Efficient Anchoring of Erianthus arundinaceus Chromatin Introgressed into Sugarcane by Specific Molecular Markers
Abstract
1. Introduction
2. Results
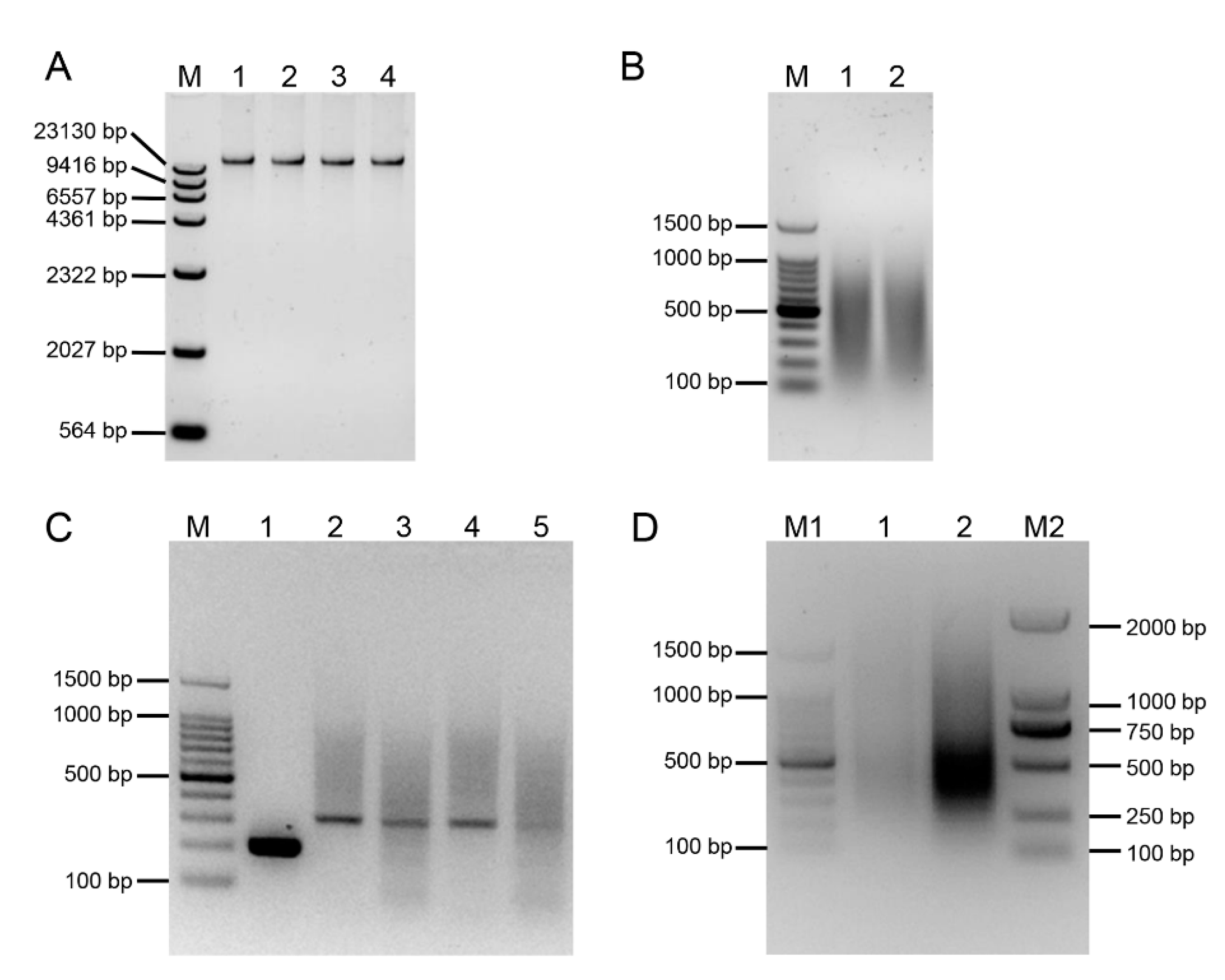
2.1. Construction of an E. arundinaceus-Derived SSH Library
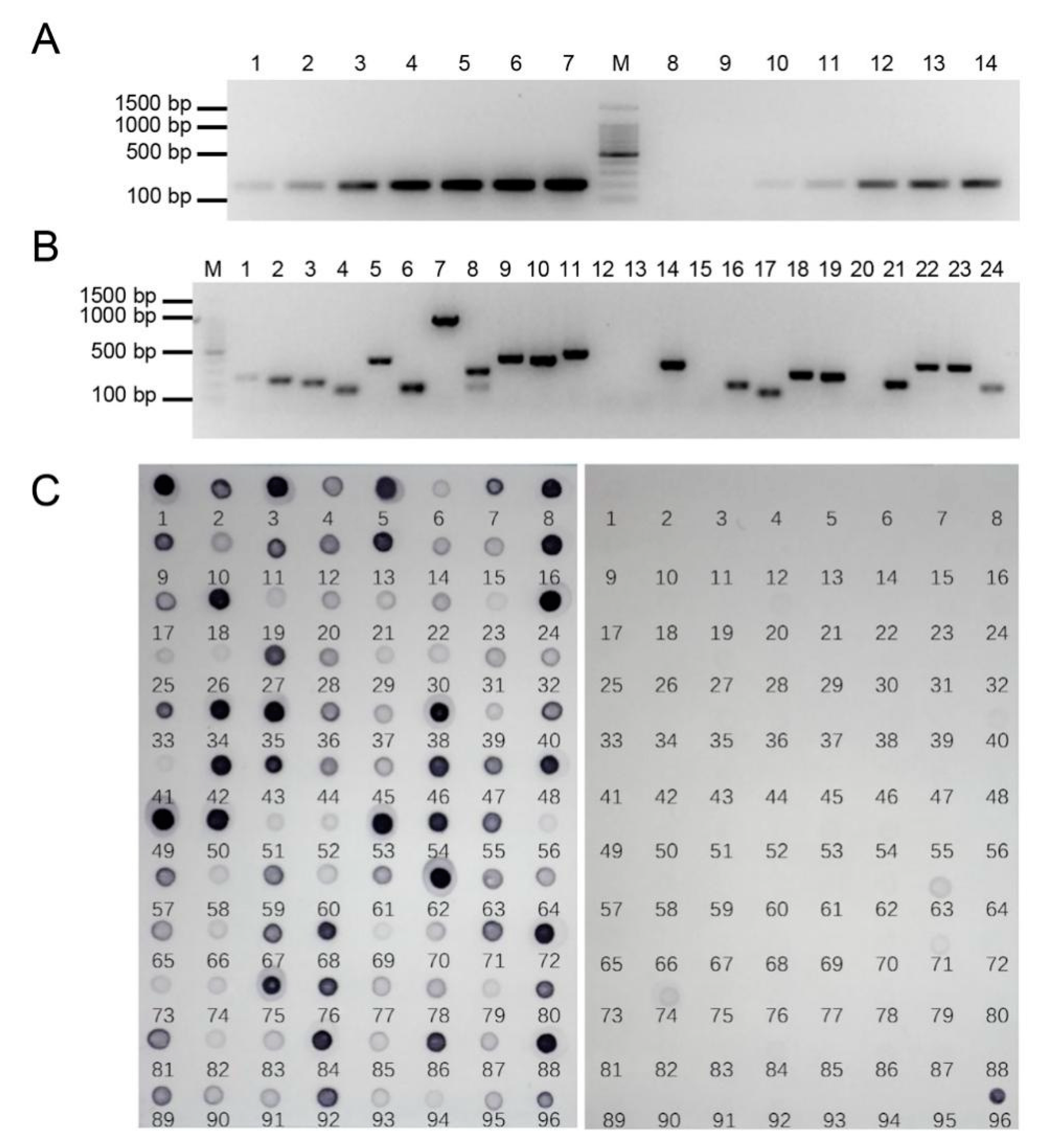
2.2. PCR Amplification and Dot-Blot Screening of E. arundinaceus-Specific Clones
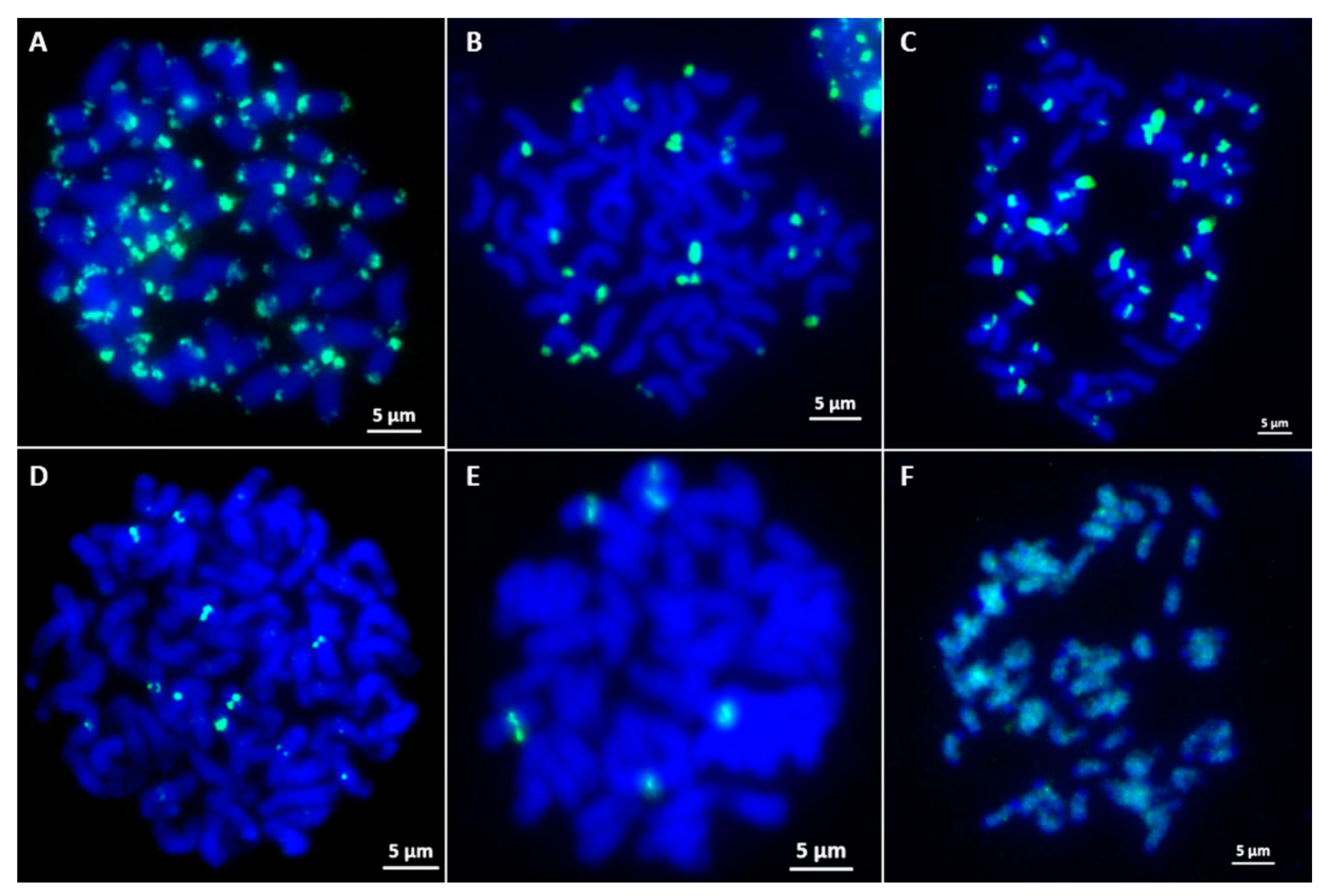
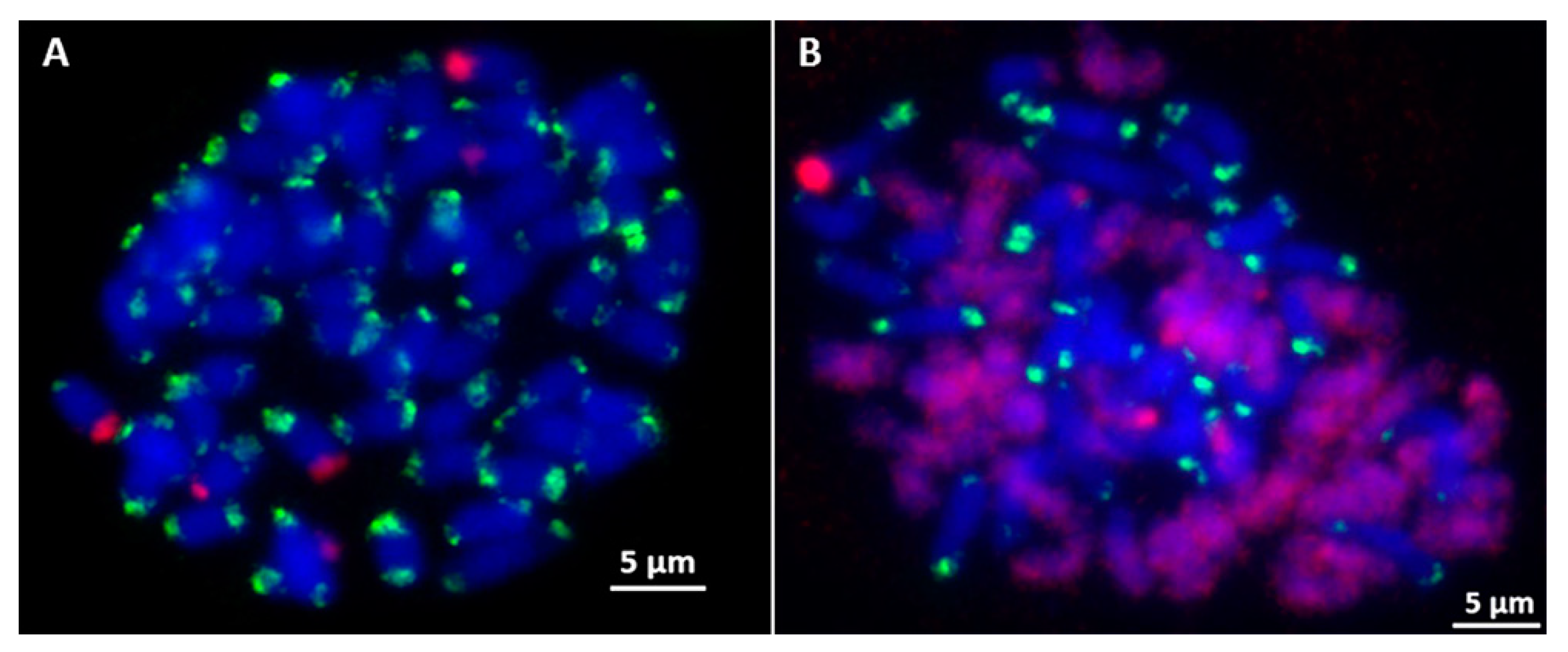
2.3. Screening and Characterization of E. arundinaceus-Specific Clones Using FISH
2.4. Development of Candidate E. arundinaceus-Specific Molecular Markers
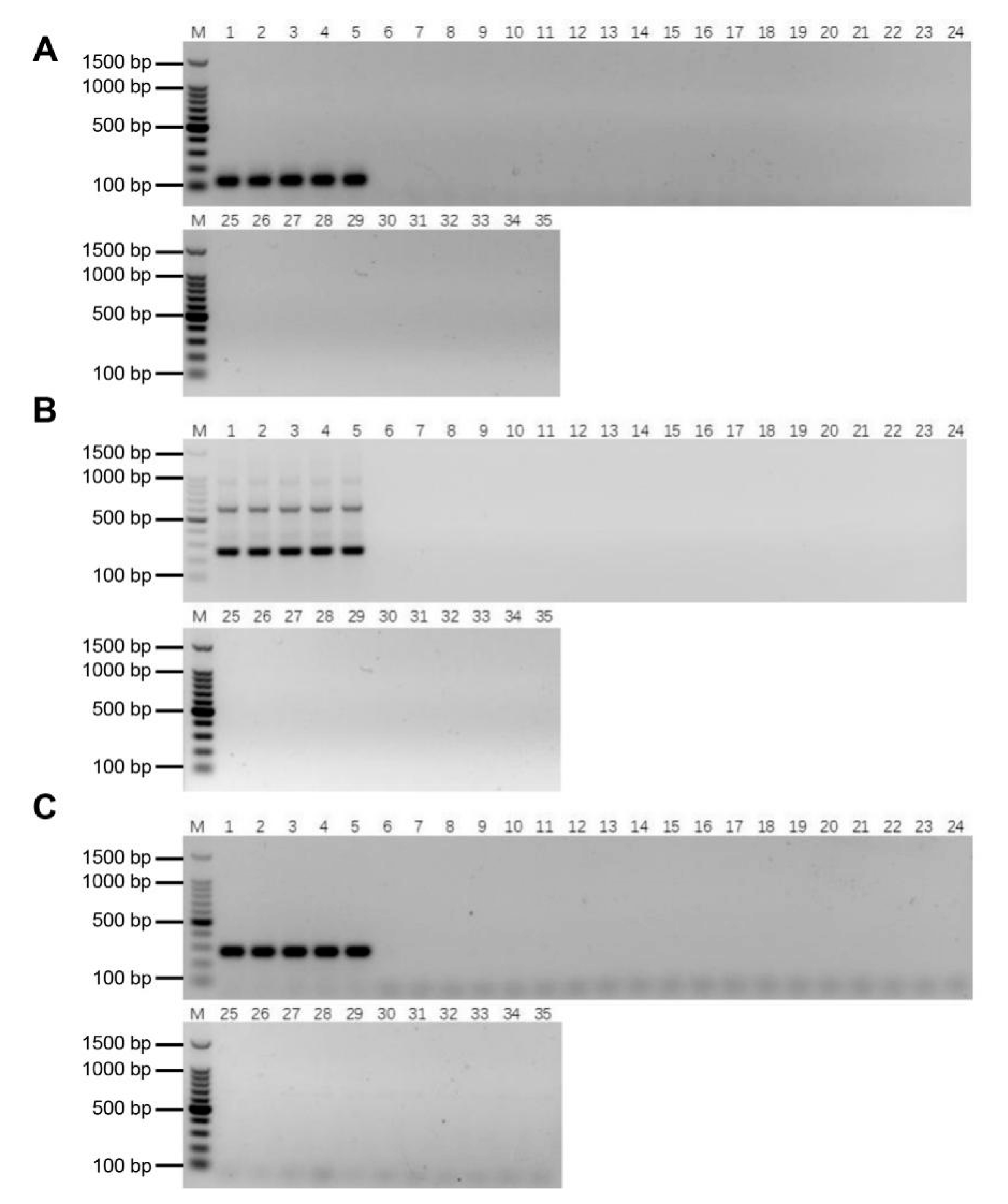
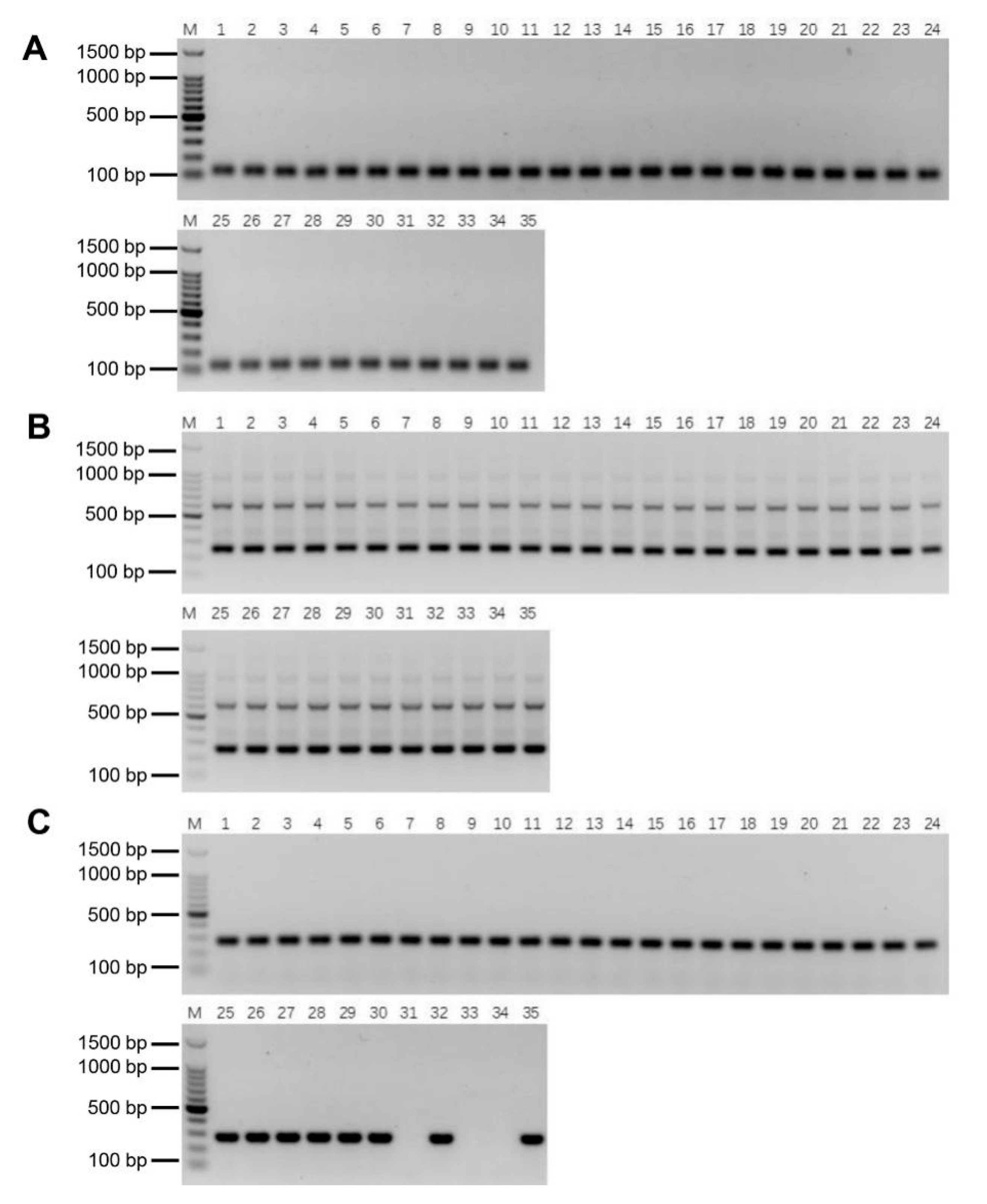
2.5. Validation of the Specificity and Stability of E. arundinaceus-Specific Markers
2.6. Authenticity of the Putative BC4 Progeny
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Suppressive Subtractive Hybridization
4.3. Preparation of Dig-Labeled gDNAs and Reverse Dot Blot (RDB)
4.4. Fluorescence In Situ Hybridization (FISH)
4.5. Development, Verification, and Detection of E. arundinaceus-Specific Molecular Markers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gallo, A.B.; Simoes-Moreira, J.R.; Costa, H.K.M.; Santos, M.M.; dos Santos, E.M. Energy storage in the energy transition context: A technology review. Renew. Sust. Energ. Rev. 2016, 65, 800–822. [Google Scholar] [CrossRef]
