Membrane-Bound EMC10 Is Required for Sperm Motility via Maintaining the Homeostasis of Cytoplasm Sodium in Sperm
Abstract
:1. Introduction
2. Results
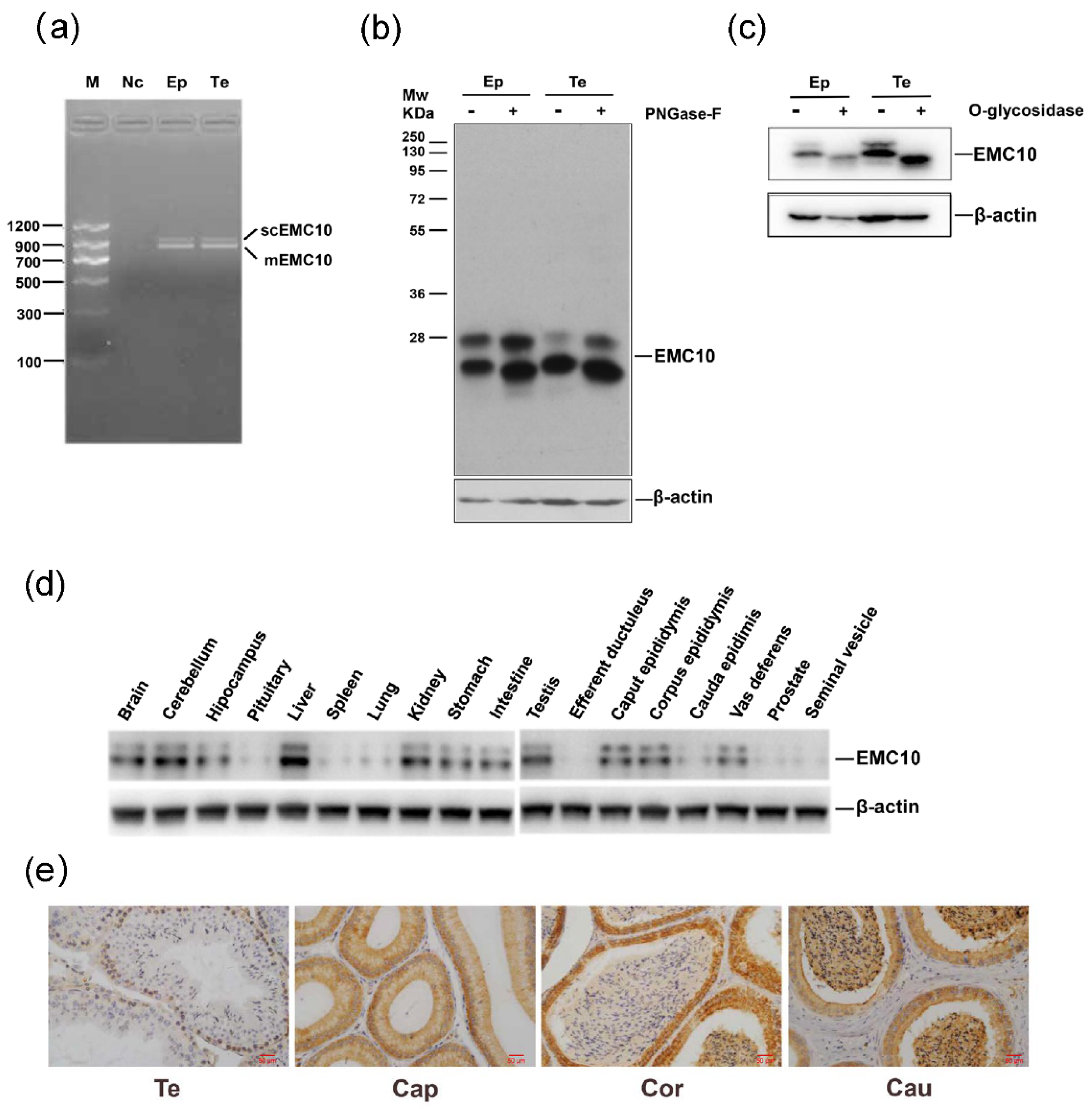
2.1. Cloning, Protein Expression Profile and Glycosylation Analysis of Mouse EMC10
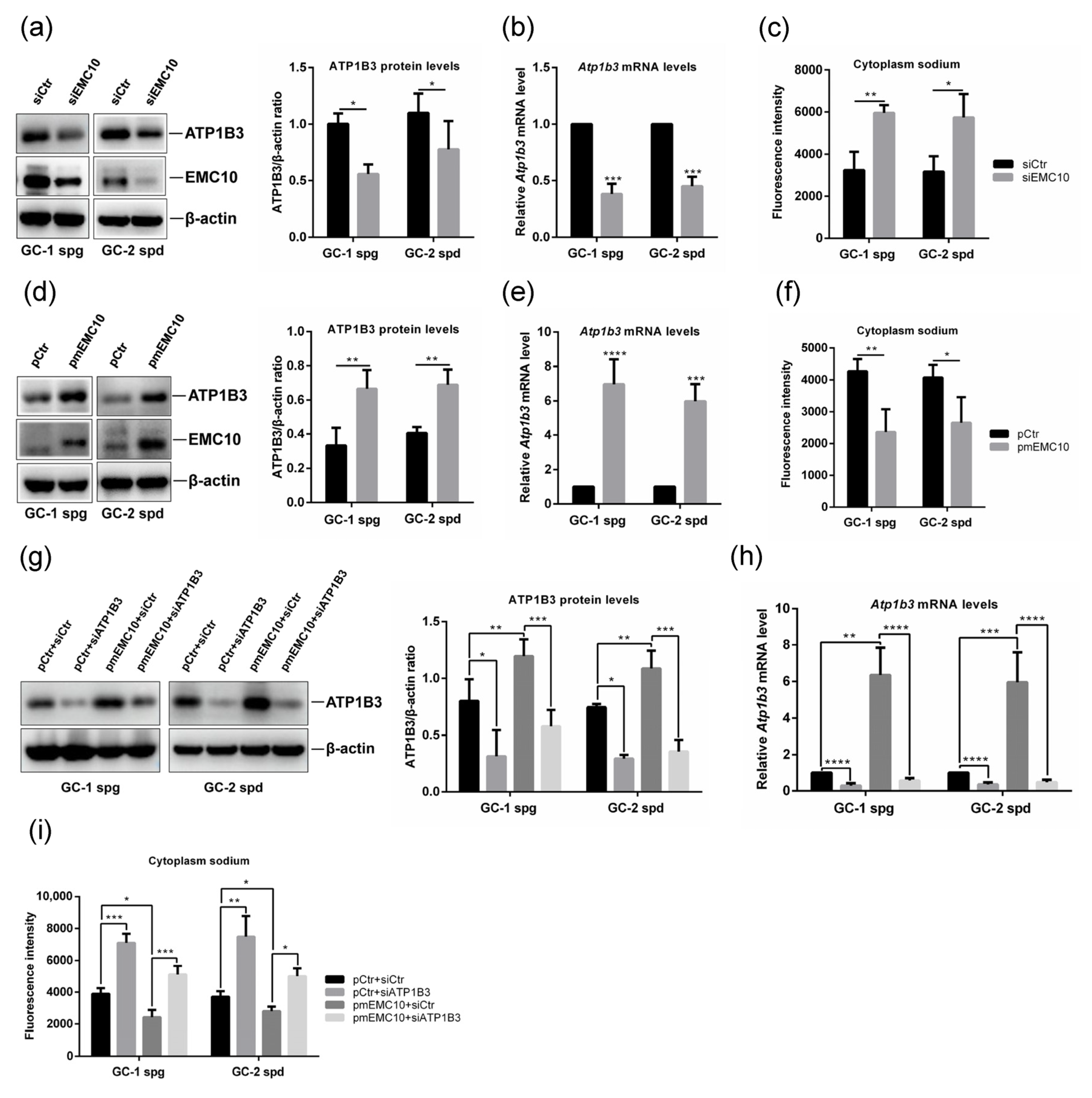
2.2. Membrane-Bound EMC10 Maintains the Homeostasis of Cytoplasm Sodium via Positively Regulating ATP1B3 in Mouse Germ Cells
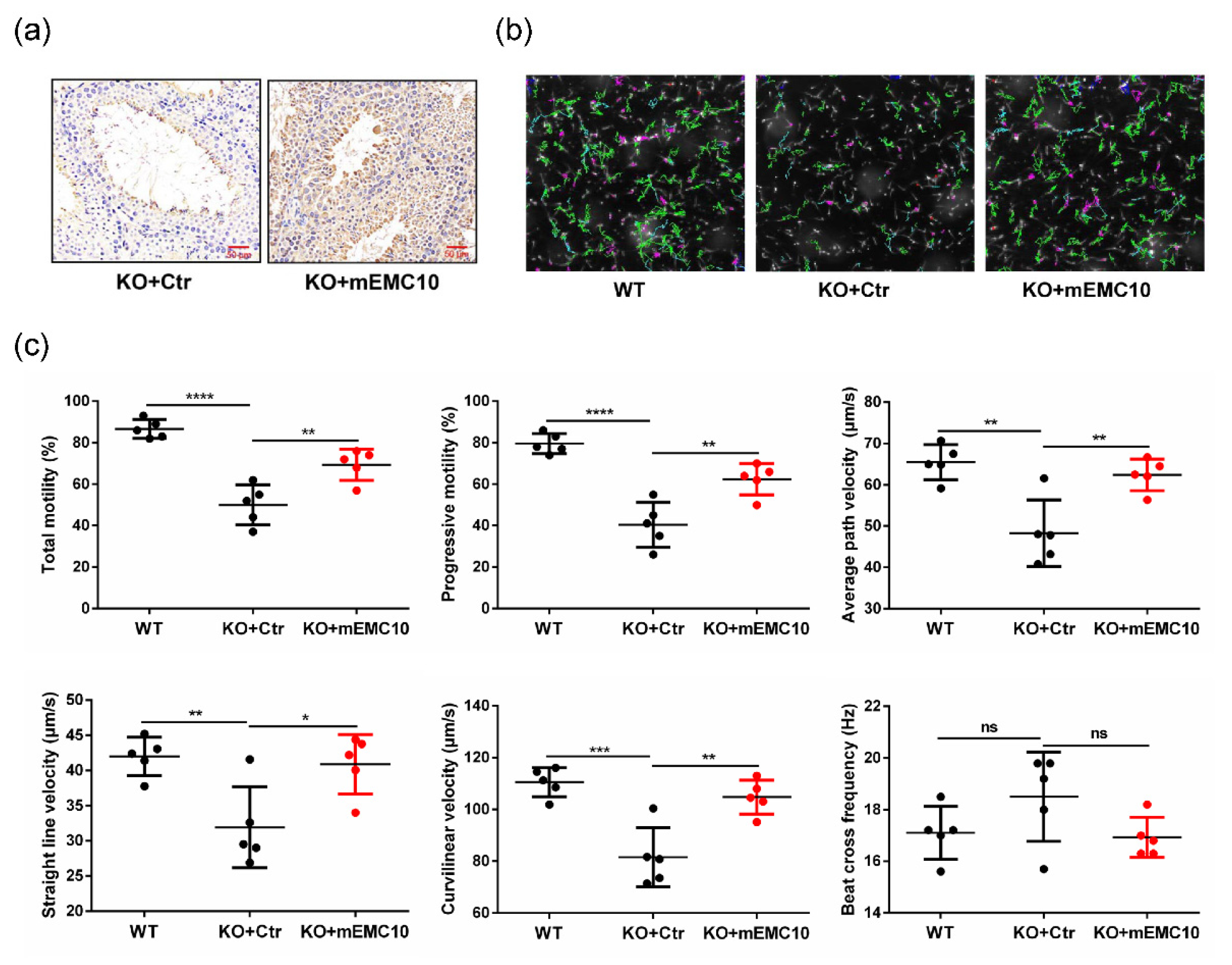
2.3. Intra-Testis Overexpression of Membrane-Bound EMC10 Improves Sperm Motility in Emc10 KO Mice
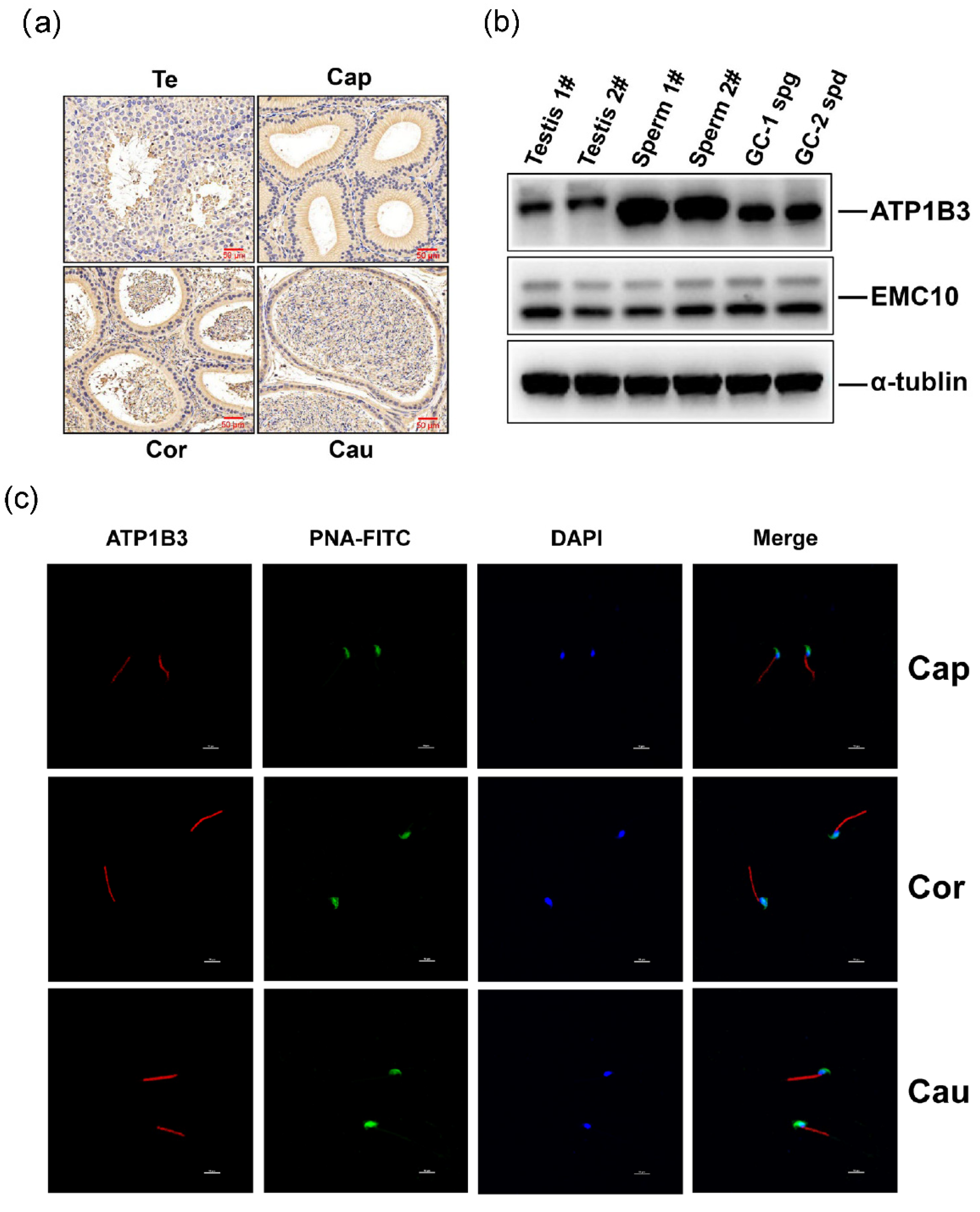
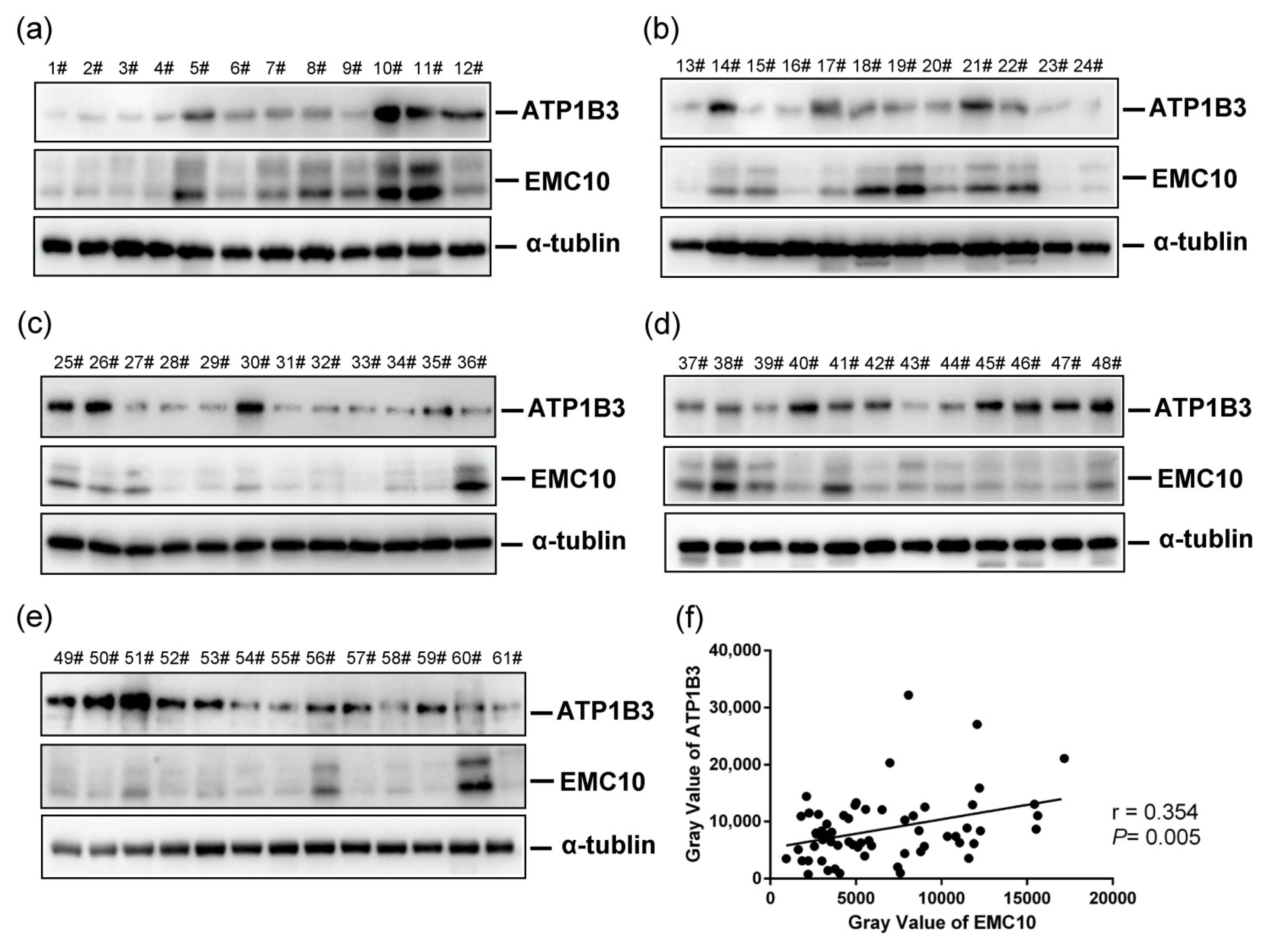
2.4. The Levels of ATP1B3 Protein Are Positively Correlated with Those of EMC10 Protein in Human Spermatozoa
2.5. Seminal Plasma scEMC10 Levels Are Not Associated with Sperm Motility in Humans
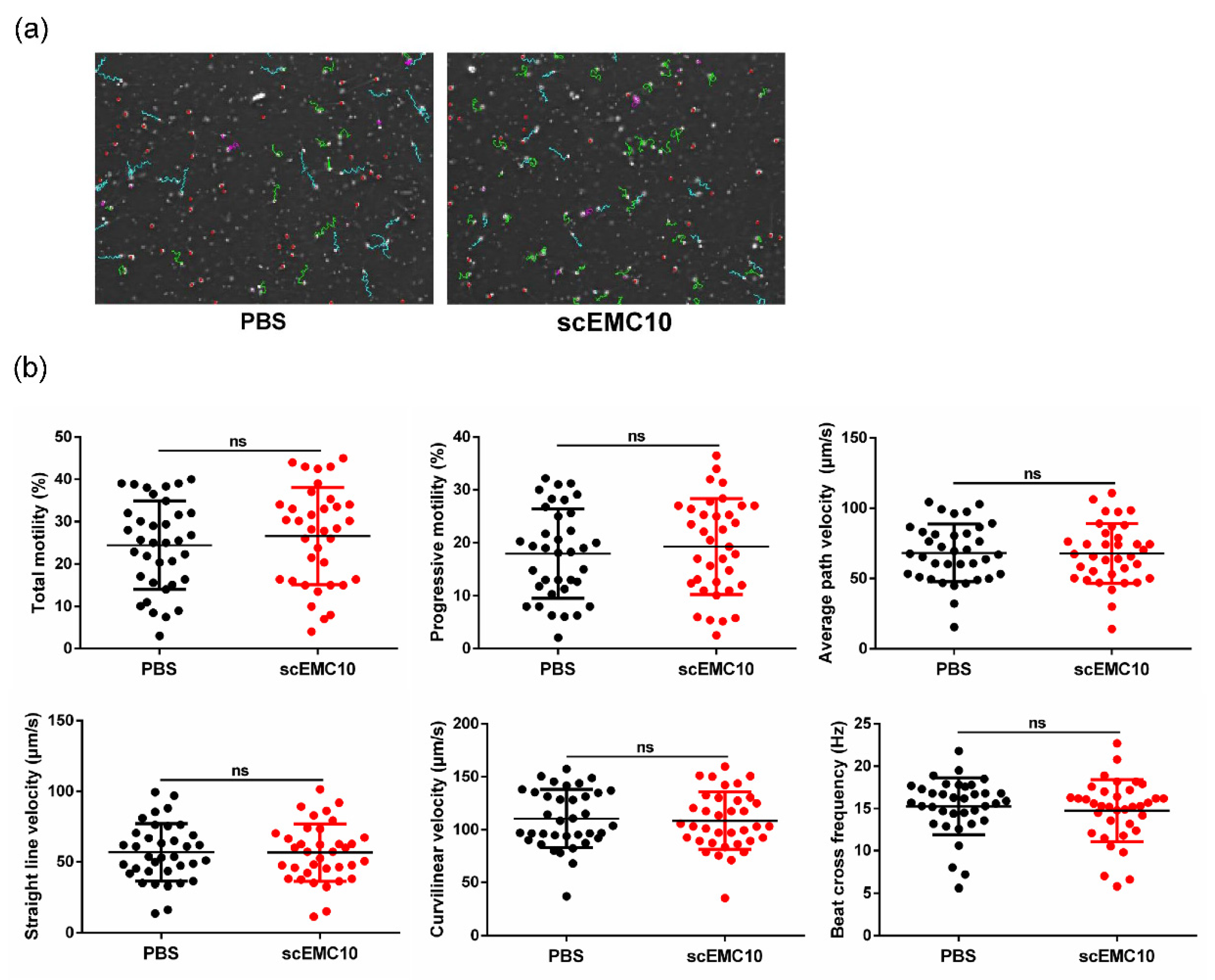
2.6. scEMC10 Is Not Capable of Improving the Motility of Asthenospermic Sperm
3. Discussion
4. Materials and Methods
4.1. Patients and Semen Samples
4.2. Measurement of scEMC10 in Seminal Plasma by CLIA
4.3. Animal Experiments
4.4. Cell Culture
4.5. Construction of Stably Transfected Cells
4.6. RNA Interference
4.7. Western Blotting Analysis
4.8. RNA Extraction and Quantitative Real-Time PCR
4.9. CoroNa Green Dye as Sodium Indicator
4.10. Immunohistochemistry
4.11. Immunofluorescence Staining
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Minhas, S.; Bettocchi, C.; Boeri, L.; Capogrosso, P.; Carvalho, J.; Cilesiz, N.C.; Cocci, A.; Corona, G.; Dimitropoulos, K.; Gul, M.; et al. European Association of Urology Guidelines on Male Sexual and Reproductive Health: 2021 Update on Male Infertility. Eur. Urol. 2021, 80, 603–620. [Google Scholar] [CrossRef] [PubMed]
