Functional Characterization of Two RNA Methyltransferase Genes METTL3 and METTL14 Uncovers the Roles of m6A in Mediating Adaptation of Plutella xylostella to Host Plants
Abstract
:1. Introduction
2. Results
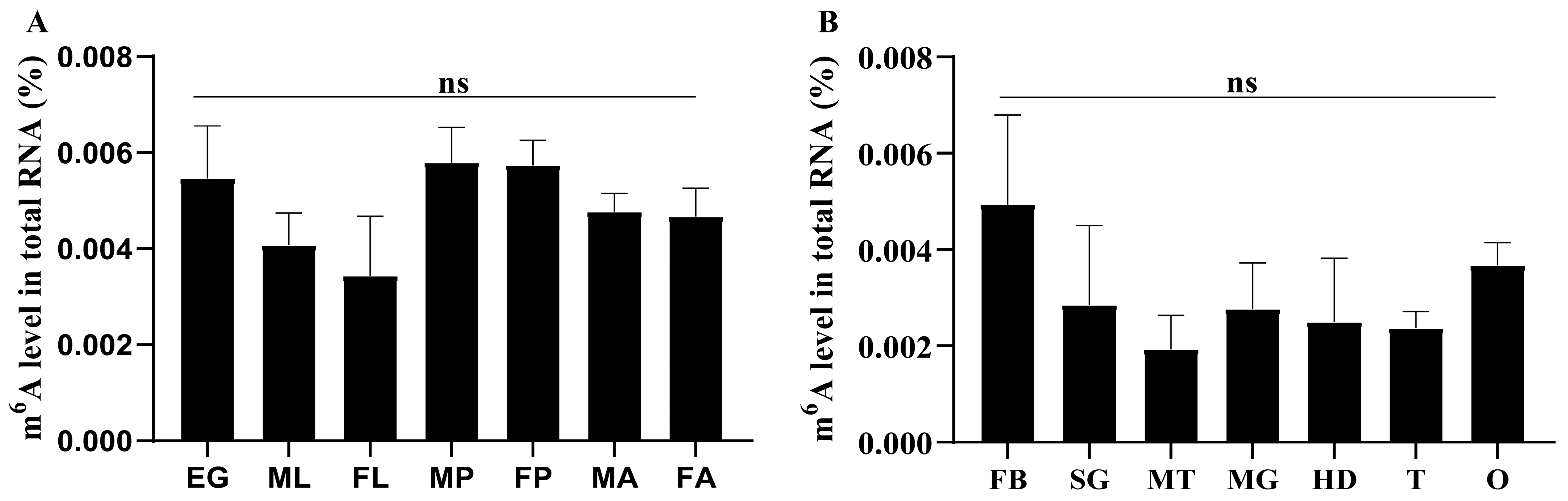
2.1. m6A Modification in P. xylostella
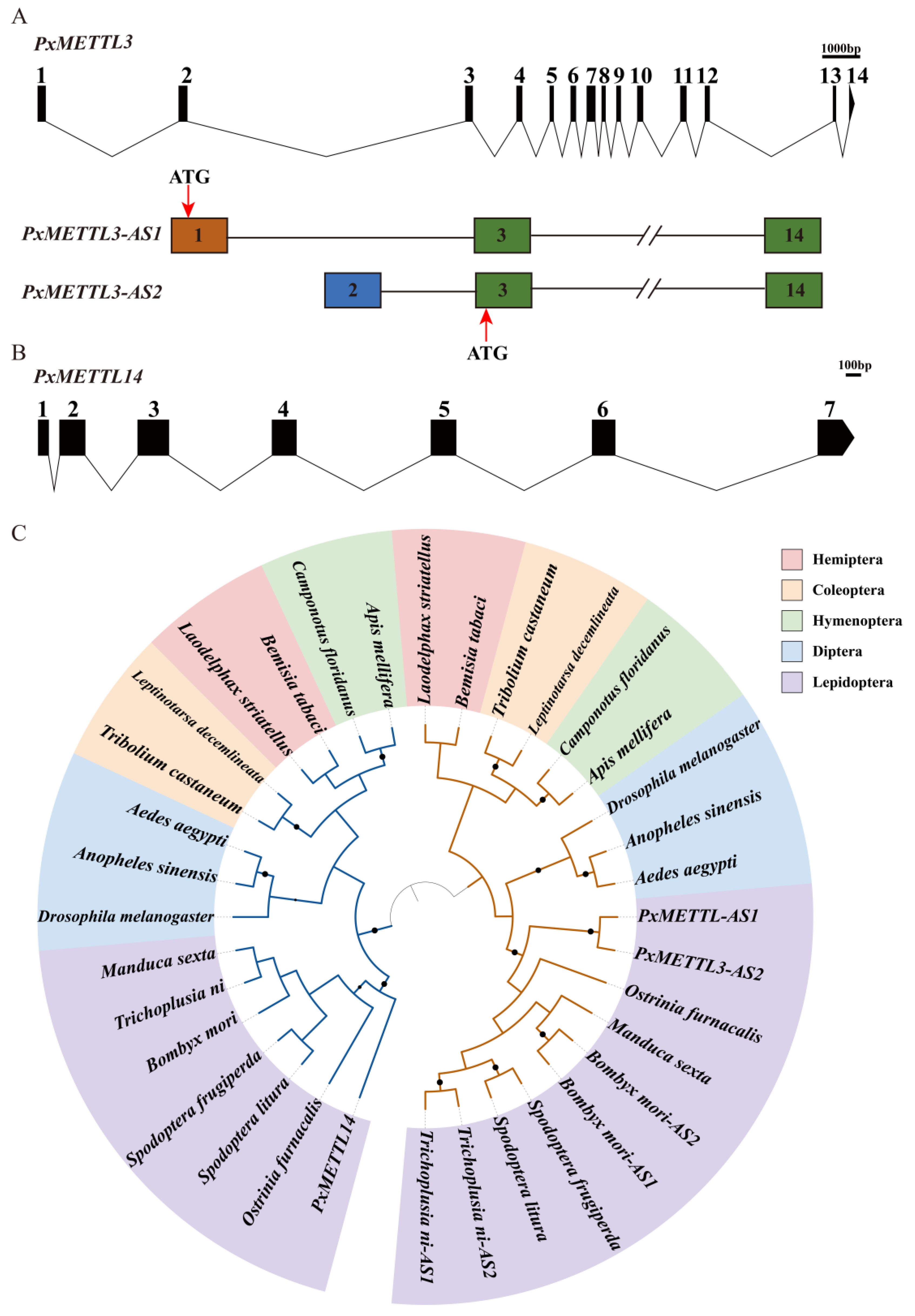
2.2. Molecular Characteristics of PxMETTL3 and PxMETTL14
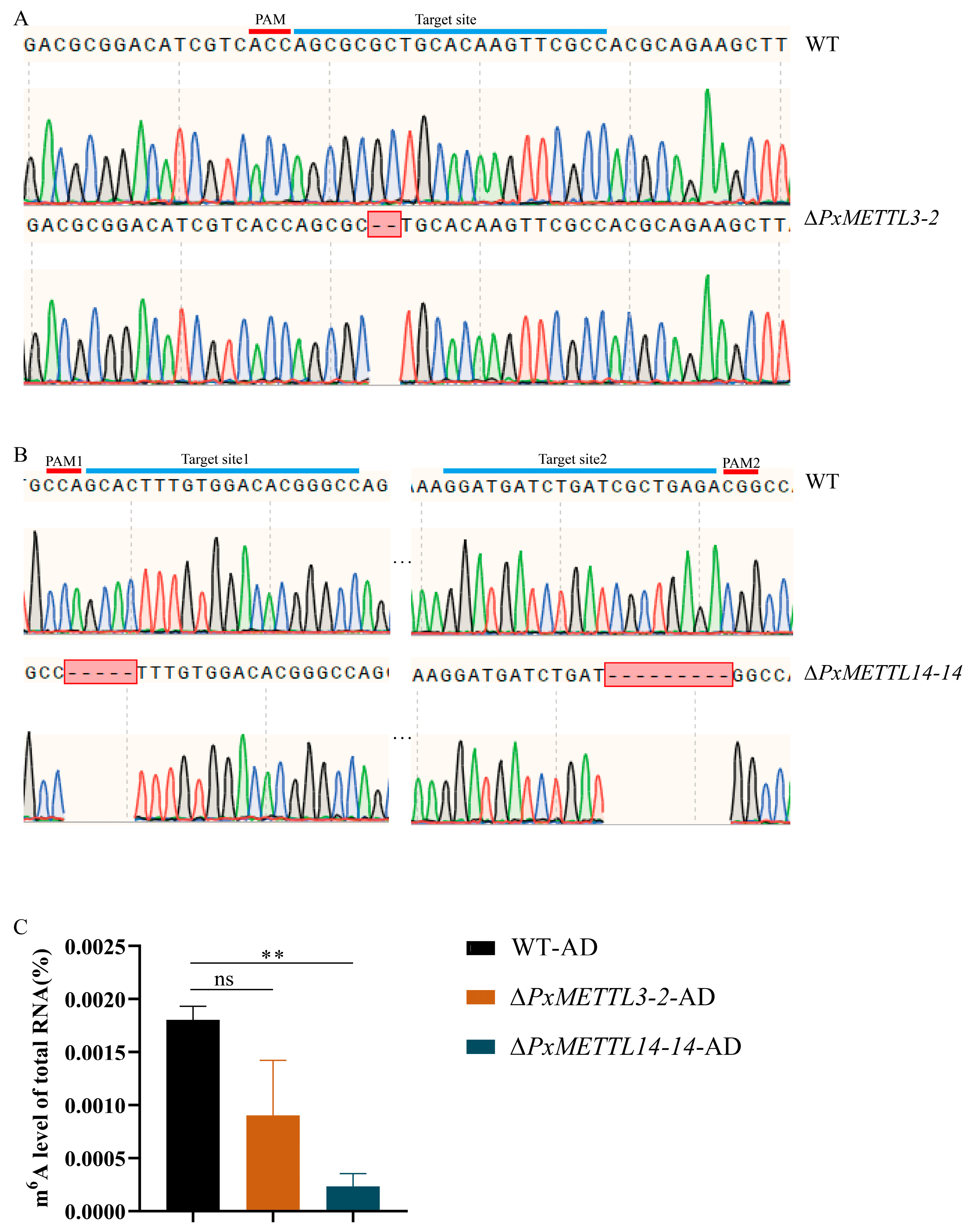
2.3. Mutant Strains of PxMETTL3 and PxMETTL14
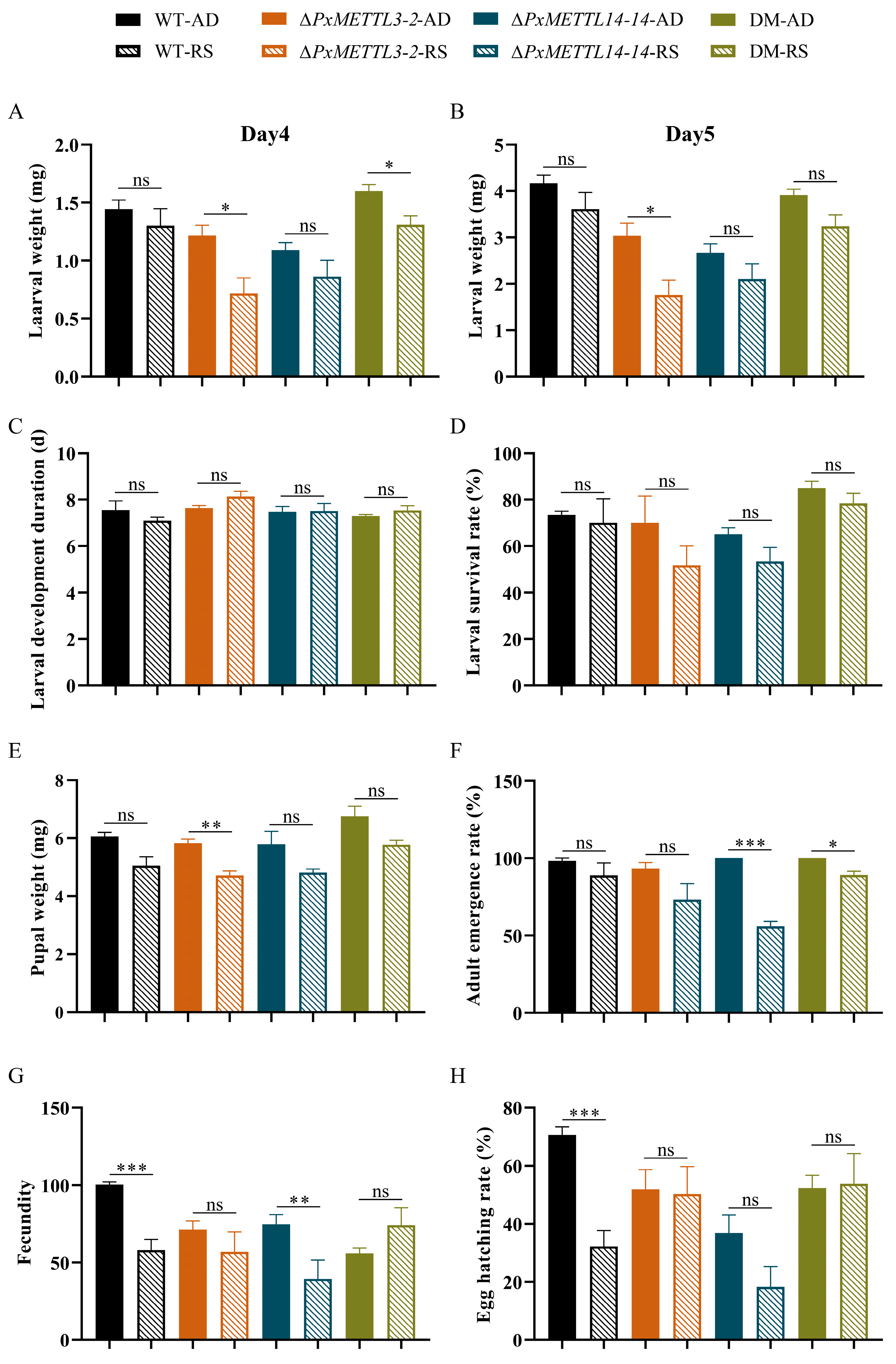
2.4. Comparison of the Performance of WT and Mutant Strains Undergoing Host Transfer
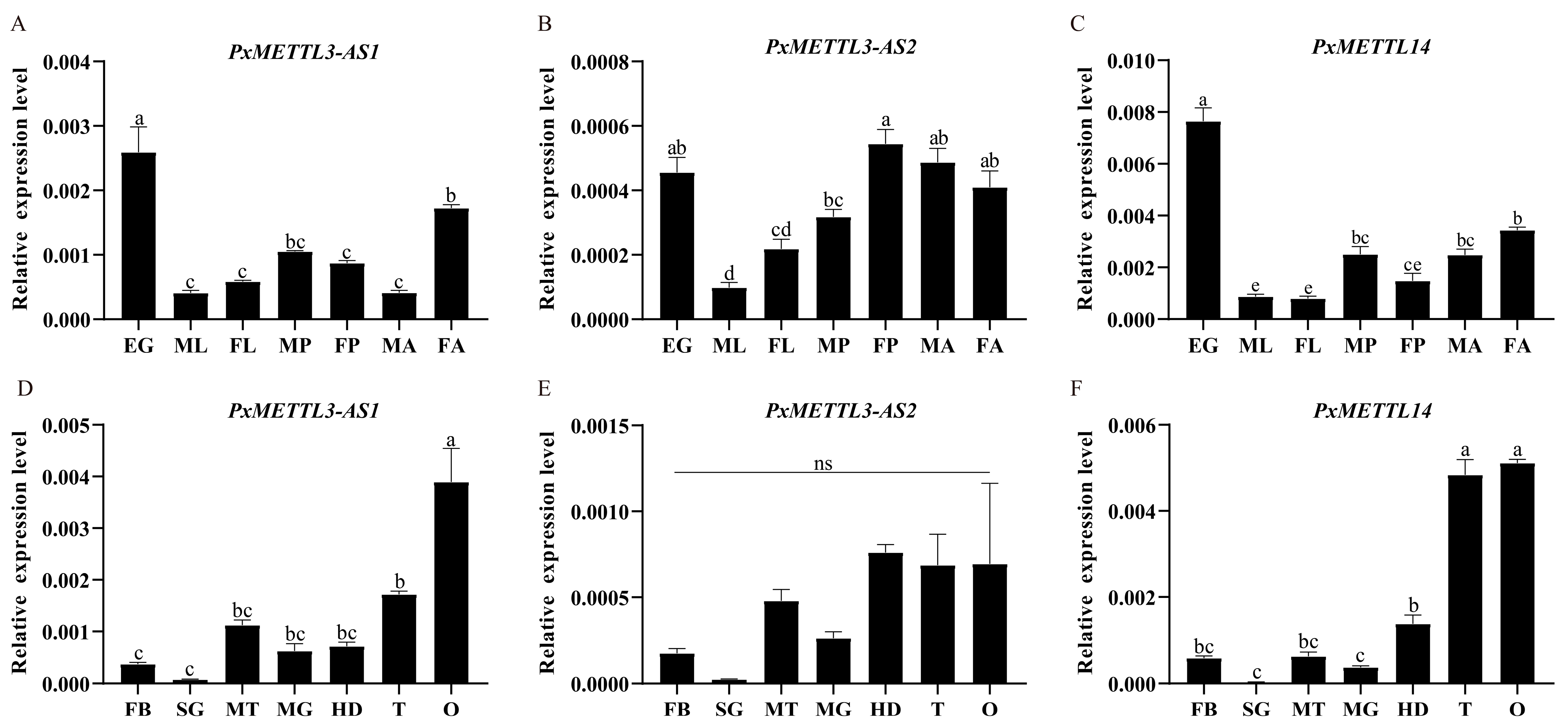
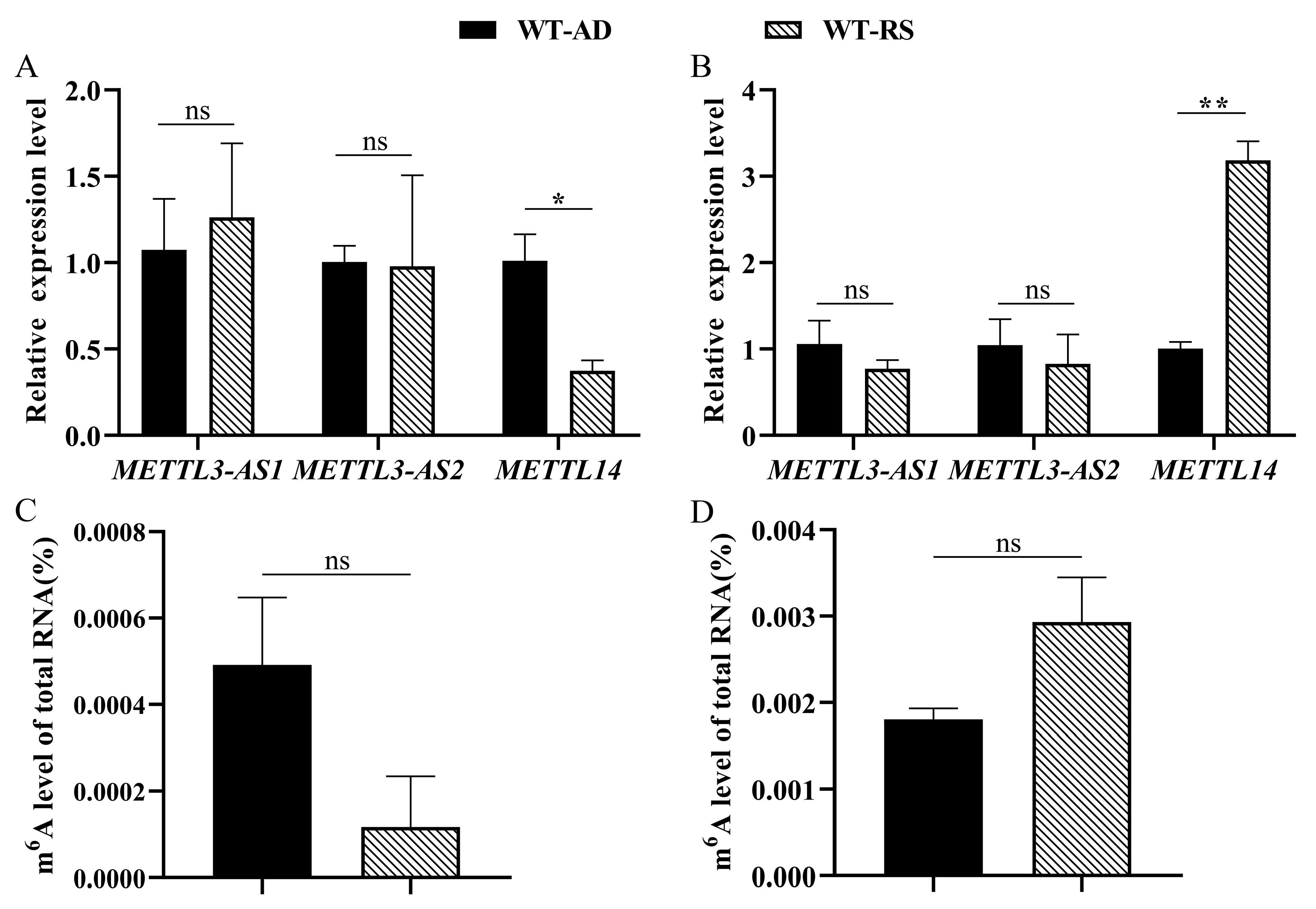
2.5. Changes in PxMETTL3 and PxMETTL14 and m6A Upon Host Transfer
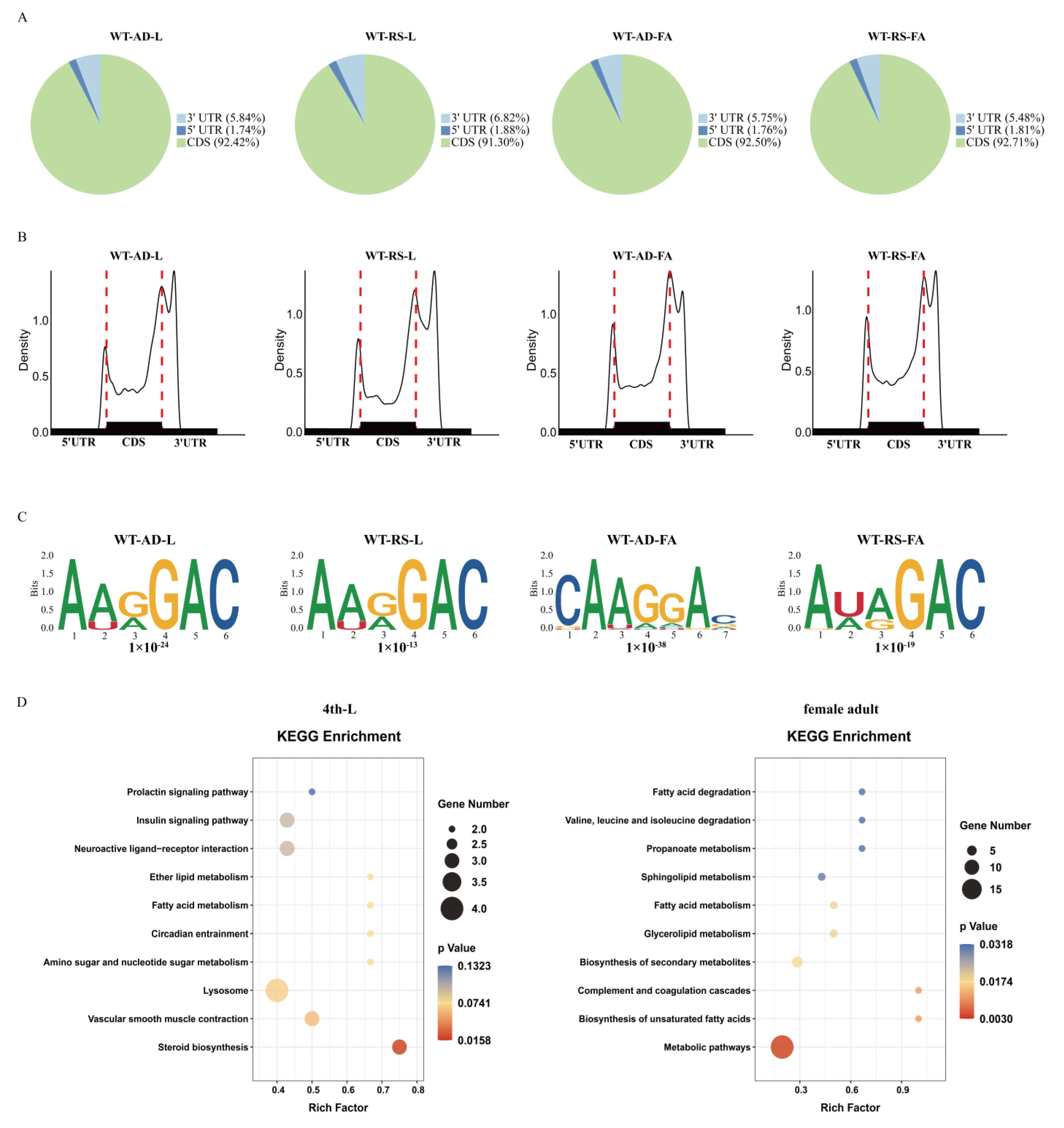
2.6. Transcriptome and Epitranscriptome Dynamics during Host Transfer
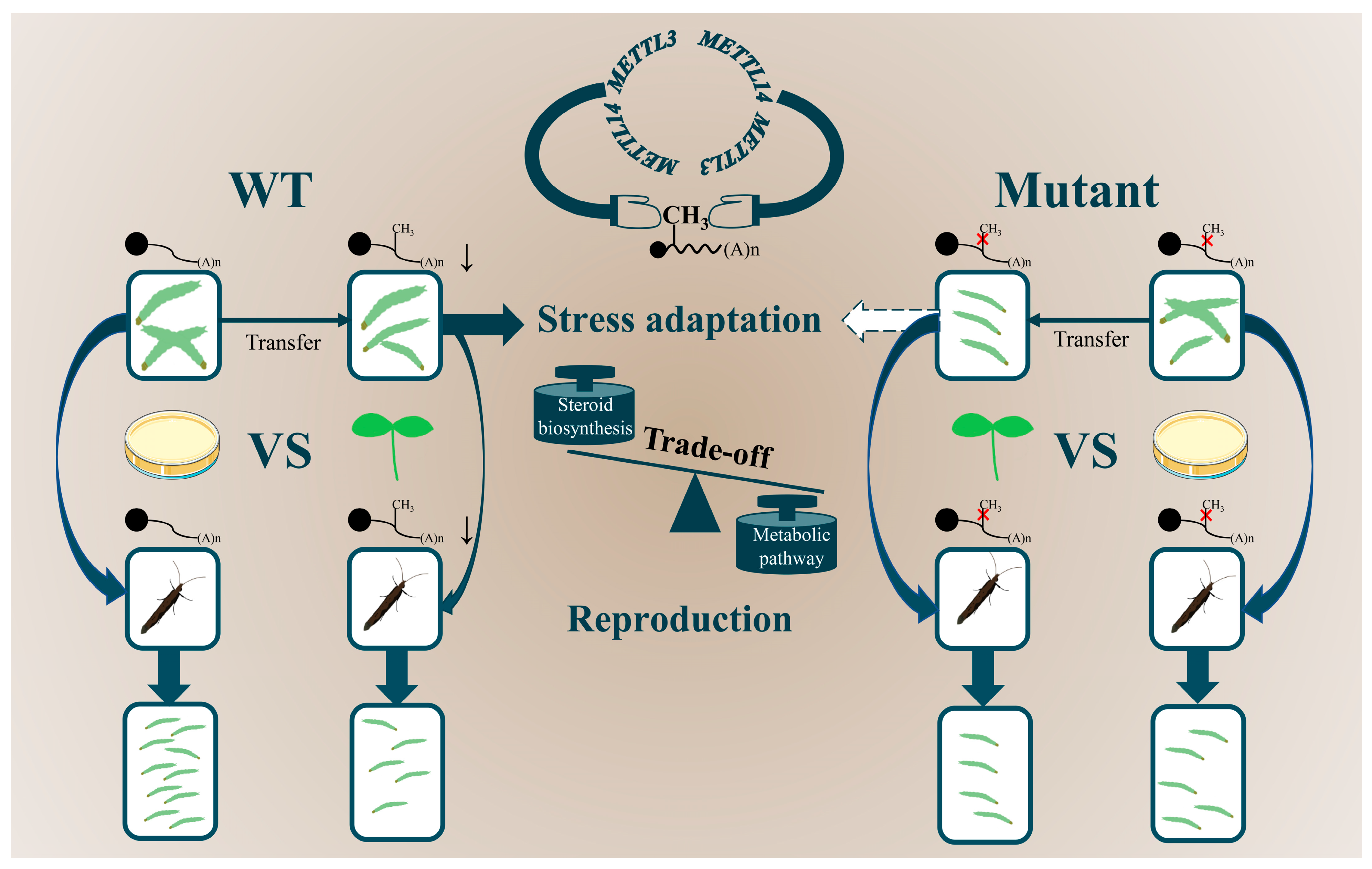
3. Discussion
4. Materials and Methods
4.1. Insect Strains and Host Plant
4.2. Extraction of RNA and Gene Cloning
4.3. Construction of Phylogenetic Tree
4.4. qRT–PCR
4.5. Design of sgRNA and Off-Target Analysis
4.6. Preparation of sgRNA and Embryo Microinjection
4.7. Establishment of Mutant Strains
4.8. Quantification of m6A
4.9. Insect Bioassays
4.10. m6A-seq and RNA-seq
4.11. Statistical Analysis
4.12. Data Availability