- Tessler, Z.D.; Vorosmarty, C.J.; Grossberg, M.; Gladkova, I.; Aizenman, H.; Syvitski, J.P.M.; Foufoula-Georgiou, E. Profiling risk and sustainability in coastal deltas of the world. Science 2015, 349, 638–643. [Google Scholar] [CrossRef]
- Aguilar-Reynosa, A.; Romani, A.; Rodriguez-Jasso, R.M.; Aguilar, C.N.; Garrote, G.; Ruiz, H.A. Microwave heating processing as alternative of pretreatment in second-generation biorefinery: An overview. Energy Convers. Manag. 2017, 136, 50–65. [Google Scholar] [CrossRef]
- Wu, J.Y.; Huang, Y.J.; Lin, Y.Q.; Fu, C.; Liu, S.M.; Deng, Z.H.; Li, Q.W.; Huang, Z.X.; Chen, R.K.; Zhang, M.Q. Unexpected Inheritance Pattern of Erianthus arundinaceus Chromosomes in the Intergeneric Progeny between Saccharum spp. and Erianthus arundinaceus. PLoS ONE 2014, 9, e110390. [Google Scholar] [CrossRef] [PubMed]
- Piperidis, N.; Chen, J.W.; Deng, H.H.; Wang, L.P.; Jackson, P.; Piperidis, G. GISH characterization of Erianthus arundinaceus chromosomes in three generations of sugarcane intergeneric hybrids. Genome 2010, 53, 331–336. [Google Scholar] [CrossRef]
- Pachakkil, B.; Terajima, Y.; Ohmido, N.; Ebina, M.; Irei, S.; Hayashi, H.; Takagi, H. Cytogenetic and agronomic characterization of intergeneric hybrids between Saccharum spp. hybrid and Erianthus arundinaceus. Sci. Rep. 2019, 9, 1748. [Google Scholar] [CrossRef]