- Vander Borght, M.; Wyns, C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018, 62, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zheng, D.; Wu, H.; Li, R.; Xu, S.; Kang, Y.; Cao, Y.; Chen, X.; Zhu, Y.; Xu, S.; et al. Epidemiology of infertility in China: A population-based study. BJOG 2018, 125, 432–441. [Google Scholar] [CrossRef]
- Brugh, V.M., III; Matschke, H.M.; Lipshultz, L.I. Male factor infertility. Endocrinol. Metab. Clin. N. Am. 2003, 32, 689–707. [Google Scholar] [CrossRef]
- Krausz, C.; Riera-Escamilla, A. Genetics of male infertility. Nat. Rev. Urol. 2018, 15, 369–384. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, F.; Zhang, M.; Xiong, Z.; Yin, Q.; Ru, Y.; Shi, H.; Li, J.; Mao, S.; Li, Y.; et al. EMC10 governs male fertility via maintaining sperm ion balance. J. Mol. Cell Biol. 2018, 10, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; You, Q.; Feng, X.; Kovach, A.; Li, H. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature 2020, 584, 475–478. [Google Scholar] [CrossRef]
- Junes-Gill, K.S.; Gallaher, T.K.; Gluzman-Poltorak, Z.; Miller, J.D.; Wheeler, C.J.; Fan, X.; Basile, L.A. hHSS1: A novel secreted factor and suppressor of glioma growth located at chromosome 19q13.33. J. Neuro Oncol. 2011, 102, 197–211. [Google Scholar] [CrossRef]
- Wang, X.; Gong, W.; Liu, Y.; Yang, Z.; Zhou, W.; Wang, M.; Yang, Z.; Wen, J.; Hu, R. Molecular cloning of a novel secreted peptide, INM02, and regulation of its expression by glucose. J. Endocrinol. 2009, 202, 355–364. [Google Scholar] [CrossRef]
