Overexpression of PgCBF3 and PgCBF7 Transcription Factors from Pomegranate Enhances Freezing Tolerance in Arabidopsis under the Promoter Activity Positively Regulated by PgICE1
Abstract
:1. Introduction
2. Results
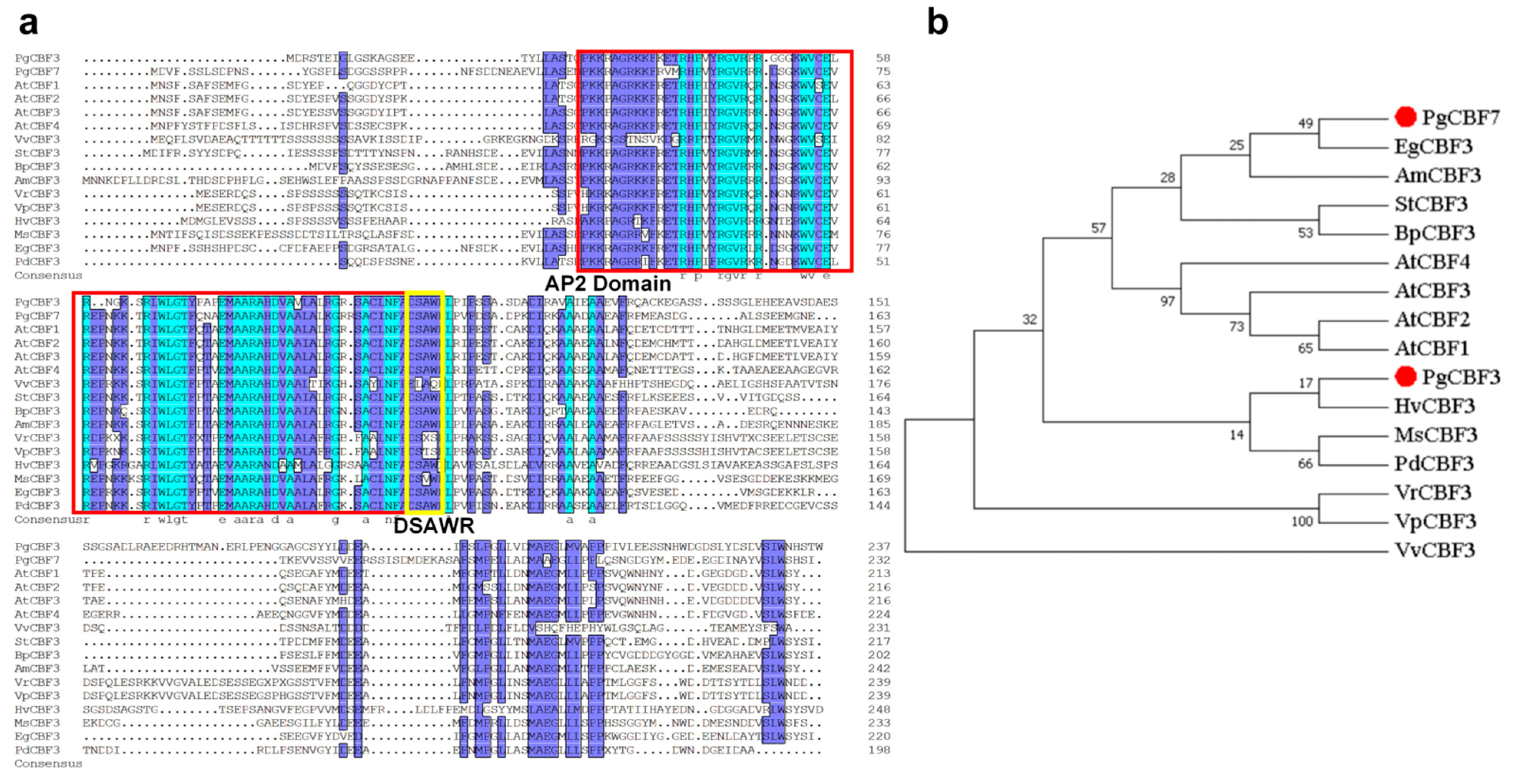
2.1. Screening and Analyzing the Candidate CBF Genes
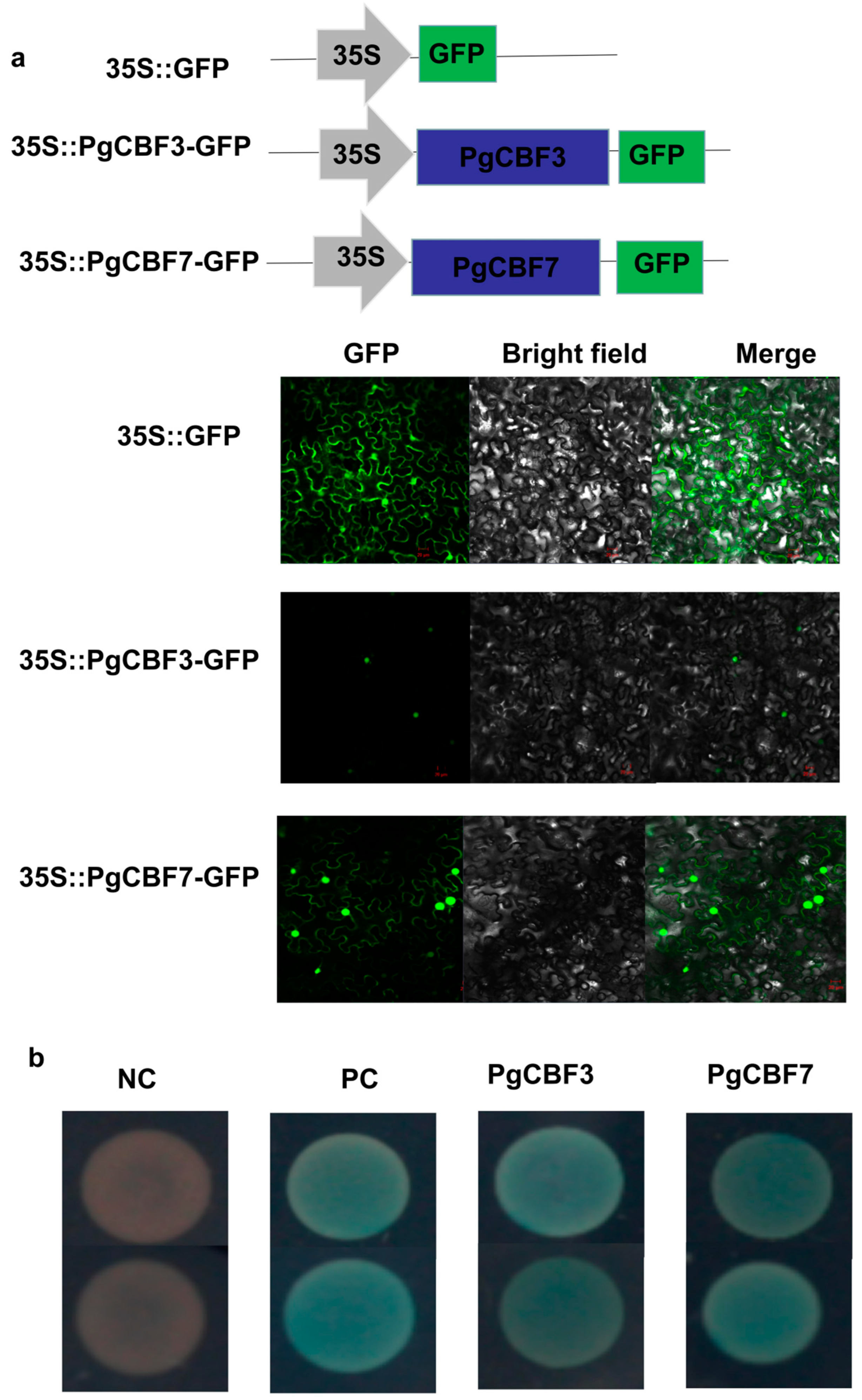
2.2. Subcellular Localization and Transcriptional Activity Analysis of PgCBF3 and PgCBF7
2.3. Validation of Activation Activity of PgICE1 on the Promoters of PgCBF3/PgCBF7
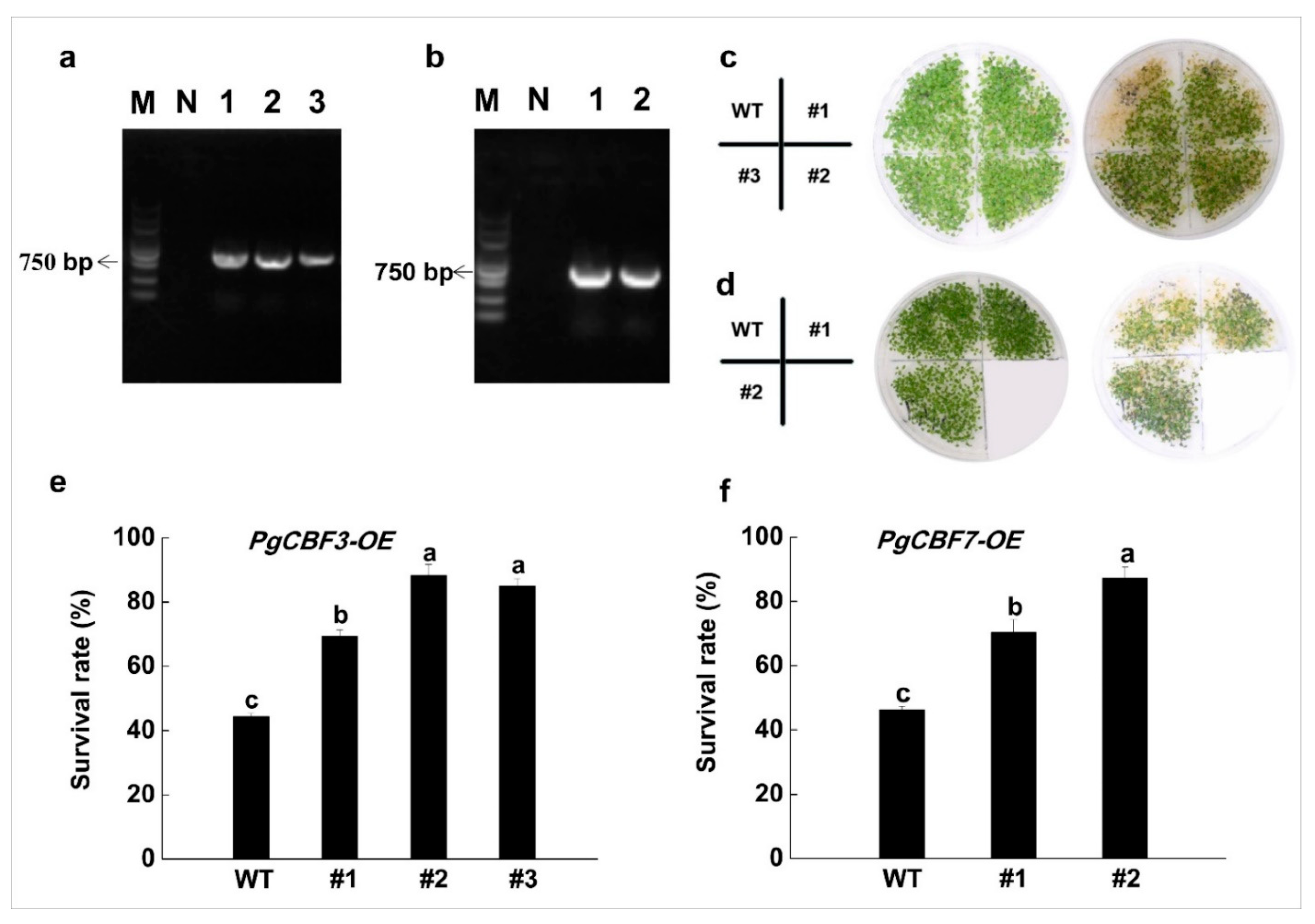
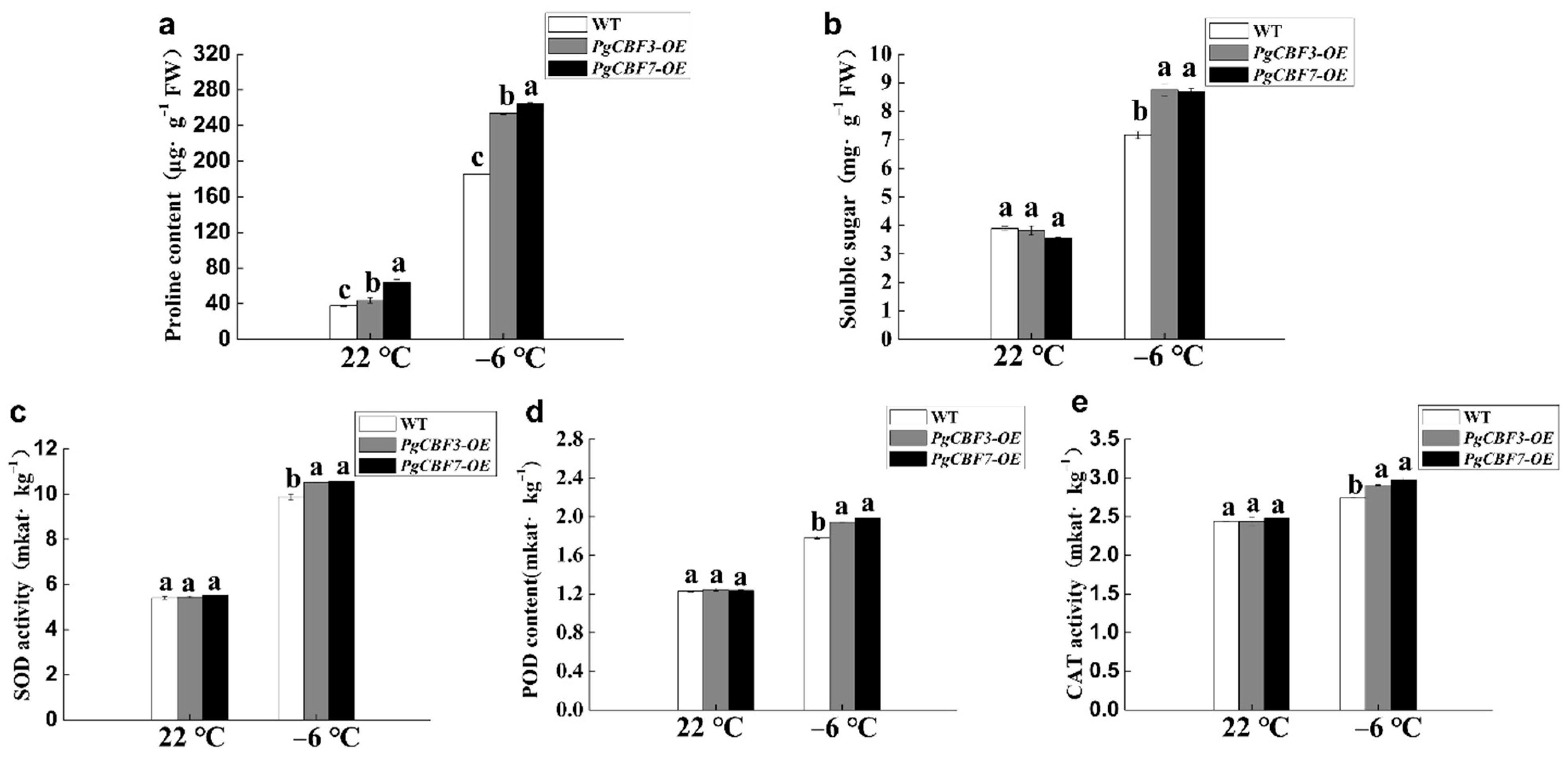
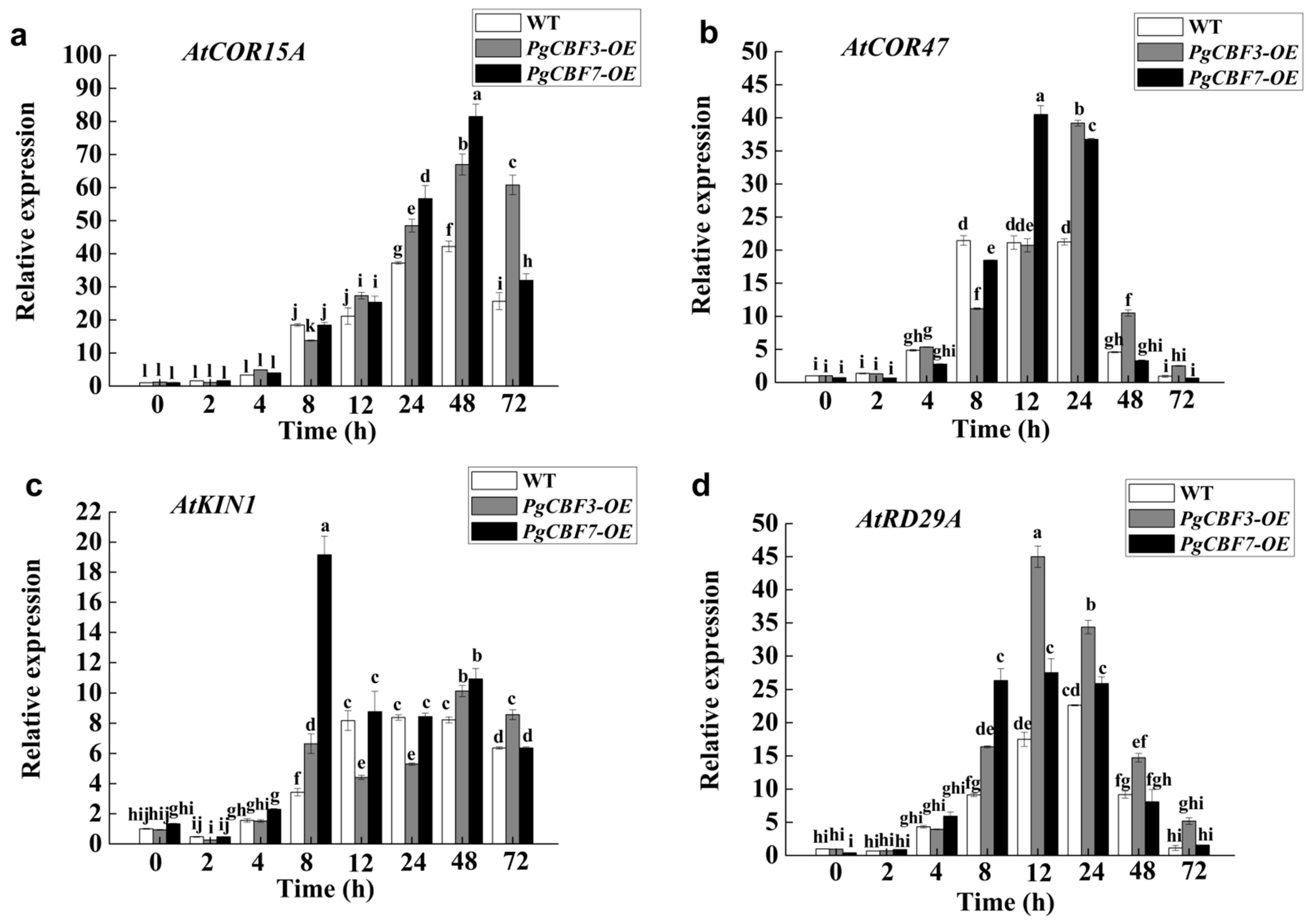
2.4. Overexpression of PgCBF3/PgCBF7 Enhanced Freezing Tolerance in A. thaliana
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cold-Tolerance Assays
4.3. RNA Extraction and Quantitative RT-PCR (qRT-PCR) Analysis
4.4. Bioinformatics Analysis of PgCBF3 and PgCBF7
4.5. Histochemical Staining
4.6. Chlorophyll Fluorescence Detection
4.7. Recombination Vectors Construction and Transgenic Plants Generation
4.8. Determination of Electrolyte Leakage, MDA and Proline Content, and Enzyme Activity
4.9. Determination of Electrolyte Leakage, MDA and Proline Content, and Enzyme Activity
4.10. Transcriptional Activation Activity Analysis
4.11. Extraction of Genomic DNA and the Cloning of the Promoters
4.12. Yeast One-Hybrid Assay
4.13. Dual-Luciferase (Dual-LUC) Reporter Assay
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.; Clément, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, F.; Kopka, J.; Sung, D.Y.; Zhao, W.; Popp, M.; Porat, R.; Guy, C.L. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007, 50, 967–981. [Google Scholar] [CrossRef]