- Yang, S.; Zeng, K.; Chen, K.; Wu, J.; Wang, Q.; Li, X.; Deng, Z.; Huang, Y.; Huang, F.; Chen, R.; et al. Chromosome transmission in BC4 progenies of intergeneric hybrids between Saccharum spp. and Erianthus arundinaceus (Retz.) Jeswiet. Sci. Rep. 2019, 9, 2528. [Google Scholar] [CrossRef] [PubMed]
- Villano, C.; Miraglia, V.; Iorizzo, M.; Aversano, R.; Carputo, D. Combined Use of Molecular Markers and High-Resolution Melting (HRM) to Assess Chromosome Dosage in Potato Hybrids. J. Hered. 2016, 107, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, X.; Huang, F.; Huang, Y.; Liu, X.; Wu, J.; Wang, Q.; Deng, Z.; Chen, R.; Zhang, M. A new method based on SNP of nrDNA-ITS to identify Saccharum spontaneum and its progeny in the genus Saccharum. PLoS ONE 2018, 13, e0197458. [Google Scholar] [CrossRef]
- Chen, J.W.; Jackson, P.A.; Lao, F.Y.; Liu, R. Identification of BC2 Progeny from Saccharum officinarum × Erianthus arundinaceus by ISSR Markers. Sugar Crops China 2008, 1, 1–3. [Google Scholar]
- Nair, N.V.; Mary, S. RAPD analysis reveals the presence of Indian and Indonesian forms of Erianthus arundinaceus (Retz.) Jeswiet. in the Andaman-Nicobar Islands, India. Curr. Sci. India 2006, 90, 1118–1122. [Google Scholar]