- Reboll, M.R.; Korf-Klingebiel, M.; Klede, S.; Polten, F.; Brinkmann, E.; Reimann, I.; Schonfeld, H.J.; Bobadilla, M.; Faix, J.; Kensah, G.; et al. EMC10 (Endoplasmic Reticulum Membrane Protein Complex Subunit 10) Is a Bone Marrow-Derived Angiogenic Growth Factor Promoting Tissue Repair After Myocardial Infarction. Circulation 2017, 136, 1809–1823. [Google Scholar] [CrossRef]
- Miller-Vedam, L.E.; Brauning, B.; Popova, K.D.; Schirle Oakdale, N.T.; Bonnar, J.L.; Prabu, J.R.; Boydston, E.A.; Sevillano, N.; Shurtleff, M.J.; Stroud, R.M.; et al. Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients. eLife 2020, 9, e62611. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Xu, S.Y.; Wu, X.Y.; Song, H.D.; Mao, Y.F.; Fan, H.Y.; Yu, F.; Mou, B.; Gu, Y.Y.; Xu, L.Q.; et al. Gene expression profiling in human insulinoma tissue: Genes involved in the insulin secretion pathway and cloning of novel full-length cDNAs. Endocr. Relat. Cancer 2004, 11, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Junes-Gill, K.S.; Lawrence, C.E.; Wheeler, C.J.; Cordner, R.; Gill, T.G.; Mar, V.; Shiri, L.; Basile, L.A. Human Hematopoietic Signal peptide-containing Secreted 1 (hHSS1) modulates genes and pathways in glioma: Implications for the regulation of tumorigenicity and angiogenesis. BMC Cancer 2014, 14, 920. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Hsu, P.K.; Stark, K.L.; Karayiorgou, M.; Gogos, J.A. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell 2013, 152, 262–275. [Google Scholar] [CrossRef]
- Diamantopoulou, A.; Sun, Z.; Mukai, J.; Xu, B.; Fenelon, K.; Karayiorgou, M.; Gogos, J.A. Loss-of-function mutation in Mirta22/Emc10 rescues specific schizophrenia-related phenotypes in a mouse model of the 22q11.2 deletion. Proc. Natl. Acad. Sci. USA 2017, 114, E6127–E6136. [Google Scholar] [CrossRef] [PubMed]