- Song, Q.; Wang, X.; Li, J.; Chen, T.H.; Liu, Y.; Yang, X. CBF1 and CBF4 in Solanum tuberosum L. Differ in their effect on low-temperature tolerance and development. Environ. Exp. Bot. 2021, 185, 104416. [Google Scholar] [CrossRef]
- Kasamo, K.; Kagita, F.; Yamanishi, H.; Sakaki, T. Low temperature-induced changes in the thermotropic properties and fatty acid composition of the plasma membrane and tonoplast of cultured rice (Oryza sativa L.) cells. Plant Cell Physiol. 1992, 33, 609–616. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [Green Version]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.-h.; Hong, X.; Agarwal, M.; Zhu, J.-K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Zarka, D.G.; Vogel, J.T.; Cook, D.; Thomashow, M.F. Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 2003, 133, 910–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, J.; Catalá, R.; Salinas, J. The CBFs: Three Arabidopsis transcription factors to cold acclimate. Plant Sci. 2011, 180, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pino, M.T.; Skinner, J.S.; Jeknić, Z.; Hayes, P.M.; Soeldner, A.H.; Thomashow, M.F.; Chen, T.H. Ectopic AtCBF1 over-expression enhances freezing tolerance and induces cold acclimation-associated physiological modifications in potato. Plant Cell Environ. 2008, 31, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Lee, J.T.; Yang, P.T.; Chiu, L.H.; Charng, Y.Y.; Wang, Y.C.; Chan, M.T. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 2002, 129, 1086–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Shi, A.; Mou, B. Genome-wide identification and expression analysis of the CBF/DREB1 gene family in lettuce. Sci. Rep. 2020, 10, 5733. