Spatial Distribution and Retention in Loblolly Pine Seedlings of Exogenous dsRNAs Applied through Roots
Abstract
1. Introduction
2. Results
2.1. Plant Material and RNA Recovery
2.2. Recovery of Exogenous dsRNA
2.2.1. Gel Recovery
2.2.2. Sequence Alignment
2.2.3. Logistic Regression Modeling
3. Discussion
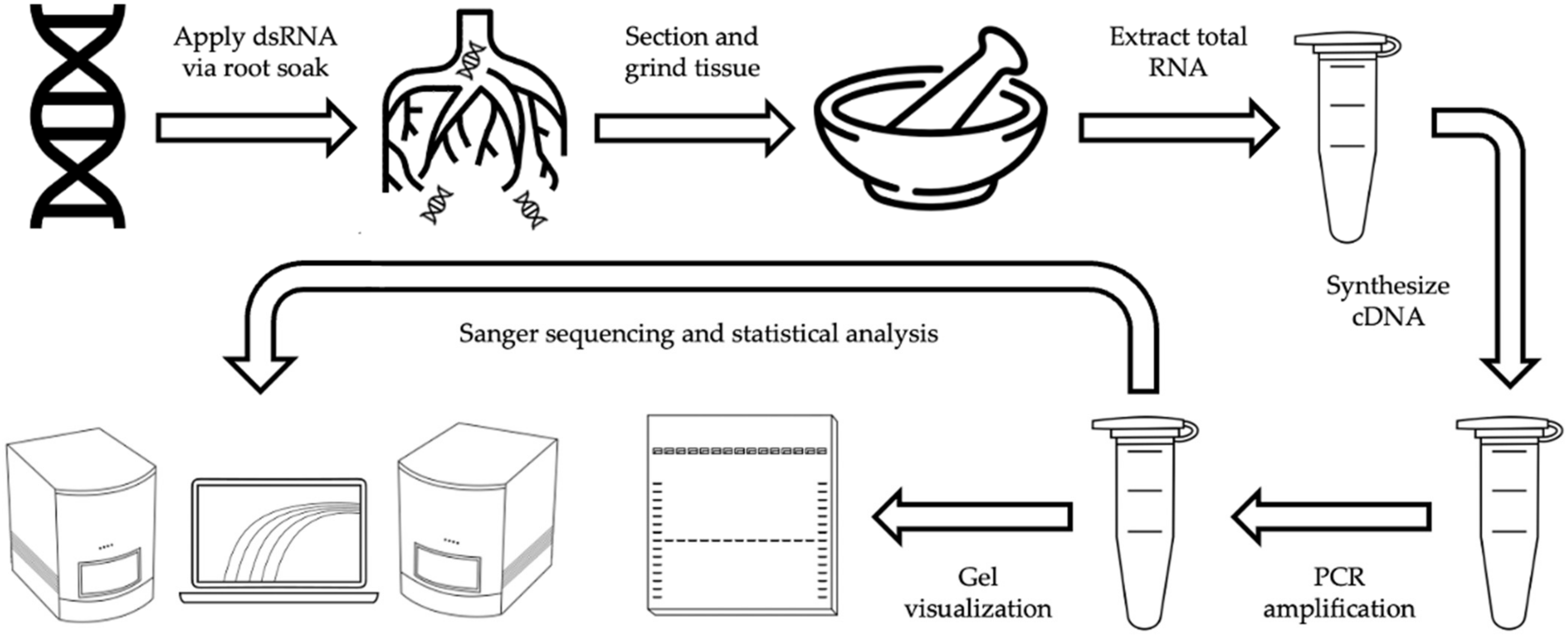
4. Materials and Methods
4.1. Plant Material
4.2. Target Gene Selection
4.3. dsRNA Synthesis
4.4. dsRNA Exposure
4.5. Plant Processing
4.5.1. Tissue Sectioning
4.5.2. Tissue Homogenization & RNA Recovery
4.5.3. RNA Quantification
4.6. Analysis of dsRNA Presence
4.6.1. cDNA Synthesis and PCR Amplification
4.6.2. Gel Electrophoresis and Visualization
4.6.3. Product Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Heigwer, F.; Port, F.; Boutros, M. RNA Interference (RNAi) Screening in Drosophila. Genetics 2018, 208, 853–874. [Google Scholar] [CrossRef]
- Jain, R.G.; Robinson, K.E.; Fletcher, S.J.; Mitter, N. RNAi-Based Functional Genomics in Hemiptera. Insects 2020, 11, 557. [Google Scholar] [CrossRef]
- Schmitt-Engel, C.; Schultheis, D.; Schwirz, J.; Ströhlein, N.; Troelenberg, N.; Majumdar, U.; Dao, V.A.; Grossmann, D.; Richter, T.; Tech, M.; et al. The iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology. Nat. Commun. 2015, 6, 7822. [Google Scholar] [CrossRef]
- Tabassum, B.; Nasir, I.; Aslam, U.; Husnain, T. How RNA Interference Combat Viruses in Plants. In Functional Genomics; Meroni, G., Petrera, F., Eds.; InTechOpen: London, UK, 2012. [Google Scholar]
- Rosa, C.; Kuo, Y.-W.; Hada, W.; Falk, B.W. RNA Interference Mechanisms and Applications in Plant Pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610. [Google Scholar] [CrossRef]
- Alamalakala, L.; Parimi, S.; Patel, N.; Char, B. Insect RNAi: Integrating a New Tool in the Crop Protection Toolkit; Springer International Publishing: Cham, Switzerland, 2018; pp. 193–232. [Google Scholar]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.-W.; Broeck, J.V. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2019, 9, 1912. [Google Scholar] [CrossRef]
- Sen, G.L.; Blau, H.M. A brief history of RNAi: The silence of the genes. FASEB J. 2006, 20, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- Whyard, S.; Singh, A.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, M.; Wang, B.; Han, Z. Comparison of the RNA interference effects triggered by dsRNA and siRNA in Tribolium castaneum. Pest Manag. Sci. 2013, 69, 781–786. [Google Scholar] [CrossRef]
- Wytinck, N.; Manchur, C.L.; Li, V.H.; Whyard, S.; Belmonte, M.F. dsRNA Uptake in Plant Pests and Pathogens: Insights into RNAi-Based Insect and Fungal Control Technology. Plants 2020, 9, 1780. [Google Scholar] [CrossRef]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef]