- Umair, M.; Ballow, M.; Asiri, A.; Alyafee, Y.; Al Tuwaijri, A.; Alhamoudi, K.M.; Aloraini, T.; Abdelhakim, M.; Althagafi, A.T.; Kafkas, S.; et al. EMC10 homozygous variant identified in a family with global developmental delay, mild intellectual disability, and speech delay. Clin. Genet. 2020, 98, 555–561. [Google Scholar] [CrossRef]
- Shao, D.D.; Straussberg, R.; Ahmed, H.; Khan, A.; Tian, S.; Hill, R.S.; Smith, R.S.; Majmundar, A.J.; Ameziane, N.; Neil, J.E.; et al. A recurrent, homozygous EMC10 frameshift variant is associated with a syndrome of developmental delay with variable seizures and dysmorphic features. Genet. Med. 2021, 23, 1158–1162. [Google Scholar] [CrossRef]
- Haddad-Eid, E.; Gur, N.; Eid, S.; Pilowsky-Peleg, T.; Straussberg, R. The phenotype of homozygous EMC10 variant: A new syndrome with intellectual disability and language impairment. Eur. J. Paediatr. Neurol. 2022, 37, 56–61. [Google Scholar] [CrossRef]
- Eichler, J. Protein glycosylation. Curr. Biol. 2019, 29, R229–R231. [Google Scholar] [CrossRef]
- Hofmann, M.C.; Narisawa, S.; Hess, R.A.; Millan, J.L. Immortalization of germ cells and somatic testicular cells using the SV40 large T antigen. Exp. Cell Res. 1992, 201, 417–435. [Google Scholar] [CrossRef]
- Hofmann, M.C.; Hess, R.A.; Goldberg, E.; Millán, J.L. Immortalized germ cells undergo meiosis in vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 5533–5537. [Google Scholar] [CrossRef] [PubMed]
- Nowicka-Bauer, K.; Szymczak-Cendlak, M. Structure and Function of Ion Channels Regulating Sperm Motility—An Overview. Int. J. Mol. Sci. 2021, 22, 3259. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.H. Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 2002, 71, 511–535. [Google Scholar] [CrossRef] [PubMed]