- Cai, Q.; Aitken, K.; Deng, H.H.; Chen, X.W.; Fu, C.; Jackson, P.A.; McIntyre, C.L. Verification of the introgression of Erianthus arundinaceus germplasm into sugarcane using molecular markers. Plant Breed. 2005, 124, 322–328. [Google Scholar] [CrossRef]
- Alix, K.; Paulet, F.; Glaszmann, J.-C.; D’Hont, A. Inter-Alu-like species-specific sequences in the Saccharum complex. Theor. Appl. Genet. 1999, 99, 962–968. [Google Scholar] [CrossRef]
- Govindaraj, P.; Balamurugan, A.; Natarajan, U.S. Identification of intergeneric hybrids between Erianthus arundinaceus and Saccharum spontaneum through STMS markers. Int. Sugar J. 2012, 114, 350–358. [Google Scholar]
- Gao, Y.J.; Liu, X.H.; Zhang, R.H.; Zhou, H.; Liao, J.X.; Duan, W.X.; Zhang, G.M. Verification of Progeny from Crosses Between Sugarcane (Saccharum spp.) and an Intergeneric Hybrid (Erianthus arundinaceus × Saccharum spontaneum) with Molecular Makers. Sugar Tech. 2015, 17, 31–35. [Google Scholar] [CrossRef]
- Devos, K.M.; Gale, M.D. The use of random amplified polymorphic DNA markers in wheat. Theor. Appl. Genet. 1992, 84, 567–572. [Google Scholar] [CrossRef]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.-X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef]
- Phung, N.T.P.; Mai, C.D.; Mournet, P.; Frouin, J.; Droc, G.; Ta, N.K.; Jouannic, S.; Lê, L.T.; Do, V.N.; Gantet, P.; et al. Characterization of a panel of Vietnamese rice varieties using DArT and SNP markers for association mapping purposes. BMC Plant Biol. 2014, 14, 371. [Google Scholar] [CrossRef]
