M.K.; Kubota, K.; Lee, D.-S.; Soares, M.J. FOSL1 is integral to establishing the maternal-fetal interface. Mol. Cell. Biol. 2011, 31, 4801–4813. [Google Scholar] [CrossRef]
- Renaud, S.J.; Kubota, K.; Rumi, M.K.; Soares, M.J. The FOS transcription factor family differentially controls trophoblast migration and invasion. J. Biol. Chem. 2014, 289, 5025–5039. [Google Scholar] [CrossRef]
- Marrs, J.A.; Sarmah, S. The Genius of the Zebrafish Model: Insights on Development and Disease. Biomedicines 2021, 9, 577. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Perriere, G.; Gouy, M. WWW-query: An on-line retrieval system for biological sequence banks. Biochimie 1996, 78, 364–369. [Google Scholar] [CrossRef]
- Nguyen, N.T.T.; Vincens, P.; Dufayard, J.F.; Crollius, H.R.; Louis, A. Genomicus in 2022: Comparative tools for thousands of genomes and reconstructed ancestors. Nucleic Acids Res. 2022, 50, D1025–D1031. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef]
- Ceol, C.J.; Houvras, Y.; Jane-Valbuena, J.; Bilodeau, S.; Orlando, D.A.; Battisti, V.; Fritsch, L.; Lin, W.M.; Hollmann, T.J.; Ferré, F.; et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011, 471, 513–517. [Google Scholar] [CrossRef]
- Weidenfeld-Baranboim, K.; Bitton-Worms, K.; Aronheim, A. TRE-dependent transcription activation by JDP2–CHOP10 association. Nucleic Acids Res. 2008, 36, 3608–3619. [Google Scholar] [CrossRef]
- Aronheim, A.; Zandi, E.; Hennemann, H.; Elledge, S.J.; Karin, M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 1997, 17, 3094–3102. [Google Scholar] [CrossRef] [PubMed]
- Bozec, A.; Bakiri, L.; Jimenez, M.; Schinke, T.; Amling, M.; Wagner, E.F. Fra-2/AP-1 controls bone formation by regulating osteoblast differentiation and collagen production. J. Cell Biol. 2010, 190, 1093–1106. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.L.; Horsfield, J.A.; Black, M.A.; Rutherford, K.; Gemmell, N.J. Identification of sex differences in zebrafish (Danio rerio) brains during early sexual differentiation and masculinization using 17α-methyltestoterone. Biol. Reprod. 2018, 99, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, L.; Sharpe, M.; Novikov, N.; Gonzalez-Rosa, J.M.; Borikova, A.; Nevis, K.; Paffett-Lugassy, N.; Zhao, L.; Adams, M.; Guner-Ataman, B.; et al. The AP-1 transcription factor component Fosl2 potentiates the rate of myocardial differentiation from the zebrafish second heart field. Development 2016, 143, 113–122. [Google Scholar] [CrossRef]
- Beisaw, A.; Kuenne, C.; Guenther, S.; Dallmann, J.; Wu, C.C.; Bentsen, M.; Looso, M.; Stainier, D.Y. AP-1 Contributes to Chromatin Accessibility to Promote Sarcomere Disassembly and Cardiomyocyte Protrusion During Zebrafish Heart Regeneration. Circ. Res. 2020, 126, 1760–1778. [Google Scholar] [CrossRef]
- Murphy, L.O.; Smith, S.; Chen, R.H.; Fingar, D.C.; Blenis, J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 2002, 4, 556–564. [Google Scholar] [CrossRef]
- Ruhl, T.; Zeymer, M.; von der Emde, G. Cannabinoid modulation of zebrafish fear learning and its functional analysis investigated by c-Fos expression. Pharmacol. Biochem. Behav. 2017, 153, 18–31. [Google Scholar] [CrossRef]
- Louie, K.W.; Saera-Vila, A.; Kish, P.E.; Colacino, J.A.; Kahana, A. Temporally distinct transcriptional regulation of myocyte dedifferentiation and Myofiber growth during muscle regeneration. BMC Genom. 2017, 18, 854. [Google Scholar] [CrossRef]