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Guo, C.; Yang, J.; Xu, M.; Zuo, J.; Bao, M.; Zhang, J. Overexpression of a Mei (Prunus mume) CBF gene confers tolerance to freezing and oxidative stress in Arabidopsis. Plant Cell Tissue Organ Cult. 2016, 126, 373–385. [Google Scholar] [CrossRef]
- Hu, Z.; Ban, Q.; Hao, J.; Zhu, X.; Cheng, Y.; Mao, J.; Lin, M.; Xia, E.; Li, Y. Genome-wide characterization of the C-repeat binding factor (CBF) gene family involved in the response to abiotic stresses in tea plant (Camellia sinensis). Front. Plant Sci. 2020, 11, 921. [Google Scholar] [CrossRef]
- Yang, X.; Wang, R.; Jing, H.; Chen, Q.; Bao, X.; Zhao, J.; Hu, G.; Liu, C.; Fu, J. Three novel c-repeat binding factor genes of Dimocarpus longan regulate cold stress response in Arabidopsis. Front. Plant Sci. 2020, 11, 1026. [Google Scholar] [CrossRef]
- Xiao, H.; Siddiqua, M.; Braybrook, S.; Nassuth, A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 2006, 29, 1410–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, H.; Tattersall, E.A.; Siddiqua, M.K.; Cramer, G.R.; Nassuth, A. CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant Cell Environ. 2008, 31, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Ebadi, A.; Mousavi, S.A.; Salami, S.A.; Zarei, A. Comparison of CBF1, CBF2, CBF3 and CBF4 expression in some grapevine cultivars and species under cold stress. Sci. Hortic. 2015, 197, 521–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Liu, X.-D.; Chi, X.-J.; Wu, C.-A.; Li, Y.-Z.; Song, L.-L.; Liu, X.-M.; Wang, Y.-F.; Wang, F.-W.; Zhang, C. Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 2011, 233, 219–229. [Google Scholar] [CrossRef]