- Yao, F.; Zhang, X.; Ye, X.; Li, J.; Long, L.; Yu, C.; Li, J.; Wang, Y.; Wu, Y.; Wang, J.; et al. Characterization of molecular diversity and genome-wide association study of stripe rust resistance at the adult plant stage in Northern Chinese wheat landraces. BMC Genet. 2019, 20, 38. [Google Scholar] [CrossRef]
- Souza, G.M.; Berges, H.; Bocs, S.; Casu, R.; D’Hont, A.; Ferreira, J.E.; Henry, R.; Ming, R.; Potier, B.; Sluys, M.A.V. The Sugarcane Genome Challenge: Strategies for Sequencing a Highly Complex Genome. Trop. Plant Biol. 2011, 4, 145–156. [Google Scholar] [CrossRef]
- Li, T.X.; Wang, J.K.; Bai, Y.F.; Sun, X.D.; Lu, Z.H. A novel method for screening species-specific gDNA probes for species identification. Nucleic Acids Res. 2004, 32, 8. [Google Scholar] [CrossRef]
- Dai, Z.Y.; Mao, X.X.; Magnuson, J.K.; Lasure, L.L. Identification of genes associated with morphology in Aspergillus niger by using suppression subtractive hybridization. Appl. Environ. Microb. 2004, 70, 2474–2485. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Harakava, R.; Gabriel, D.W. Genetic differences between two strains of Xylella fastidiosa revealed by suppression subtractive hybridization. Appl. Environ. Microb. 2003, 69, 1315–1319. