Coronatine Enhances Chilling Tolerance of Tomato Plants by Inducing Chilling-Related Epigenetic Adaptations and Transcriptional Reprogramming
Abstract
:1. Introduction
2. Results
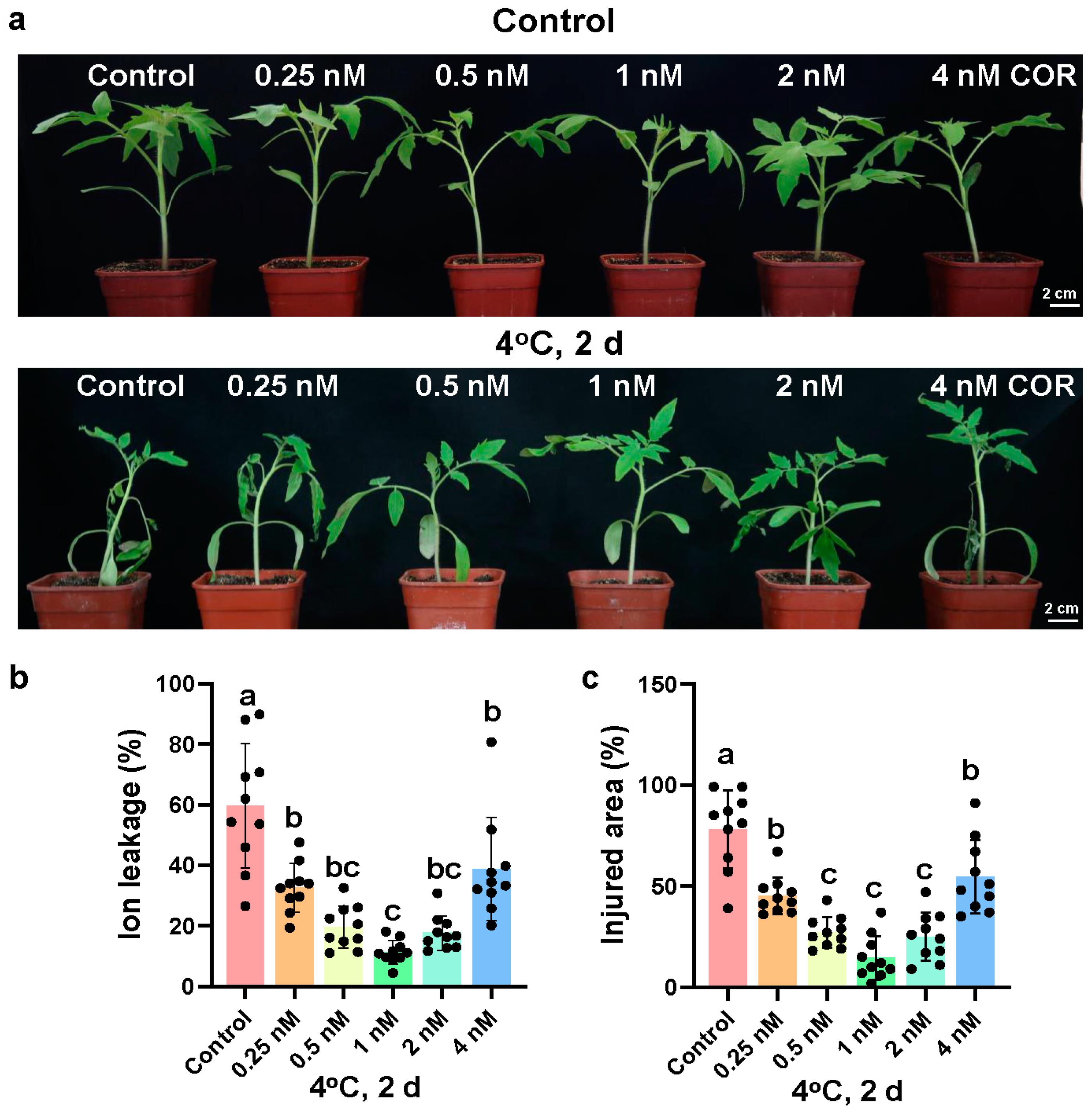
2.1. Coronatine Increases the Chilling Tolerance of Tomato Plants Dependent on CBF Pathway
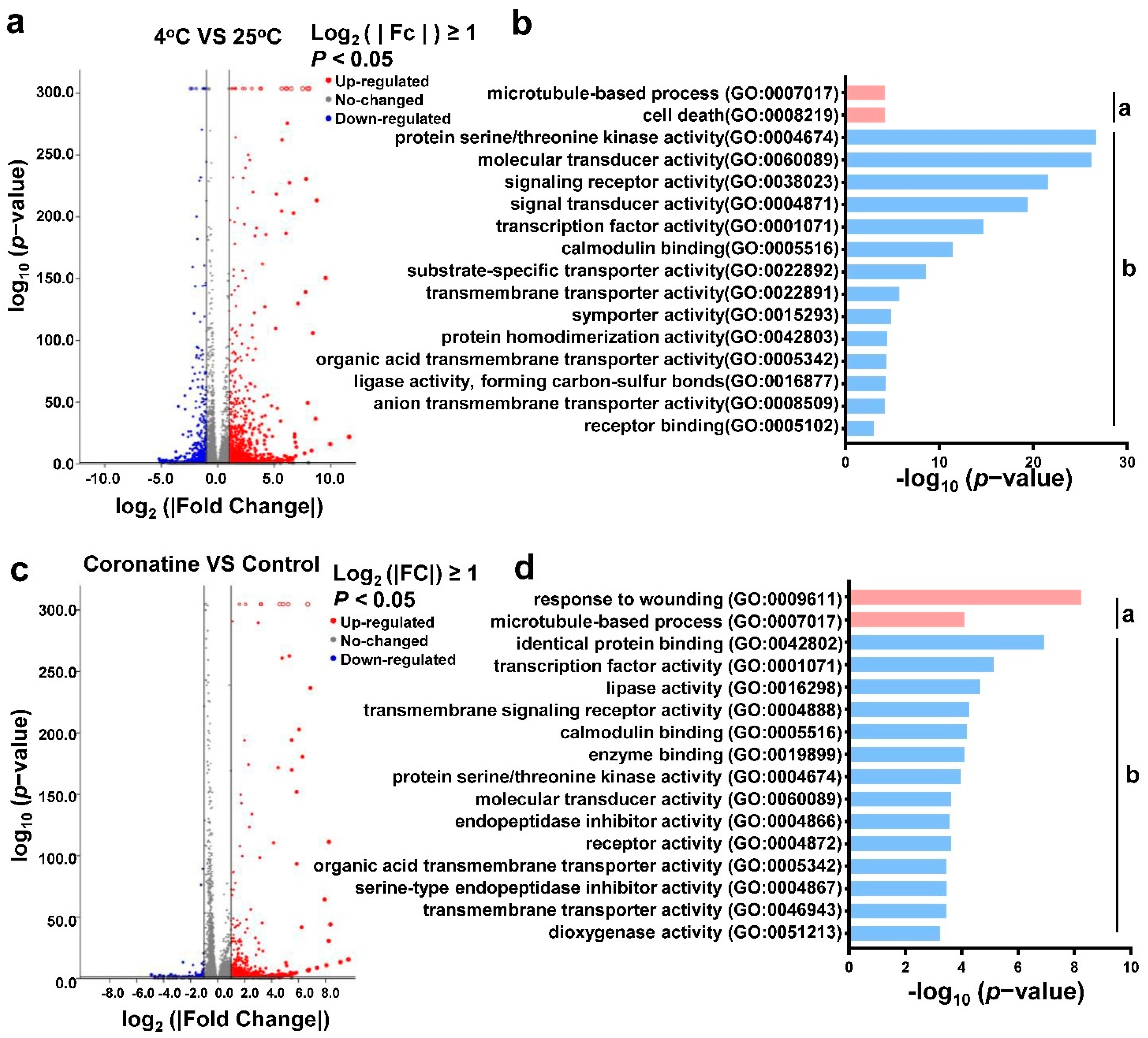
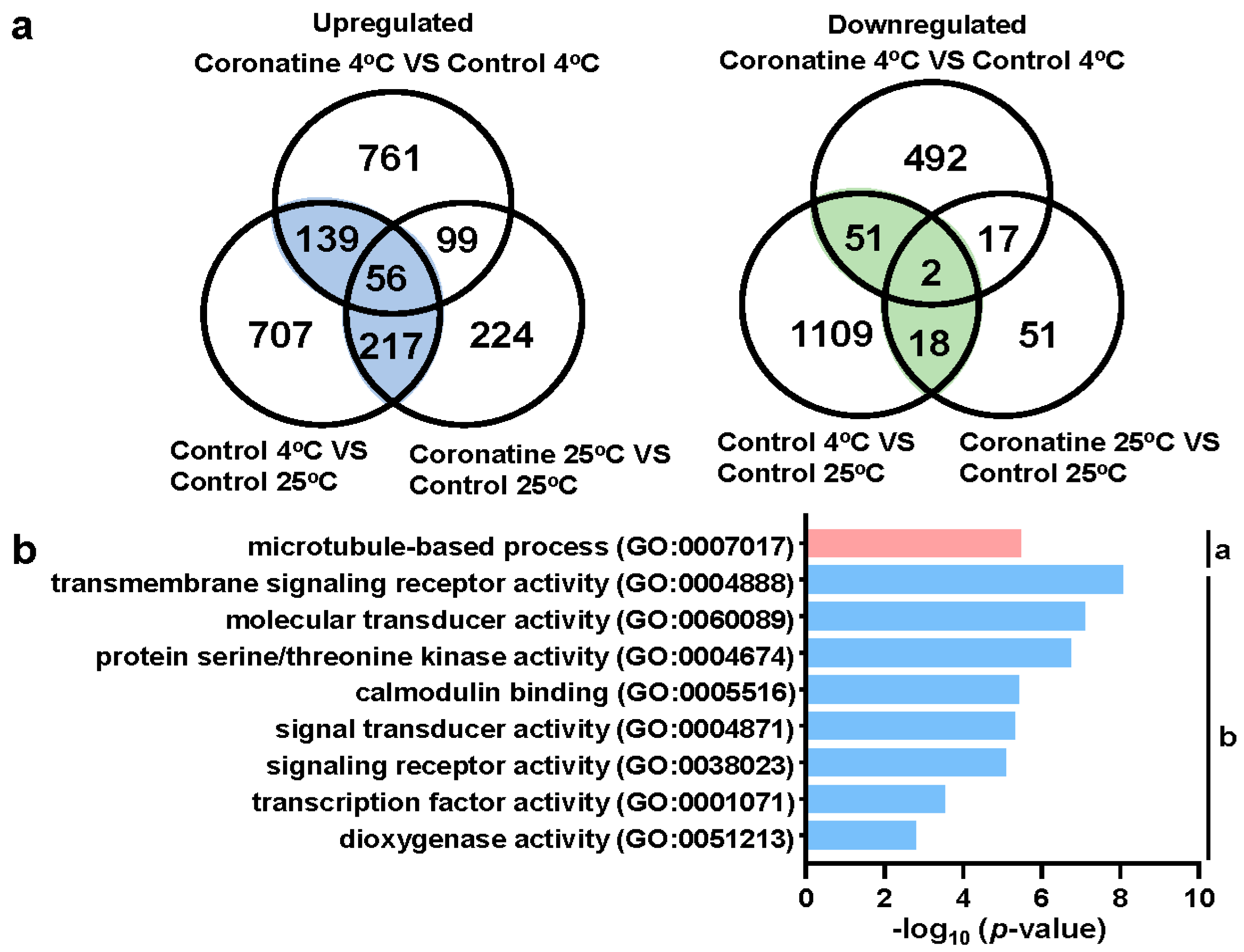
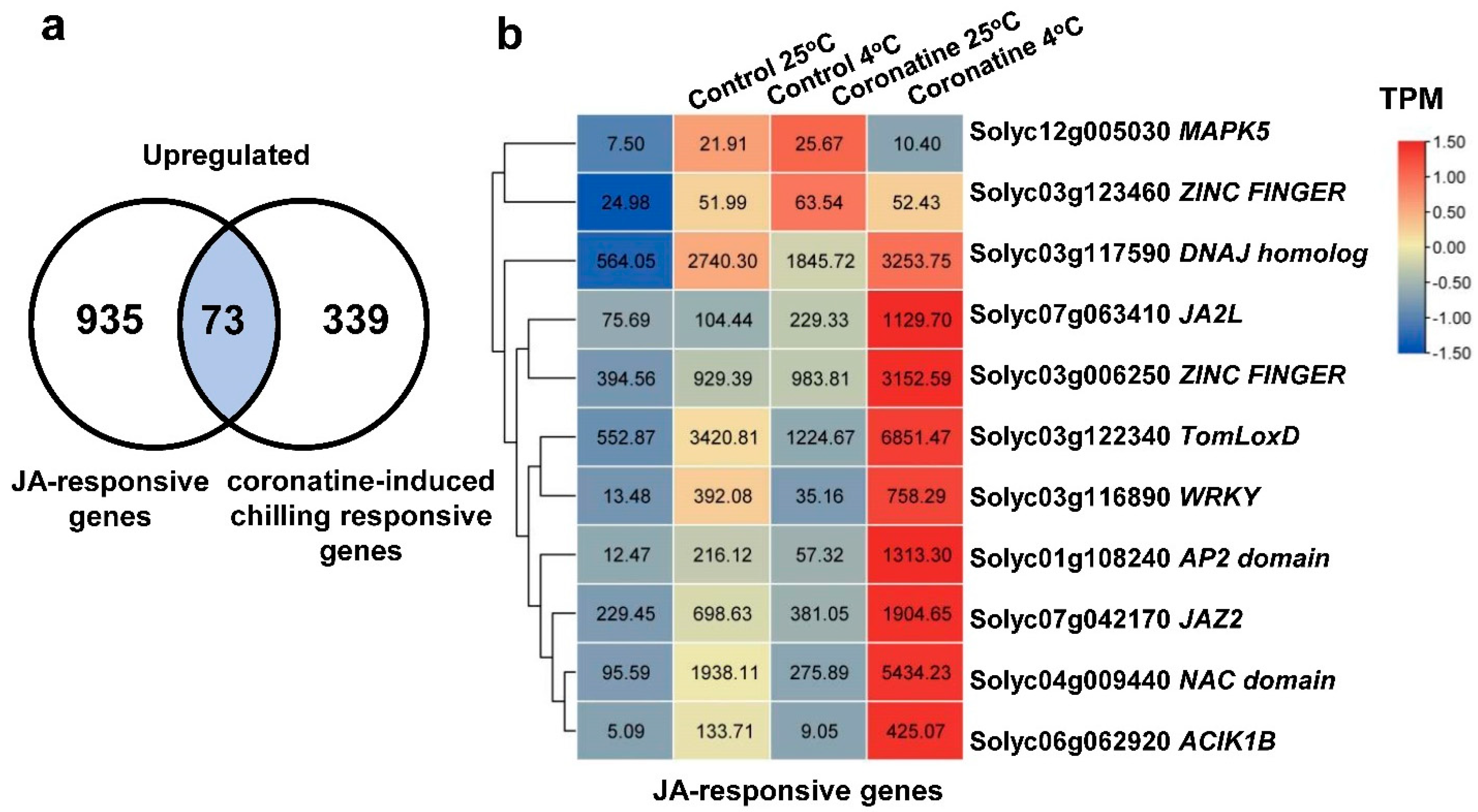
2.2. Coronatine Promotes the Expression of a Wide Range of Chilling-Responsive Genes through Transcriptional Reprogramming
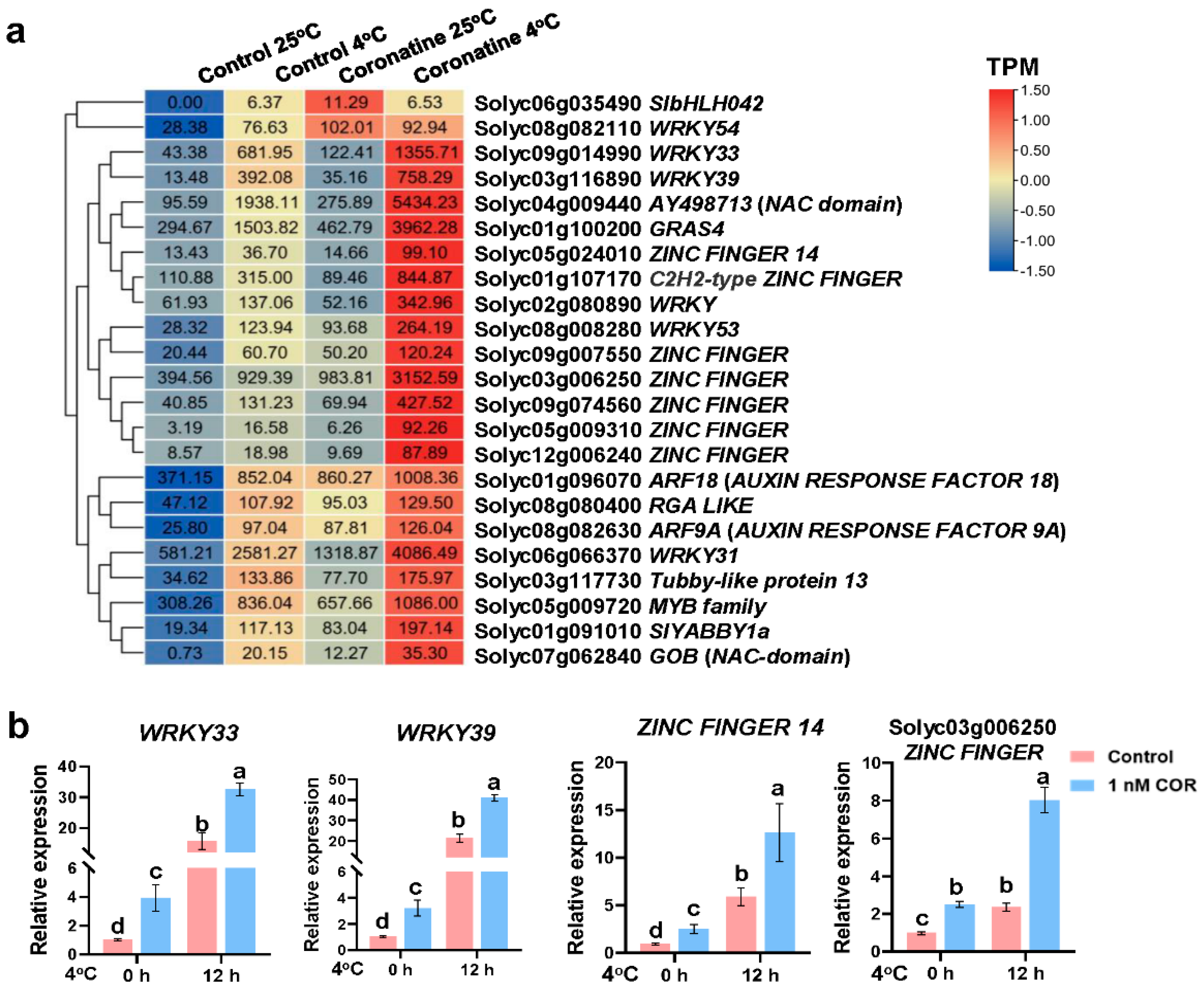
2.3. Chilling-Induced CBFs, Other First-Wave TF Genes, and JA-Responsive Genes Play Essential Roles in Coronatine-Mediated Chilling Tolerance
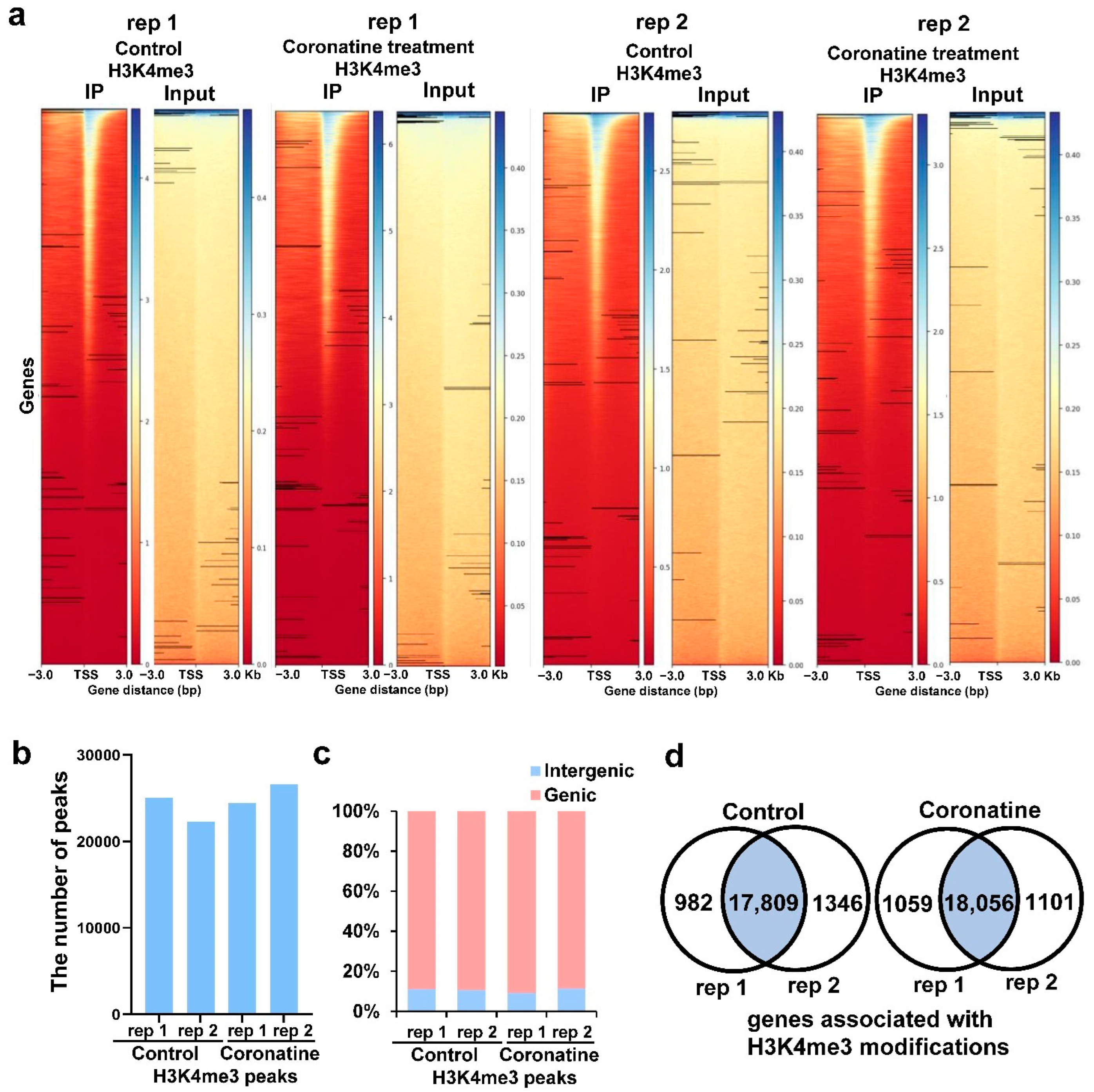
2.4. Identification of the Genome-Wide H3K4me3 Profiles in Tomato
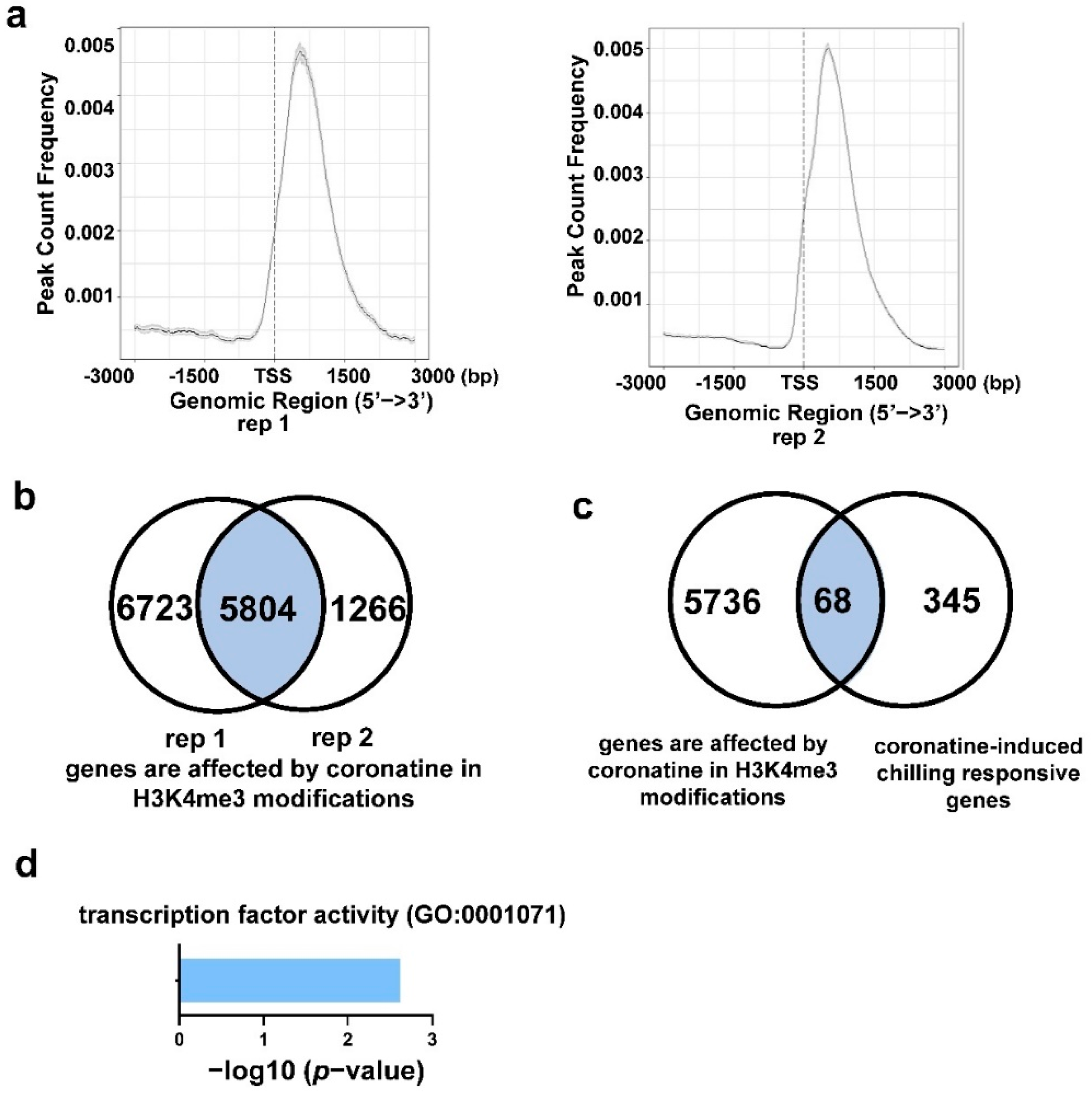
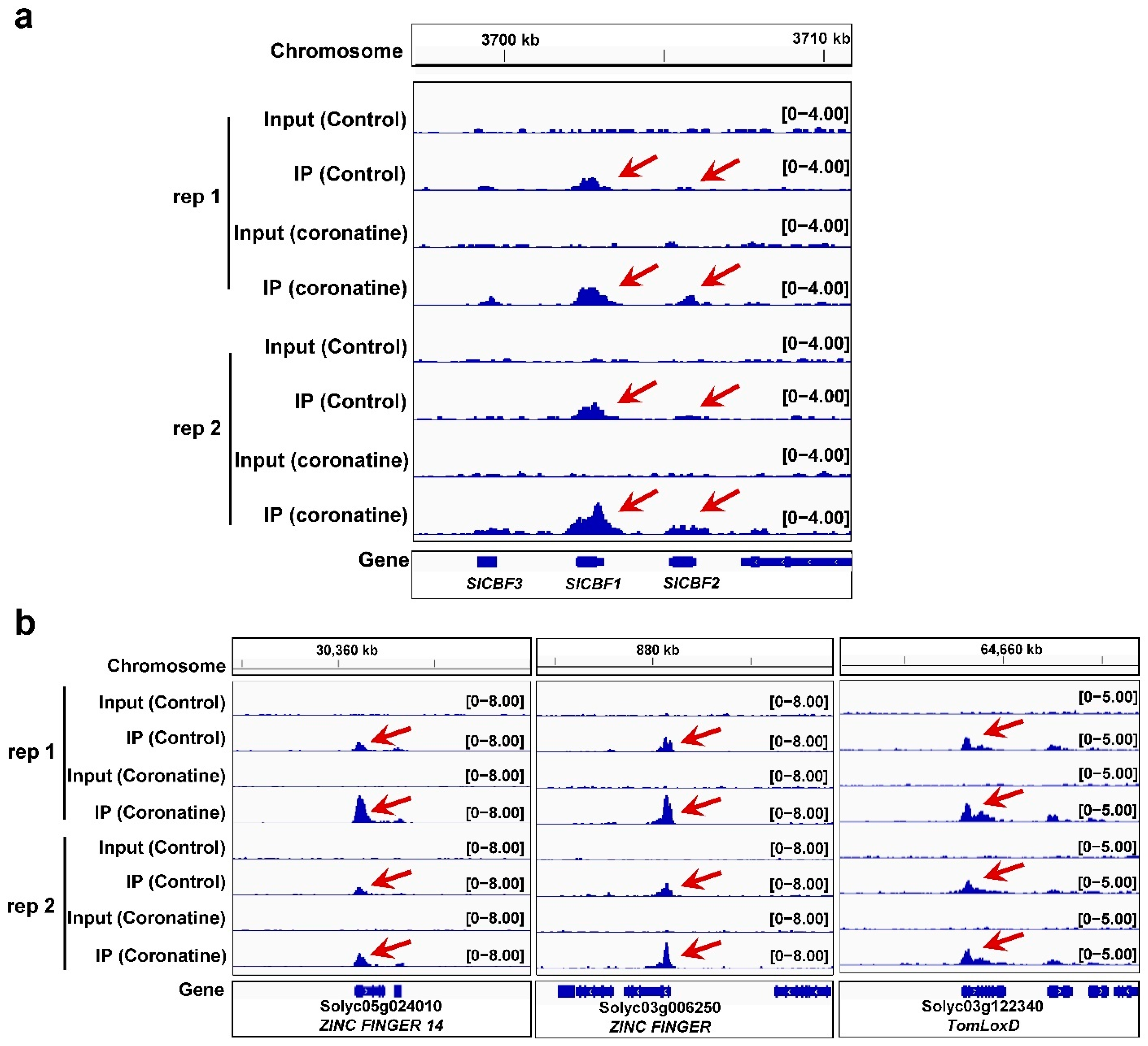
2.5. Coronatine-Enhanced H3K4me3 Modifications Are Associated with Transcriptional Reprogramming
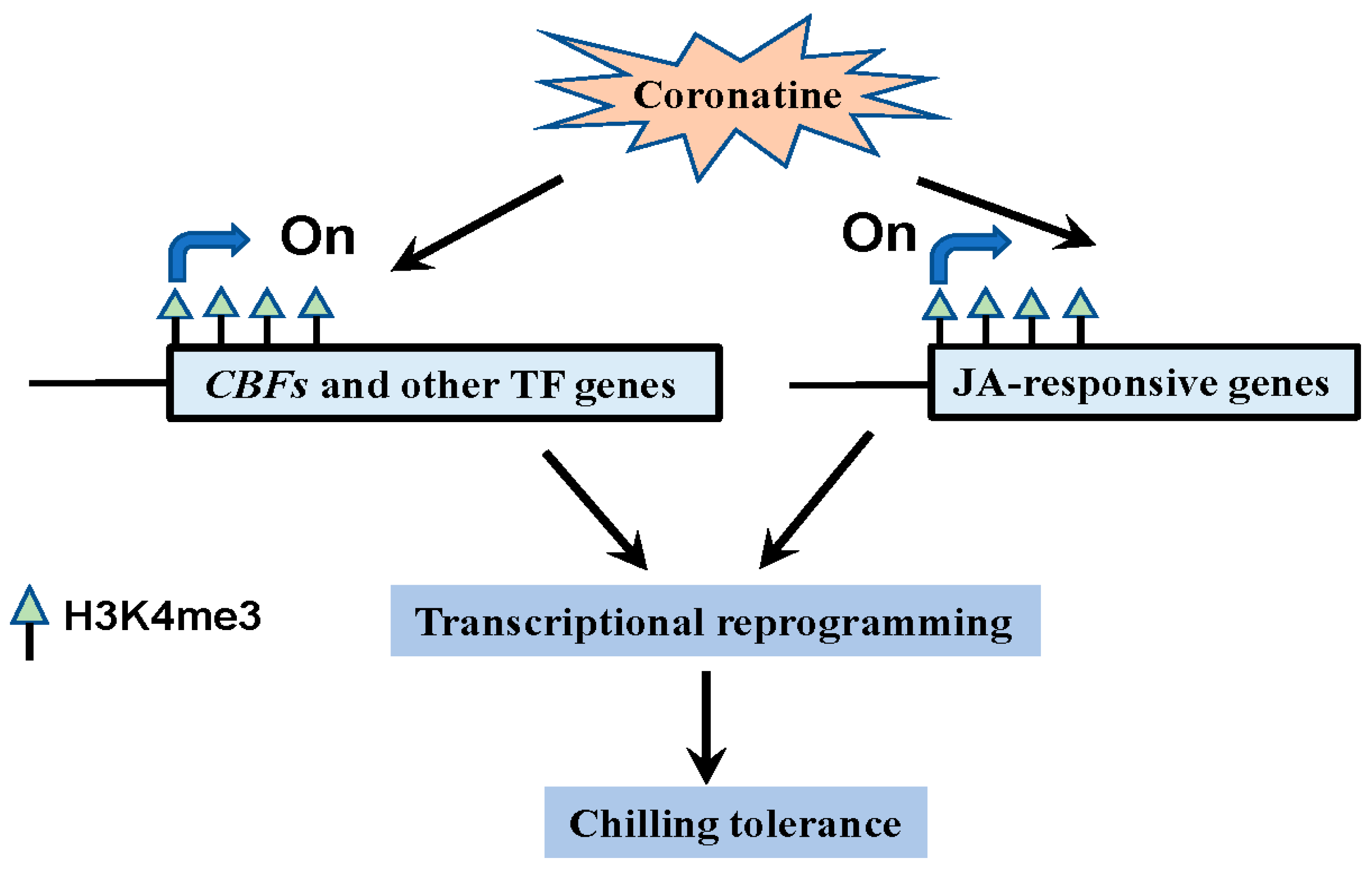
3. Discussion
3.1. Coronatine Is Involved in JA- and Oxidative Reactions-Mediated Chilling Tolerance
3.2. Coronatine Is Implicated in the Early Events of Chilling Response
3.3. Epigenetic Modifications Play Critical Roles in the Coronatine-Mediated Chilling Tolerance
3.4. CBFs and Other First-Wave Transcription Factors Play Essential Roles in Coronatine-Mediated Chilling Tolerance Similar to “Chilling Acclimation”
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Production and Preparation of Coronatine
4.3. Plant Treatment
4.4. Ion Leakage Assays
4.5. RT-qPCR Assays
4.6. RNA-Seq Analysis
4.7. ChIP-Seq Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data availability Statement
Conflicts of Interest
References
- Thomashow, M.F. PLANT COLD ACCLIMATION: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Zhang, X.; Fowler, S.G.; Cheng, H.; Lou, Y.; Rhee, S.Y.; Stockinger, E.J.; Thomashow, M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 2004, 39, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, X.; Li, M.; Yang, H.; Fu, D.; Lv, J.; Ding, Y.; Gong, Z.; Shi, Y.; Yang, S. The direct targets of CBFs: In cold stress response and beyond. J. Integr. Plant Biol. 2021, 63, 1874–1887. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef] [Green Version]