- Bachman, P.M.; Huizinga, K.M.; Jensen, P.D.; Mueller, G.; Tan, J.; Uffman, J.P.; Levine, S.L. Ecological risk assessment for DvSnf7 RNA: A plant-incorporated protectant with targeted activity against western corn rootworm. Regul. Toxicol. Pharmacol. 2016, 81, 77–88. [Google Scholar] [CrossRef]
- Haller, S.; Widmer, F.; Siegfried, B.D.; Zhuo, X.; Romeis, J. Responses of two ladybird beetle species (Coleoptera: Coccinellidae) to dietary RNAi. Pest Manag. Sci. 2019, 75, 2652–2662. [Google Scholar] [CrossRef]
- Hollowell, H.; Rieske, L.K. Southern pine beetle-specific RNA interference exhibits no effect on model nontarget insects. J. Pest Sci. 2022, 95, 1429–1441. [Google Scholar] [CrossRef]
- Pampolini, F.; Rieske, L.K. Emerald Ash Borer Specific Gene Silencing Has No Effect on Non-target Organisms. Front. Agron. 2020, 2, 608827. [Google Scholar] [CrossRef]
- Fletcher, S.J.; Reeves, P.T.; Hoang, B.T.; Mitter, N. A Perspective on RNAi-Based Biopesticides. Front. Plant Sci. 2020, 11, 51. [Google Scholar] [CrossRef]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2009, 56, 227–235. [Google Scholar] [CrossRef]
- Killiny, N.; Hajeri, S.; Tiwari, S.; Gowda, S.; Stelinski, L.L. Double-Stranded RNA Uptake through Topical Application, Mediates Silencing of Five CYP4 Genes and Suppresses Insecticide Resistance in Diaphorina citri. PLoS ONE 2014, 9, e110536. [Google Scholar] [CrossRef] [PubMed]
- Miguel, K.; Scott, J. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci. 2015, 72, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef] [PubMed]
- Petek, M.; Coll, A.; Ferenc, R.; Razinger, J.; Gruden, K. Validating the Potential of Double-Stranded RNA Targeting Colorado Potato Beetle Mesh Gene in Laboratory and Field Trials. Front. Plant Sci. 2020, 11, 1250. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Jarausch, W.; Buchholz, G.; Bassler, A.; Braun, M.; Manthey, T.; Krczal, G.; Wassenegger, M. Delivery of Hairpin RNAs and Small RNAs Into Woody and Herbaceous Plants by Trunk Injection and Petiole Absorption. Front. Plant Sci. 2018, 9, 1253. [Google Scholar] [CrossRef]
- Biedenkopf, D.; Will, T.; Knauer, T.; Jelonek, L.; Furch, A.C.U.; Busche, T.; Koch, A. Systemic spreading of exogenous applied RNA biopesticides in the crop plant Hordeum vulgare. ExRNA 2020, 2, 12. [Google Scholar] [CrossRef]
- Hunter, W.B.; Glick, E.; Paldi, N.; Bextine, B.R. Advances in RNA interference: dsRNA Treatment in Trees and Grapevines for Insect Pest Suppression. Southwest. Entomol. 2012, 37, 85–87. [Google Scholar] [CrossRef]