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hsieh, W.J.; Pan, M.J. Identification Leptospira santarosai serovar shermani specific sequences by suppression subtractive hybridization. FEMS Microbiol. Lett. 2004, 235, 117–124. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Diatchenko, L.; Lau, Y.F.; Campbell, A.P.; Chenchik, A.; Moqadam, F.; Huang, B.; Lukyanov, S.; Lukyanov, K.; Gurskaya, N.; Sverdlov, E.D.; et al. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 1996, 93, 6025–6030. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Chen, S.; Gao, Y.; Gao, Y.; Zhu, X.; Huang, Z.; Chen, J. Development of genome-specific molecular markers for Lophopyrum elongatum based on suppression subtractive hybridization. Acta Agron. Sin. 2012, 38, 1818–1826. [Google Scholar] [CrossRef]
- Huang, Y.J.; Wu, J.Y.; Wang, P.; Lin, Y.Q.; Fu, C.; Deng, Z.H.; Wang, Q.N.; Li, Q.W.; Chen, R.K.; Zhang, M.Q. Characterization of Chromosome Inheritance of the Intergeneric BC2 and BC3 Progeny between Saccharum spp. and Erianthus arundinaceus. PLoS ONE 2015, 10, e133722. [Google Scholar] [CrossRef]
- Fukuhara, S.; Terajima, Y.; Irei, S.; Sakaigaichi, T.; Ujihara, K.; Sugimoto, A.; Matsuoka, M. Identification and characterization of intergeneric hybrid of commercial sugarcane (Saccharum spp. hybrid) and Erianthus arundinaceus (Retz.) Jeswiet. Euphytica 2013, 189, 321–327. [Google Scholar] [CrossRef]