- Kitashiba, H.; Ishizaka, T.; Isuzugawa, K.; Nishimura, K.; Suzuki, T. Expression of a sweet cherry DREB1/CBF ortholog in Arabidopsis confers salt and freezing tolerance. J. Plant Physiol. 2004, 161, 1171–1176. [Google Scholar] [CrossRef]
- Wisniewski, M.; Norelli, J.; Bassett, C.; Artlip, T.; Macarisin, D. Ectopic expression of a novel peach (Prunus persica) CBF transcription factor in apple (Malus × domestica) results in short-day induced dormancy and increased cold hardiness. Planta 2011, 233, 971–983. [Google Scholar] [CrossRef]
- Barros, P.M.; Gonçalves, N.; Saibo, N.J.; Oliveira, M.M. Functional characterization of two almond C-repeat-binding factors involved in cold response. Tree Physiol. 2012, 32, 1113–1128. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, Y.; Yu, B.; Song, Q.; Liu, Y.; Chen, T.H.; Li, G.; Yang, X. Ectopic expression of StCBF1 and ScCBF1 have different functions in response to freezing and drought stresses in Arabidopsis. Plant Sci. 2018, 270, 221–233. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wu, Y.; Tian, Y.; Dai, T.; Xie, G.; Xu, Y.; Chen, F. Overexpressing Jatropha curcas CBF2 in Nicotiana benthamiana improved plant tolerance to drought stress. Gene 2020, 742, 144588. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Sadeghi, R.; Kokini, J. Pomegranate as a promising opportunity in medicine and nanotechnology. Trends Food Sci. Technol. 2017, 69, 59–73. [Google Scholar] [CrossRef]
- Hegazi, N.M.; El-Shamy, S.; Fahmy, H.; Farag, M.A. Pomegranate juice as a super-food: A comprehensive review of its extraction, analysis, and quality assessment approaches. J. Food Compos. Anal. 2021, 97, 103773. [Google Scholar] [CrossRef]