- Goldman, J.A.; Kuzu, G.; Lee, N.; Karasik, J.; Gemberling, M.; Foglia, M.J.; Karra, R.; Dickson, A.L.; Sun, F.; Tolstorukov, M.Y.; et al. Resolving Heart Regeneration by Replacement Histone Profiling. Dev. Cell 2017, 40, 392–404.e5. [Google Scholar] [CrossRef]
- Sawada, A.; Shinya, M.; Jiang, Y.J.; Kawakami, A.; Kuroiwa, A.; Takeda, H. Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development 2001, 128, 4873–4880. [Google Scholar] [CrossRef]
- Furthauer, M.; Van Celst, J.; Thisse, C.; Thisse, B. Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development 2004, 131, 2853–2864. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.; Maegawa, S.; Kiang, A.; Habas, R.; Weinberg, E.; Dawid, I.B. A role for MKP3 in axial patterning of the zebrafish embryo. Development 2004, 131, 2769–2779. [Google Scholar] [CrossRef] [PubMed]
- Shinya, M.; Koshida, S.; Sawada, A.; Kuroiwa, A.; Takeda, H. Fgf signalling through MAPK cascade is required for development of the subpallial telencephalon in zebrafish embryos. Development 2001, 128, 4153–4164. [Google Scholar] [CrossRef] [PubMed]
- Sari, D.W.K.; Akiyama, R.; Naoki, H.; Ishijima, H.; Bessho, Y.; Matsui, T. Time-lapse observation of stepwise regression of Erk activity in zebrafish presomitic mesoderm. Sci. Rep. 2018, 8, 4335. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Comandante-Lou, N.; Baumann, D.G.; Fallahi-Sichani, M. AP-1 transcription factor network explains diverse patterns of cellular plasticity in melanoma cells. Cell Rep. 2022, 40, 111147. [Google Scholar] [CrossRef]
- Ramsdale, R.; Jorissen, R.N.; Li, F.Z.; Al-Obaidi, S.; Ward, T.; Sheppard, K.E.; Bukczynska, P.E.; Young, R.J.; Boyle, S.E.; Shackleton, M.; et al. The transcription cofactor c-JUN mediates phenotype switching and BRAF inhibitor resistance in melanoma. Sci. Signal. 2015, 8, ra82. [Google Scholar] [CrossRef]
- Caramel, J.; Papadogeorgakis, E.; Hill, L.; Browne, G.J.; Richard, G.; Wierinckx, A.; Saldanha, G.; Osborne, J.; Hutchinson, P.; Tse, G.; et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 2013, 24, 466–480. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubra, K.; Gaddu, G.K.; Liongue, C.; Heidary, S.; Ward, A.C.; Dhillon, A.S.; Basheer, F. Phylogenetic and Expression Analysis of Fos Transcription Factors in Zebrafish. Int. J. Mol. Sci. 2022, 23, 10098. https://doi.org/10.3390/ijms231710098
Kubra K, Gaddu GK, Liongue C, Heidary S, Ward AC, Dhillon AS, Basheer F. Phylogenetic and Expression Analysis of Fos Transcription Factors in Zebrafish. International Journal of Molecular Sciences. 2022; 23(17):10098. https://doi.org/10.3390/ijms231710098
Chicago/Turabian StyleKubra, Khadizatul, Gurveer K. Gaddu, Clifford Liongue, Somayyeh Heidary, Alister C. Ward, Amardeep S. Dhillon, and Faiza Basheer. 2022. "Phylogenetic and Expression Analysis of Fos Transcription Factors in Zebrafish" International Journal of Molecular Sciences 23, no. 17: 10098. https://doi.org/10.3390/ijms231710098
APA StyleKubra, K., Gaddu, G. K., Liongue, C., Heidary, S., Ward, A. C., Dhillon, A. S., & Basheer, F. (2022). Phylogenetic and Expression Analysis of Fos Transcription Factors in Zebrafish. International Journal of Molecular Sciences, 23(17), 10098. https://doi.org/10.3390/ijms231710098