- Choi, D.W.; Rodriguez, E.M.; Close, T.J. Barley Cbf3 gene identification, expression pattern, and map location. Plant Physiol. 2002, 129, 1781–1787. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Barrero-Gil, J.; Huertas, R.; Rambla, J.L.; Granell, A.; Salinas, J. Tomato plants increase their tolerance to low temperature in a chilling acclimation process entailing comprehensive transcriptional and metabolic adjustments. Plant Cell Environ. 2016, 39, 2303–2318. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Shiba, H.; Ohta, M.; Kang, S.W.; Sato, A.; Yuasa, T.; Iwaya-Inoue, M.; Kamada, H.; Ezura, H. SlICE1 encoding a MYC-type transcription factor controls cold tolerance in tomato, Solanum lycopersicum. Plant Biotechnol. 2012, 29, 253–260. [Google Scholar] [CrossRef]
- Li, X.D.; Zhuang, K.Y.; Liu, Z.M.; Yang, D.Y.; Ma, N.N.; Meng, Q.W. Overexpression of a novel NAC-type tomato transcription factor, SlNAM1, enhances the chilling stress tolerance of transgenic tobacco. J. Plant Physiol. 2016, 204, 54–65. [Google Scholar] [CrossRef]
- Min, D.; Li, F.; Zhang, X.; Cui, X.; Shu, P.; Dong, L.; Ren, C. SlMYC2 Involved in Methyl Jasmonate-Induced Tomato Fruit Chilling Tolerance. J. Agric. Food Chem. 2018, 66, 3110–3117. [Google Scholar] [CrossRef]
- Kaplan, F.; Guy, C.L. Beta-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004, 135, 1674–1684. [Google Scholar] [CrossRef]
- Kaplan, F.; Kopka, J.; Sung, D.Y.; Zhao, W.; Popp, M.; Porat, R.; Guy, C.L. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007, 50, 967–981. [Google Scholar] [CrossRef]
- He, J.; Han, W.; Wang, J.; Qian, Y.; Saito, M.; Bai, W.; Song, J.; Lv, G. Functions of oligosaccharides in improving tomato seeding growth and chilling resistance. J. Plant Growth Regul. 2021, 41, 535–545. [Google Scholar] [CrossRef]