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Mason, C.E. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genom. Hum. Genet. 2014, 15, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Bodi, Z.; Button, J.D.; Grierson, D.; Fray, R.G. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010, 38, 5327–5335. [Google Scholar] [CrossRef]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Liu, S.; Yu, L.; Xiao, Y.; Zhang, S.; Wang, X.; Xu, Y.; Yu, H.; Li, Y.; Yang, J.; et al. RNA demethylation increases the yield and biomass of rice and potato plants in field trials. Nat. Biotechnol. 2021, 39, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Tang, H.-W.; Li, J.; Perrimon, N.; Yan, D. Xio is a component of the Drosophila sex determination pathway and RNA N6-methyladenosine methyltransferase complex. Proc. Natl. Acad. Sci. USA 2018, 115, 3674–3679. [Google Scholar] [CrossRef]
- Hu, L.; Wang, J.; Huang, H.; Yu, Y.; Ding, J.; Yu, Y.; Li, K.; Wei, D.; Ye, Q.; Wang, F.; et al. YTHDF1 regulates pulmonary hypertension through translational control of MAGED1. Am. J. Respir. Crit. Care Med. 2021, 203, 1158–1172. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, X.; Zhang, P.; Li, F.; Zhang, L.; Li, X.; Huang, T.; Zheng, Y.; Yu, T.; Zhang, T.; et al. Transcriptome-wide dynamics of m6A mRNA methylation during porcine spermatogenesis. Genom. Proteom. Bioinform. 2021, in press. [CrossRef]
- Yang, X.; Shao, F.; Guo, D.; Wang, W.; Wang, J.; Zhu, R.; Gao, Y.; He, J.; Lu, Z. WNT/beta-catenin-suppressed FTO expression increases m6A of c-Myc mRNA to promote tumor cell glycolysis and tumorigenesis. Cell Death Dis. 2021, 12, 462. [Google Scholar] [CrossRef]