- Blanco, G.; Melton, R.J.; Sanchez, G.; Mercer, R.W. Functional characterization of a testes-specific alpha-subunit isoform of the sodium/potassium adenosinetriphosphatase. Biochemistry 1999, 38, 13661–13669. [Google Scholar] [CrossRef] [PubMed]
- Wagoner, K.; Sanchez, G.; Nguyen, A.N.; Enders, G.C.; Blanco, G. Different expression and activity of the alpha1 and alpha4 isoforms of the Na,K-ATPase during rat male germ cell ontogeny. Reproduction 2005, 130, 627–641. [Google Scholar] [CrossRef]
- Darszon, A.; Acevedo, J.J.; Galindo, B.E.; Hernandez-Gonzalez, E.O.; Nishigaki, T.; Trevino, C.L.; Wood, C.; Beltran, C. Sperm channel diversity and functional multiplicity. Reproduction 2006, 131, 977–988. [Google Scholar] [CrossRef]
- Jimenez, T.; McDermott, J.P.; Sanchez, G.; Blanco, G. Na,K-ATPase alpha4 isoform is essential for sperm fertility. Proc. Natl. Acad. Sci. USA 2011, 108, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Syeda, S.S.; Sanchez, G.; McDermott, J.P.; Hong, K.H.; Blanco, G.; Georg, G.I. The Na+ and K+ transport system of sperm (ATP1A4) is essential for male fertility and an attractive target for male contraceptiondagger. Biol. Reprod. 2020, 103, 343–356. [Google Scholar] [CrossRef]
- McDermott, J.P.; Numata, S.; Blanco, G. Na,K-ATPase Atp1a4 isoform is important for maintaining sperm flagellar shape. J. Assist. Reprod. Genet. 2021, 38, 1493–1505. [Google Scholar] [CrossRef]