- Gao, C.; Sheteiwy, M.S.; Han, J.; Dong, Z.; Pan, R.; Guan, Y.; Alhaj Hamoud, Y.; Hu, J. Polyamine biosynthetic pathways and their relation with the cold tolerance of maize (Zea mays L.) seedlings. Plant Signal. Behav. 2020, 15, 1807722. [Google Scholar] [CrossRef]
- Sheteiwy, M.; Shen, H.Q.; Xu, J.G.; Guan, Y.J.; Song, W.J.; Hu, J. Seed polyamines metabolism induced by seed priming with spermidine and 5-aminolevulinic acid for chilling tolerance improvement in rice (Oryza sativa L.) seedlings. Environ. Exp. Bot. 2017, 137, 58–72. [Google Scholar] [CrossRef]
- Kaniuga, Z. Chilling response of plants: Importance of galactolipase, free fatty acids and free radicals. Plant Biol. 2008, 10, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Hajihashemi, S.; Brestic, M.; Landi, M.; Skalicky, M. Resistance of Fritillaria imperialis to freezing stress through gene expression, osmotic adjustment and antioxidants. Sci. Rep. 2020, 10, 10427. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.; Sharma, N.; Sahu, P.P.; Prasad, M. Chromatin-based epigenetic regulation of plant abiotic stress response. Curr. Genom. 2016, 17, 490–498. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 2016, 537, 558–562. [Google Scholar] [CrossRef]
- Dong, X.; Zhang, M.; Chen, J.; Peng, L.; Zhang, N.; Wang, X.; Lai, J. Dynamic and antagonistic allele-specific epigenetic modifications controlling the expression of imprinted genes in maize endosperm. Mol. Plant 2017, 10, 442–455. [Google Scholar] [CrossRef]
- Motallebipour, M.; Ameur, A.; Reddy Bysani, M.S.; Patra, K.; Wallerman, O.; Mangion, J.; Barker, M.A.; McKernan, K.J.; Komorowski, J.; Wadelius, C. Differential binding and co-binding pattern of FOXA1 and FOXA3 and their relation to H3K4me3 in HepG2 cells revealed by ChIP-seq. Genome Biol. 2009, 10, R129. [Google Scholar] [CrossRef]
- Zhang, X.; Bernatavichute, Y.V.; Cokus, S.; Pellegrini, M.; Jacobsen, S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009, 10, R62. [Google Scholar] [CrossRef]
- Wang, Z.; Casas-Mollano, J.A.; Xu, J.; Riethoven, J.J.; Zhang, C.; Cerutti, H. Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 8487–8492. [Google Scholar] [CrossRef] [Green Version]