- Ni, W.; Yao, S.; Zhou, Y.; Liu, Y.; Huang, P.; Zhou, A.; Liu, J.; Che, L.; Li, J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol. Cancer 2019, 18, 143. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Alarcon, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Li, L.; Huang, Y.; Ma, J.; Min, J. Readers, writers and erasers of N6-methylated adenosine modification. Curr. Opin. Struct. Biol. 2017, 47, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Jia, G.; Pang, X.; Wang, R.N.; Wang, X.; Li, C.J.; Smemo, S.; Dai, Q.; Bailey, K.A.; Nobrega, M.A.; et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013, 4, 1798. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, R.G.; Soller, M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016, 540, 301–304. [Google Scholar] [CrossRef] [Green Version]
- Lence, T.; Akhtar, J.; Bayer, M.; Schmid, K.; Spindler, L.; Ho, C.H.; Kreim, N.; Andrade-Navarro, M.A.; Poeck, B.; Helm, M.; et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 2016, 540, 242–247. [Google Scholar] [CrossRef]
- Kan, L.; Grozhik, A.V.; Vedanayagam, J.; Patil, D.P.; Pang, N.; Lim, K.-S.; Huang, Y.-C.; Joseph, B.; Lin, C.-J.; Despic, V.; et al. The m6A pathway facilitates sex determination in Drosophila. Nat. Commun. 2017, 8, 15737. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wu, N.; Zhang, L.; Wang, X. RNA N6-methyladenosine modification suppresses replication of rice black streaked dwarf virus and is associated with virus persistence in its insect vector. Plant Biotechnol. J. 2021, 22, 1070–1081. [Google Scholar] [CrossRef]