- Jimenez, T.; Sanchez, G.; McDermott, J.P.; Nguyen, A.N.; Kumar, T.R.; Blanco, G. Increased expression of the Na,K-ATPase alpha4 isoform enhances sperm motility in transgenic mice. Biol. Reprod. 2011, 84, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Bjorndahl, L.; Kirkman Brown, J.; other Editorial Board Members of the WHO Laboratory Manual for the Examination and Processing of Human Semen. The sixth edition of the WHO Laboratory Manual for the Examination and Processing of Human Semen: Ensuring quality and standardization in basic examination of human ejaculates. Fertil. Steril. 2022, 117, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Guzick, D.S.; Overstreet, J.W.; Factor-Litvak, P.; Brazil, C.K.; Nakajima, S.T.; Coutifaris, C.; Carson, S.A.; Cisneros, P.; Steinkampf, M.P.; Hill, J.A.; et al. Sperm morphology, motility, and concentration in fertile and infertile men. N. Engl. J. Med. 2001, 345, 1388–1393. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Emery, B.; Carrell, D.T. Sperm DNA Fragmentation: Consequences for Reproduction. Adv. Exp. Med. Biol. 2019, 1166, 87–105. [Google Scholar] [CrossRef]
- Barati, E.; Nikzad, H.; Karimian, M. Oxidative stress and male infertility: Current knowledge of pathophysiology and role of antioxidant therapy in disease management. Cell. Mol. Life Sci. 2020, 77, 93–113. [Google Scholar] [CrossRef]
- Roshdy, N.; Mostafa, T. Seminal plasma survivin in fertile and infertile males. J. Urol. 2009, 181, 1269–1272. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ru, Y.; Shi, H.; Wang, Y.; Wu, B.; Upur, H.; Zhang, Y. Cholecystokinin receptors regulate sperm protein tyrosine phosphorylation via uptake of HCO3. Reproduction 2015, 150, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, D.; Curtis, E.F.; Miller, R.G. Specific labelling by peanut agglutinin of the outer acrosomal membrane of the human spermatozoon. J. Reprod. Fertil. 1987, 81, 127–135. [Google Scholar] [CrossRef]