- van Dijk, K.; Ding, Y.; Malkaram, S.; Riethoven, J.J.; Liu, R.; Yang, J.; Laczko, P.; Chen, H.; Xia, Y.; Ladunga, I.; et al. Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Wani, S.H.; Roychoudhury, A. Epigenetic control of plant cold responses. Front. Plant Sci. 2017, 8, 1643. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.S.; Lee, D.; Choi, G.; Chung, W.I. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 2009, 60, 112–121. [Google Scholar] [CrossRef]
- Cintas, N.A.; Koike, S.T.; Bull, C.T. A new pathovar, Pseudomonas syringae pv. alisalensis pv. nov., proposed for the causal agent of bacterial blight of broccoli and broccoli raab. Plant Dis. 2002, 86, 992–998. [Google Scholar] [CrossRef] [PubMed]
- Bender, C.L.; Alarcon-Chaidez, F.; Gross, D.C. Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 1999, 63, 266–292. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.M.; Hernandez-Guzman, G.; Kloek, A.P.; Alarcon-Chaidez, F.; Sreedharan, A.; Rangaswamy, V.; Penaloza-Vazquez, A.; Bender, C.L.; Kunkel, B.N. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 2004, 17, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Uppalapati, S.R.; Ayoubi, P.; Weng, H.; Palmer, D.A.; Mitchell, R.E.; Jones, W.; Bender, C.L. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 2005, 42, 201–217. [Google Scholar] [CrossRef]
- Uppalapati, S.R.; Ishiga, Y.; Wangdi, T.; Urbanczyk-Wochniak, E.; Ishiga, T.; Mysore, K.S.; Bender, C.L. Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: Phenotypic and gene expression analyses of the virulence function of coronatine. Mol. Plant Microbe Interact. 2008, 21, 383–395. [Google Scholar] [CrossRef] [Green Version]
- Hao, L.; Wang, Y.; Zhang, J.; Xie, Y.; Zhang, M.; Duan, L.; Li, Z. Coronatine enhances drought tolerance via improving antioxidative capacity to maintaining higher photosynthetic performance in soybean. Plant Sci. 2013, 210, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, W.J.; Wang, Q.; Eneji, A.E.; Li, Z.H.; Duan, L.S. Coronatine enhances chilling tolerance in cucumber (Cucumis sativus L.) seedlings by improving the antioxidative defence system. J. Agron. Crop Sci. 2009, 195, 377–383. [Google Scholar] [CrossRef]