- Yang, X.; Wei, X.; Yang, J.; Du, T.; Yin, C.; Fu, B.; Huang, M.; Liang, J.; Gong, P.; Liu, S.; et al. Epitranscriptomic regulation of insecticide resistance. Sci. Adv. 2021, 7, eabe5903. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xiao, Y.; Li, Y.; Wang, X.; Qi, S.; Wang, Y.; Zhao, L.; Wang, K.; Peng, W.; Luo, G.Z.; et al. RNA m6A modification functions in larval development and caste differentiation in honeybee (Apis mellifera). Cell Rep. 2021, 34, 108580. [Google Scholar] [CrossRef]
- Jiang, T.; Li, J.; Qian, P.; Xue, P.; Xu, J.; Chen, Y.; Zhu, J.; Tang, S.; Zhao, Q.; Qian, H.; et al. The role of N6-methyladenosine modification on diapause in silkworm (Bombyx mori) strains that exhibit different voltinism. Mol. Reprod. Dev. 2019, 86, 1981–1992. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Dai, K.; Liang, Z.; Zhu, M.; Pan, J.; Zhang, M.; Yan, B.; Zhu, H.; Zhang, Z.; et al. N6-methyladenosine level in silkworm midgut/ovary cell line is associated with Bombyx mori nucleopolyhedrovirus infection. Front. Microbiol. 2020, 10, 2988. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, B.; Lai, Y.; Yang, F.; You, M.; He, W. Knockout of single allele of fl(2)d significantly decreases the fecundity and fertility in Plutella xylostella. Sci. Agric. Sin. 2021, 54, 3029–3042. [Google Scholar] [CrossRef]
- Sarfraz, M.; Dosdall, L.M.; Keddie, B.A. Diamondback moth–host plant interactions: Implications for pest management. Crop Prot. 2006, 25, 625–639. [Google Scholar] [CrossRef]