- Yu, F.; Huang, Y.; Luo, L.; Li, X.; Wu, J.; Chen, R.; Zhang, M.; Deng, Z. An improved suppression subtractive hybridization technique to develop species-specific repetitive sequences from Erianthus arundinaceus (Saccharum complex). BMC Plant Biol. 2018, 18, 269. [Google Scholar] [CrossRef]
- Naito, K.; Zhang, F.; Tsukiyama, T.; Saito, H.; Hancock, C.N.; Richardson, A.O.; Okumoto, Y.; Tanisaka, T.; Wessler, S.R. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 2009, 461, 1130–1134. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.; Schwarzacher, T. Organisation of the plant genome in chromosomes. Plant J. Cell Mol. Biol. 2011, 66, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genom. Proteom. Bioinf. 2014, 12, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Liu, H.; He, Q.; Pu, M.; Chen, J.; Lai, J.; Li, X.; Jin, W. Differential genome evolution and speciation of Coix lacryma-jobi L. and Coix aquatica Roxb. hybrid guangxi revealed by repetitive sequence analysis and fine karyotyping. BMC Genom. 2014, 15, 1025. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Cai, Z.; Hu, T.; Liu, H.; Bao, C.; Mao, W.; Jin, W. Repetitive sequence analysis and karyotyping reveals centromere-associated DNA sequences in radish (Raphanus sativus L.). BMC Plant Biol. 2015, 15, 105. [Google Scholar] [CrossRef]