- Xie, Z.; Duan, L.; Tian, X.; Wang, B.; Eneji, A.E.; Li, Z. Coronatine alleviates salinity stress in cotton by improving the antioxidative defense system and radical-scavenging activity. J. Plant Physiol. 2008, 165, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wilkerson, C.G.; Kuchar, J.A.; Phinney, B.S.; Howe, G.A. Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc. Natl. Acad. Sci. USA 2005, 102, 19237–19242. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhao, J.; Tzeng, D.T.W.; Liu, Y.; Deng, L.; Yang, T.; Zhai, Q.; Wu, F.; Huang, Z.; Zhou, M.; et al. MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Y.; McCaig, B.C.; Wingerd, B.A.; Wang, J.; Whalon, M.E.; Pichersky, E.; Howe, G.A. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 2004, 16, 126–143. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef]
- Ding, C.K.; Wang, C.Y.; Gross, K.C.; Smith, D.L. Jasmonate and salicylate induce the expression of pathogenesis-related-protein genes and increase resistance to chilling injury in tomato fruit. Planta 2002, 214, 895–901. [Google Scholar] [CrossRef]
- Gonzalez-Aguilar, G.A.; Fortiz, J.; Cruz, R.; Baez, R.; Wang, C.Y. Methyl jasmonate reduces chilling injury and maintains postharvest quality of mango fruit. J. Agric. Food Chem. 2000, 48, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.L.; Wang, J.N.; Shan, W.; Fan, J.G.; Kuang, J.F.; Wu, K.Q.; Li, X.P.; Chen, W.X.; He, F.Y.; Chen, J.Y.; et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 2013, 36, 30–51. [Google Scholar] [CrossRef]
- Ding, F.; Ren, L.; Xie, F.; Wang, M.; Zhang, S. Jasmonate and melatonin act synergistically to potentiate cold tolerance in tomato plants. Front. Plant Sci. 2021, 12, 763284. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, H.H.; Hua, S.; Lin, K.H.; Chen, N.; Zhang, Y.; You, Z.; Kuo, Y.W.; Chen, S.P. Cloning and overexpression of the ascorbate peroxidase gene from the yam (Dioscorea alata) enhances chilling and flood tolerance in transgenic Arabidopsis. J. Plant Res. 2019, 132, 857–866. [Google Scholar] [CrossRef]