- Furlong, M.J.; Wright, D.J.; Dosdall, L.M. Diamondback moth ecology and management: Problems, progress, and prospects. Annu. Rev. Entomol. 2013, 58, 517–541. [Google Scholar] [CrossRef]
- Yang, F.-Y.; Chen, J.-H.; Ruan, Q.-Q.; Wang, B.-B.; Jiao, L.; Qiao, Q.-X.; He, W.-Y.; You, M.-S. Fitness comparison of Plutella xylostella on original and marginal hosts using age-stage, two-sex life tables. Ecol. Evol. 2021, 11, 9765–9775. [Google Scholar] [CrossRef]
- Chen, W.; Dong, Y.; Lin, L.; Saqib, H.S.A.; Ma, X.; Xu, X.; Zhang, L.; Jing, X.; Peng, L.; Wang, Y.; et al. Implication for DNA methylation involved in the host transfer of diamondback moth, Plutella xylostella (L.). Arch. Insect Biochem. Physiol. 2019, 102, e21600. [Google Scholar] [CrossRef] [PubMed]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.-Z.; MacQueen, A.; Zheng, G.; Duan, H.; Dore, L.C.; Lu, Z.; Liu, J.; Chen, K.; Jia, G.; Bergelson, J.; et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014, 5, 5630. [Google Scholar] [CrossRef]
- Yue, H.; Nie, X.; Yan, Z.; Weining, S. N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol. J. 2019, 17, 1194–1208. [Google Scholar] [CrossRef]
- Liu, C.; Cao, J.; Zhang, H.; Wu, J.; Yin, J. Profiling of transcriptome-wide N6-Methyladenosine (m6A) modifications and identifying m6a associated regulation in sperm tail formation in Anopheles sinensis. Int. J. Mol. Sci. 2022, 23, 4630. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef]
- Schwenke, R.A.; Lazzaro, B.P.; Wolfner, M.F. Reproduction–immunity trade-offs in insects. Annu. Rev. Entomol. 2016, 61, 239–256. [Google Scholar] [CrossRef]