- Akyıldız, A.; Karaca, E.; Ağçam, E.; Dündar, B.; Çınkır, N.İ. Changes in quality attributes during production steps and frozen-storage of pomegranate juice concentrate. J. Food Compos. Anal. 2020, 92, 103548. [Google Scholar] [CrossRef]
- Soloklui, A.A.G.; Ershadi, A.; Fallahi, E. Evaluation of cold hardiness in seven Iranian commercial pomegranate (Punica granatum L.) cultivars. HortScience 2012, 47, 1821–1825. [Google Scholar] [CrossRef] [Green Version]
- Sayyari, M.; Babalar, M.; Kalantari, S.; Serrano, M.; Valero, D. Effect of salicylic acid treatment on reducing chilling injury in stored pomegranates. Postharvest Biol. Technol. 2009, 53, 152–154. [Google Scholar] [CrossRef]
- Kashash, Y.; Mayuoni-Kirshenbaum, L.; Goldenberg, L.; Choi, H.J.; Porat, R. Effects of harvest date and low-temperature conditioning on chilling tolerance of ‘Wonderful’ pomegranate fruit. Sci. Hortic. 2016, 209, 286–292. [Google Scholar] [CrossRef]
- Jannatizadeh, A. Exogenous melatonin applying confers chilling tolerance in pomegranate fruit during cold storage. Sci. Hortic. 2019, 246, 544–549. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, S.; Wu, W.; Tong, R.; Wang, S.; Li, M.; Jian, Z.; Wan, R.; Hu, Q. Exogenous arginine treatment maintains the appearance and nutraceutical properties of hard-and soft-seed pomegranates in cold storage. Front. Nutr. 2022, 9, 828946. [Google Scholar] [CrossRef]