- Roxas, V.P.; Lodhi, S.A.; Garrett, D.K.; Mahan, J.R.; Allen, R.D. Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol. 2000, 41, 1229–1234. [Google Scholar] [CrossRef]
- Sato, Y.; Masuta, Y.; Saito, K.; Murayama, S.; Ozawa, K. Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep. 2011, 30, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Molecular regulation of plant responses to environmental temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jia, Y.; Ding, Y.; Shi, Y.; Li, Z.; Guo, Y.; Gong, Z.; Yang, S. Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 2017, 66, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ding, Y.; Shi, Y.; Zhang, X.; Zhang, S.; Gong, Z.; Yang, S. MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642. [Google Scholar] [CrossRef]
- Hsu, C.C.; Zhu, Y.; Arrington, J.V.; Paez, J.S.; Wang, P.; Zhu, P.; Chen, I.H.; Zhu, J.K.; Tao, W.A. Universal plant phosphoproteomics workflow and its application to tomato signaling in response to cold stress. Mol. Cell Proteom. 2018, 17, 2068–2080. [Google Scholar] [CrossRef]
- Xu, W.; Gao, S.; Song, J.; Yang, Q.; Wang, T.; Zhang, Y.; Zhang, J.; Li, H.; Yang, C.; Ye, Z. NDW, encoding a receptor-like protein kinase, regulates plant growth, cold tolerance and susceptibility to Botrytis cinerea in tomato. Plant Sci. 2020, 301, 110684. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Hu, Z.; Mao, Z.; Mei, Y.; Feng, S.; Shi, K. A novel leucine-rich repeat receptor-like kinase MRK1 regulates resistance to multiple stresses in tomato. Hortic. Res. 2022, 9, uhab088. [Google Scholar] [CrossRef]
- Ensminger, I.; Busch, F.; Huner, N.P.A. Photostasis and cold acclimation: Sensing low temperature through photosynthesis. Physiol. Plant 2006, 126, 28–44. [Google Scholar] [CrossRef]
- Hajihashemi, S.; Noedoost, F.; Geuns, J.M.C.; Djalovic, I.; Siddique, K.H.M. Effect of cold stress on photosynthetic traits, carbohydrates, morphology, and anatomy in nine cultivars of Stevia rebaudiana. Front. Plant Sci. 2018, 9, 1430. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F.; Artus, N.; Gilmour, S.; Stockinger, E.; Wilhelm, K.; Zarka, D.; Joseph, R.A.; Uemura, M.; Steponkus, P.L. Function and regulation of Arabidopsis thaliana COR (cold-regulated) genes. Plant Physiol. 1996, 111, 81003. [Google Scholar] [CrossRef]