- Zust, T.; Agrawal, A.A. Trade-offs between plant growth and defense against insect herbivory: An emerging mechanistic synthesis. Annu. Rev. Plant Biol. 2017, 68, 513–534. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Liu, W.; Wang, G.-L. Balancing immunity and yield in crop plants. Trends Plant Sci. 2017, 22, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Jozwiak, A.; Holoidovsky, L.; Meijler, M.M.; Meir, S.; Rogachev, I.; Aharoni, A. Glycosylation of N-hydroxy-pipecolic acid equilibrates between systemic acquired resistance response and plant growth. Mol. Plant 2021, 14, 440–455. [Google Scholar] [CrossRef] [PubMed]
- Sharp, G.L.; Martin, J.M.; Lanning, S.P.; Blake, N.K.; Brey, C.W.; Sivamani, E.; Qu, R.; Talbert, L.E. Field evaluation of transgenic and classical sources of Wheat streak mosaic virus resistance. Crop Sci. 2002, 42, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, E.T.; Moeller, M.E.; Yamanaka, N.; Ou, Q.; Laursen, J.M.; Soenderholm, C.; Zhuo, R.; Phelps, B.; Tang, K.; Zeng, J.; et al. A Drosophila genome-wide screen identifies regulators of steroid hormone production and developmental timing. Dev. Cell 2016, 37, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Q.; Zhang, J.-B.; Zheng, Y.; Zhang, W.-D.; Guo, H.-X.; Cong, S.; Ding, Y.; Yuan, B. Comprehensive analysis of differences in N6-methyladenosine RNA methylomes in the rat adenohypophysis after GnRH treatment. FASEB Journal 2022, 36, e22204. [Google Scholar] [CrossRef]