| Normal-Motile Spermatozoa | Low-Motile Spermatozoa | p | |
|---|---|---|---|
| Number (n) | 122 | 67 | |
| Age (years) | 32.8 ± 6.7 | 32.7 ± 7.3 | 0.961 |
| Semen volume (mL) | 2.70 ± 1.40 | 3.08 ± 1.54 | 0.099 |
| Sperm count (×106/mL) | 194.76 ± 160.17 | 140.37 ± 148.79 | 0.021 |
| Total motility (%) | 58.10 ± 11.31 | 25.15 ± 10.52 | <0.0001 |
| PR (%) | 55.30 ± 11.47 | 22.18 ± 10.65 | <0.0001 |
| VAP (μm/s) | 46.65 ± 10.07 | 38.05 ± 14.25 | <0.0001 |
| VCL (μm/s) | 79.23 ± 15.91 | 64.60 ± 22.05 | <0.0001 |
| VSL (μm/s) | 39.02 ± 9.50 | 31.76 ± 13.13 | <0.0001 |
| BCF (Hz) | 8.88 ± 3.17 | 7.49 ± 3.83 | 0.014 |
| scEMC10 (ng/mL) | 6.95 ± 3.50 | 7.12 ± 2.73 | 0.706 |
| Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | p | |
|---|---|---|---|---|---|
| scEMC10 (ng/mL) | 0.83–4.14 | 4.17–6.08 | 6.12–8.16 | 8.17–17.56 | |
| Number (n) | 47 | 50 | 45 | 47 | |
| Age (years) | 32.4 ± 5.9 | 32.6 ± 5.5 | 30.6 ± 6.4 | 35.2 ± 8.9 | 0.027 |
| Semen volume (mL) | 2.5 ± 1.4 | 3.2 ± 1.5 | 2.8 ± 1.3 | 2.8 ± 1.6 | 0.123 |
| Sperm count (×106/mL) | 185.7 ± 179.1 | 213.4 ± 156.9 | 151.5 ± 131.0 | 147.8 ± 155.9 | 0.064 |
| Total motility (%) | 48.4 ± 22.5 | 50.5 ± 19.0 | 42.3 ± 17.2 | 44.0 ± 17.3 | 0.101 |
| PR (%) | 45.4 ± 22.3 | 47.2 ± 19.9 | 39.9 ± 17.0 | 41.3 ± 17.5 | 0.149 |
| VAP (μm/s) | 43.4 ± 11.7 | 43.2 ± 12.1 | 44.7 ± 10.5 | 43.2 ± 15.1 | 0.95 |
| VCL (μm/s) | 75.1 ± 17.2 | 75.0 ± 20.1 | 75.3 ± 17.2 | 70.9 ± 23.2 | 0.659 |
| VSL (μm/s) | 35.3 ± 11.4 | 36.0 ± 10.8 | 37.7 ± 9.2 | 36.8 ± 14.0 | 0.659 |
| BCF (Hz) | 7.8 ± 3.4 | 8.8 ± 3.6 | 8.3 ± 3.0 | 8.7 ± 3.8 | 0.651 |
| Asthenospermia (%) | 29.8% | 30.0% | 48.9% | 34% | 0.367 |