- Thalhammer, A.; Bryant, G.; Sulpice, R.; Hincha, D.K. Disordered cold regulated15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in vivo. Plant Physiol. 2014, 166, 190–201. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ding, Y.; Shi, Y.; Ma, L.; Wang, Y.; Song, C.; Wilkins, K.A.; Davies, J.M.; Knight, H.; Knight, M.R.; et al. The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 2021, 40, e104559. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Gai, W.X.; Li, Y.; Yu, Y.N.; Ali, M.; Gong, Z.H. The CBL-interacting protein kinase CaCIPK13 positively regulates defence mechanisms against cold stress in pepper. J. Exp. Bot. 2022, 73, 1655–1667. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Li, H.; Chen, X.; Xiang, X.; Guo, Z.; Yu, J.; Zhou, Y. The role of calcium-dependent protein kinase in hydrogen peroxide, nitric oxide and ABA-dependent cold acclimation. J. Exp. Bot. 2018, 69, 4127–4139. [Google Scholar] [CrossRef] [PubMed]
- Hereme, R.; Galleguillos, C.; Morales-Navarro, S.; Molina-Montenegro, M.A. What if the cold days return? Epigenetic mechanisms in plants to cold tolerance. Planta 2021, 254, 46. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, P.; Hou, H.; Zhang, H.; Wang, Y.; Yan, S.; Huang, Y.; Li, H.; Tan, J.; Hu, A.; et al. Transcriptional regulation of cell cycle genes in response to abiotic stresses correlates with dynamic changes in histone modifications in maize. PLoS ONE 2014, 9, e106070. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, L.; He, S.; Huang, M.; Tan, J.; Zhao, L.; Yan, S.; Li, H.; Zhou, K.; Liang, Y.; et al. Cold stress selectively unsilences tandem repeats in heterochromatin associated with accumulation of H3K9ac. Plant Cell Environ. 2012, 35, 2130–2142. [Google Scholar] [CrossRef]
- Yang, R.; Hong, Y.; Ren, Z.; Tang, K.; Zhang, H.; Zhu, J.K.; Zhao, C. A role for PICKLE in the regulation of cold and salt stress tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 900. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, W.; Marand, A.; Zhu, B.; Buell, C.; Jiang, J. Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biol. 2019, 20, 123. [Google Scholar] [CrossRef]