- Arrese, E.L.; Soulages, J.L. Insect fat body: Energy, metabolism, and regulation. Annu. Rev. Entomol. 2010, 55, 207–225. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Y.; Zeng, B.; Liu, Z.; Xu, X.; Meng, Q.; Huang, Y.; Yang, G.; Vasseur, L.; Gurr, G.M.; et al. Functional characterization of Pol III U6 promoters for gene knockdown and knockout in Plutella xylostella. Insect Biochem. Mol. Biol. 2017, 89, 71–78. [Google Scholar] [CrossRef]
- You, M.; Yue, Z.; He, W.; Yang, X.; Yang, G.; Xie, M.; Zhan, D.; Baxter, S.W.; Vasseur, L.; Gurr, G.M.; et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013, 45, 220–225. [Google Scholar] [CrossRef] [Green Version]
- He, W.; You, M.; Vasseur, L.; Yang, G.; Xie, M.; Cui, K.; Bai, J.; Liu, C.; Li, X.; Xu, X.; et al. Developmental and insecticide-resistant insights from the de novo assembled transcriptome of the diamondback moth, Plutella xylostella. Genomics 2012, 99, 169–177. [Google Scholar] [CrossRef]
- Xue, W.-H.; Xu, N.; Yuan, X.-B.; Chen, H.-H.; Zhang, J.-L.; Fu, S.-J.; Zhang, C.-X.; Xu, H.-J. CRISPR/Cas9-mediated knockout of two eye pigmentation genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Biochem. Mol. Biol. 2018, 93, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Dong, Y.; Saqib, H.S.A.; Vasseur, L.; Zhou, W.; Zheng, L.; Lai, Y.; Ma, X.; Lin, L.; Xu, X.; et al. Functions of duplicated glucosinolate sulfatases in the development and host adaptation of Plutella xylostella. Insect Biochem. Mol. Biol. 2020, 119, 103316. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.-B.; Lai, Y.-F.; Li, F.