- Zhang, J.Z. Overexpression analysis of plant transcription factors. Curr. Opin. Plant Biol. 2003, 6, 430–440. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Lee, M.; Lee, J.-H.; Lee, H.-J.; Park, C.-M. The unified ICE–CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 2015, 89, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Fursova, O.V.; Pogorelko, G.V.; Tarasov, V.A. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 2009, 429, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ye, K.; Shi, Y.; Cheng, J.; Zhang, X.; Yang, S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 2017, 10, 545–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009, 50, 447–462. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; He, Y.; Li, J.; Li, L.; Liu, Y.; Chen, H. An eggplant SmICE1a gene encoding MYC-type ICE1-like transcription factor enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Biol. 2020, 22, 450–458. [Google Scholar] [CrossRef]
- Xiong, Y.; Fei, S.-Z. Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 2006, 224, 878–888. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Li, J.; Liu, Q.; Li, Y.-Q.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 2004, 45, 1042–1052. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hua, J. A moderate decrease in temperature induces COR15a expression through the CBF signaling cascade and enhances freezing tolerance. Plant J. 2009, 60, 340–349. [Google Scholar] [CrossRef]
- Campos, P.S.; nia Quartin, V.; chicho Ramalho, J.; Nunes, M.A. Electrolyte leakage and lipid degradation account for cold sensitivity in leaves of Coffea sp. plants. J. Plant Physiol. 2003, 160, 283–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, C.; Huang, X.-S.; Li, K.-Q.; Yin, H.; Li, L.-T.; Yao, Z.-H.; Zhang, S.