- Kato, A.; Lamb, J.C.; Birchler, J.A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 2004, 101, 13554–13559. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Zhang, M.; Liu, J.; Huang, Y.; Xu, L.; Deng, Z.; Zhao, X. Efficient Anchoring of Erianthus arundinaceus Chromatin Introgressed into Sugarcane by Specific Molecular Markers. Int. J. Mol. Sci. 2022, 23, 9435. https://doi.org/10.3390/ijms23169435
Wu J, Zhang M, Liu J, Huang Y, Xu L, Deng Z, Zhao X. Efficient Anchoring of Erianthus arundinaceus Chromatin Introgressed into Sugarcane by Specific Molecular Markers. International Journal of Molecular Sciences. 2022; 23(16):9435. https://doi.org/10.3390/ijms23169435
Chicago/Turabian StyleWu, Jiayun, Mingxiao Zhang, Jiarui Liu, Yongji Huang, Liangnian Xu, Zuhu Deng, and Xinwang Zhao. 2022. "Efficient Anchoring of Erianthus arundinaceus Chromatin Introgressed into Sugarcane by Specific Molecular Markers" International Journal of Molecular Sciences 23, no. 16: 9435. https://doi.org/10.3390/ijms23169435
APA StyleWu, J., Zhang, M., Liu, J., Huang, Y., Xu, L., Deng, Z., & Zhao, X. (2022). Efficient Anchoring of Erianthus arundinaceus Chromatin Introgressed into Sugarcane by Specific Molecular Markers. International Journal of Molecular Sciences, 23(16), 9435. https://doi.org/10.3390/ijms23169435