- Zeng, C.; Chen, Z.; Xia, J.; Zhang, K.; Chen, X.; Zhou, Y.; Bo, W.; Song, S.; Deng, D.; Guo, X.; et al. Chilling acclimation provides immunity to stress by altering regulatory networks and inducing genes with protective functions in cassava. BMC Plant Biol. 2014, 14, 207. [Google Scholar] [CrossRef] [Green Version]
- Zeng, R.; Li, Z.; Shi, Y.; Fu, D.; Yin, P.; Cheng, J.; Jiang, C.; Yang, S. Natural variation in a type-A response regulator confers maize chilling tolerance. Nat. Commun. 2021, 12, 4713. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. Agrigo v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Li, Z.; Wu, S.; Yu, C.; Wang, X.; Wang, Y.; Peng, Z.; Gao, Y.; Li, R.; Shen, Y.; et al. Coronatine Enhances Chilling Tolerance of Tomato Plants by Inducing Chilling-Related Epigenetic Adaptations and Transcriptional Reprogramming. Int. J. Mol. Sci. 2022, 23, 10049. https://doi.org/10.3390/ijms231710049
Liu Z, Li Z, Wu S, Yu C, Wang X, Wang Y, Peng Z, Gao Y, Li R, Shen Y, et al. Coronatine Enhances Chilling Tolerance of Tomato Plants by Inducing Chilling-Related Epigenetic Adaptations and Transcriptional Reprogramming. International Journal of Molecular Sciences. 2022; 23(17):10049. https://doi.org/10.3390/ijms231710049
Chicago/Turabian StyleLiu, Ziyan, Zhuoyang Li, Shifeng Wu, Chunxin Yu, Xi Wang, Ye Wang, Zhen Peng, Yuerong Gao, Runzhi Li, Yuanyue Shen, and et al. 2022. "Coronatine Enhances Chilling Tolerance of Tomato Plants by Inducing Chilling-Related Epigenetic Adaptations and Transcriptional Reprogramming" International Journal of Molecular Sciences 23, no. 17: 10049. https://doi.org/10.3390/ijms231710049
APA StyleLiu, Z., Li, Z., Wu, S., Yu, C., Wang, X., Wang, Y., Peng, Z., Gao, Y., Li, R., Shen, Y., & Duan, L. (2022). Coronatine Enhances Chilling Tolerance of Tomato Plants by Inducing Chilling-Related Epigenetic Adaptations and Transcriptional Reprogramming. International Journal of Molecular Sciences, 23(17), 10049. https://doi.org/10.3390/ijms231710049