| Male infertility (%) | 21.2% | 18% | 20% | 19.1% | 0.981 |
| scEMC10 Level | p | |
|---|---|---|
| Sperm count (×106/mL) | −0.106 | 0.148 |
| Sperm motility (%) | −0.142 | 0.052 |
| PR (%) | −0.129 | 0.076 |
| VAP (μm/s) | −0.011 | 0.879 |
| VCL (μm/s) | −0.134 | 0.066 |
| VSL (μm/s) | 0.044 | 0.545 |
| BCF (Hz) | 0.012 | 0.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Mao, S.; Chen, K.; Dai, J.; Jin, S.; Chen, L.; Wang, Y.; Guo, L.; Yang, Y.; Zhan, C.; et al. Membrane-Bound EMC10 Is Required for Sperm Motility via Maintaining the Homeostasis of Cytoplasm Sodium in Sperm. Int. J. Mol. Sci. 2022, 23, 10069. https://doi.org/10.3390/ijms231710069
Liu L, Mao S, Chen K, Dai J, Jin S, Chen L, Wang Y, Guo L, Yang Y, Zhan C, et al. Membrane-Bound EMC10 Is Required for Sperm Motility via Maintaining the Homeostasis of Cytoplasm Sodium in Sperm. International Journal of Molecular Sciences. 2022; 23(17):10069. https://doi.org/10.3390/ijms231710069
Chicago/Turabian StyleLiu, Lijie, Shanhua Mao, Kuangyang Chen, Jiarong Dai, Shuoshuo Jin, Lijiao Chen, Yahao Wang, Lina Guo, Yiting Yang, Chongwen Zhan, and et al. 2022. "Membrane-Bound EMC10 Is Required for Sperm Motility via Maintaining the Homeostasis of Cytoplasm Sodium in Sperm" International Journal of Molecular Sciences 23, no. 17: 10069. https://doi.org/10.3390/ijms231710069
APA StyleLiu, L., Mao, S., Chen, K., Dai, J., Jin, S., Chen, L., Wang, Y., Guo, L., Yang, Y., Zhan, C., Xiong, Z., Diao, H., Zhou, Y., Ding, Q., & Wang, X. (2022). Membrane-Bound EMC10 Is Required for Sperm Motility via Maintaining the Homeostasis of Cytoplasm Sodium in Sperm. International Journal of Molecular Sciences, 23(17), 10069. https://doi.org/10.3390/ijms231710069