-L. Overexpression of a bHLH1 transcription factor of Pyrus ussuriensis confers enhanced cold tolerance and increases expression of stress-responsive genes. Front. Plant Sci. 2016, 7, 441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, M.; Li, J.; Yang, Q.; Jamil, W.; Teng, Y.; Bai, S. Phylogenetic, molecular, and functional characterization of PpyCBF proteins in Asian pears (Pyrus pyrifolia). Int. J. Mol. Sci. 2019, 20, 2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.A.; Bhattacharjee, S.; Armin, S.-M.; Qian, P.; Xin, W.; Li, H.-Y.; Burritt, D.J.; Fujita, M.; Tran, L.-S.P. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: Insights from ROS detoxification and scavenging. Front. Plant Sci. 2015, 6, 420. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of CBF expression–C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Li, H.; Wu, Z.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.; Li, K.; Poudel, K.; Zhao, D. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft-and hard-seeded cultivars. Plant Biotechnol. J. 2020, 18, 955–968. [Google Scholar] [CrossRef] [Green Version]
- Daudi, A.; O’Brien, J.A. Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio-Protocol 2012, 2, e263. [Google Scholar] [CrossRef] [Green Version]
- Grellet Bournonville, C.F.; Díaz-Ricci, J.C. Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochem. Anal. 2011, 22, 268–271. [Google Scholar] [CrossRef]
- Guo, Z.; Lv, J.; Zhang, H.; Hu, C.; Qin, Y.; Dong, H.; Zhang, T.; Dong, X.; Du, N.; Piao, F. Red and blue light function antagonistically to regulate cadmium tolerance by modulating the photosynthesis, antioxidant defense system and Cd uptake in cucumber (Cucumis sativus L.). J. Hazard. Mater. 2022, 429, 128412. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanjo, T.; Kobayashi, M.; Yoshiba, Y.