-F.; Jiao, L.; Qiao, Q.-X.; Li, S.-Y.; Xiang, X.-J.; Liao, H.; You, M.-S.; He, W.-Y. Functional Characterization of Two RNA Methyltransferase Genes METTL3 and METTL14 Uncovers the Roles of m6A in Mediating Adaptation of Plutella xylostella to Host Plants. Int. J. Mol. Sci. 2022, 23, 10013. https://doi.org/10.3390/ijms231710013
Wang B-B, Lai Y-F, Li F-F, Jiao L, Qiao Q-X, Li S-Y, Xiang X-J, Liao H, You M-S, He W-Y. Functional Characterization of Two RNA Methyltransferase Genes METTL3 and METTL14 Uncovers the Roles of m6A in Mediating Adaptation of Plutella xylostella to Host Plants. International Journal of Molecular Sciences. 2022; 23(17):10013. https://doi.org/10.3390/ijms231710013
Chicago/Turabian StyleWang, Bei-Bei, Ying-Fang Lai, Fei-Fei Li, Lu Jiao, Qing-Xuan Qiao, Shan-Yu Li, Xiu-Juan Xiang, Huang Liao, Min-Sheng You, and Wei-Yi He. 2022. "Functional Characterization of Two RNA Methyltransferase Genes METTL3 and METTL14 Uncovers the Roles of m6A in Mediating Adaptation of Plutella xylostella to Host Plants" International Journal of Molecular Sciences 23, no. 17: 10013. https://doi.org/10.3390/ijms231710013
APA StyleWang, B.-B., Lai, Y.-F., Li, F.-F., Jiao, L., Qiao, Q.-X., Li, S.-Y., Xiang, X.-J., Liao, H., You, M.-S., & He, W.-Y. (2022). Functional Characterization of Two RNA Methyltransferase Genes METTL3 and METTL14 Uncovers the Roles of m6A in Mediating Adaptation of Plutella xylostella to Host Plants. International Journal of Molecular Sciences, 23(17), 10013. https://doi.org/10.3390/ijms231710013