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett. 1999, 461, 205–210. [Google Scholar] [CrossRef] [Green Version]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Wang, S.; Tong, R.; Wang, S.; Yao, J.; Jiao, J.; Wan, R.; Wang, M.; Shi, J.; Zheng, X. Overexpression of PgCBF3 and PgCBF7 Transcription Factors from Pomegranate Enhances Freezing Tolerance in Arabidopsis under the Promoter Activity Positively Regulated by PgICE1. Int. J. Mol. Sci. 2022, 23, 9439. https://doi.org/10.3390/ijms23169439
Wang L, Wang S, Tong R, Wang S, Yao J, Jiao J, Wan R, Wang M, Shi J, Zheng X. Overexpression of PgCBF3 and PgCBF7 Transcription Factors from Pomegranate Enhances Freezing Tolerance in Arabidopsis under the Promoter Activity Positively Regulated by PgICE1. International Journal of Molecular Sciences. 2022; 23(16):9439. https://doi.org/10.3390/ijms23169439
Chicago/Turabian StyleWang, Lei, Sa Wang, Ruiran Tong, Sen Wang, Jianan Yao, Jian Jiao, Ran Wan, Miaomiao Wang, Jiangli Shi, and Xianbo Zheng. 2022. "Overexpression of PgCBF3 and PgCBF7 Transcription Factors from Pomegranate Enhances Freezing Tolerance in Arabidopsis under the Promoter Activity Positively Regulated by PgICE1" International Journal of Molecular Sciences 23, no. 16: 9439. https://doi.org/10.3390/ijms23169439
APA StyleWang, L., Wang, S., Tong, R., Wang, S., Yao, J., Jiao, J., Wan, R., Wang, M., Shi, J., & Zheng, X. (2022). Overexpression of PgCBF3 and PgCBF7 Transcription Factors from Pomegranate Enhances Freezing Tolerance in Arabidopsis under the Promoter Activity Positively Regulated by PgICE1. International Journal of Molecular Sciences, 23(16), 9439. https://doi.org/10.3390/ijms23169439