- Zha, W.; Peng, X.; Chen, R.; Du, B.; Zhu, L.; He, G. Knockdown of Midgut Genes by dsRNA-Transgenic Plant-Mediated RNA Interference in the Hemipteran Insect Nilaparvata lugens. PLoS ONE 2011, 6, e20504. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Kassa, A.; Hu, X.; Robeson, J.; McMahon, M.; Richtman, N.M.; Steimel, J.P.; Kernodle, B.M.; Crane, V.C.; Sandahl, G.; et al. Control of Western Corn Rootworm (Diabrotica virgifera virgifera) Reproduction through Plant-Mediated RNA Interference. Sci. Rep. 2017, 7, 12591. [Google Scholar] [CrossRef]
- Head, G.P.; Carroll, M.W.; Evans, S.P.; Rule, D.M.; Willse, A.R.; Clark, T.L.; Storer, N.P.; Flannagan, R.D.; Samuel, L.W.; Meinke, L.J. Evaluation of SmartStax and SmartStax PRO maize against western corn rootworm and northern corn rootworm: Efficacy and resistance management. Pest Manag. Sci. 2017, 73, 1883–1899. [Google Scholar] [CrossRef] [PubMed]
- Pallis, S.; Alyokhin, A.; Manley, B.; Rodrigues, T.B.; Buzza, A.; Barnes, E.; Narva, K. Toxicity of a novel dsRNA-based insecticide to the Colorado potato beetle in laboratory and field trials. Pest Manag. Sci. 2022, 78, 3836–3848. [Google Scholar] [CrossRef]
- Mat Jalaluddin, N.S.; Othman, R.Y.; Harikrishna, J.A. Global trends in research and commercialization of exogenous and endogenous RNAi technologies for crops. Crit. Rev. Biotechnol. 2019, 39, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.B.; Mishra, S.K.; Sridharan, K.; Barnes, E.R.; Alyokhin, A.; Tuttle, R.; Kokulapalan, W.; Garby, D.; Skizim, N.J.; Tang, Y.-W. First Sprayable Double-Stranded RNA-Based Biopesticide Product Targets Proteasome Subunit Beta Type-5 in Colorado Potato Beetle (Leptinotarsa decemlineata). Front. Plant Sci. 2021, 12, 728652. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K. RNAi in Plants: Recent Developments and Applications in Agriculture. In Gene Silencing: Theory, Techniques and Applications; Catalano, A.J., Ed.; Nova Science Publishers, Inc.: Hauppauge, New York, NY, USA, 2010; pp. 183–199. [Google Scholar]
- Xiong, Y.; Zeng, H.; Zhang, Y.; Xu, D.; Qiu, D. Silencing the HaHR3 gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. Int. J. Biol. Sci. 2013, 9, 370–381. [Google Scholar] [CrossRef]
- Price, D.R.G.; Gatehouse, J.A. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008, 26, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 2011, 67, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.B.; Duan, J.J.; Palli, S.R.; Rieske, L.K. Identification of highly effective target genes for RNAi-mediated control of emerald ash borer, Agrilus planipennis. Sci. Rep. 2018, 8, 5020. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.B.; Dhandapani, R.K.; Palli, S.R. RNA interference in the Asian Longhorned Beetle: Identification of Key RNAi genes and reference genes for RT-qPCR. Sci. Rep. 2017, 7, 8913. [Google Scholar] [CrossRef]
- Kyre, B.R.; Bentz, B.J.; Rieske, L.K. Susceptibility of mountain pine beetle (Dendroctonus ponderosae Hopkins) to gene silencing through RNAi provides potential as a novel management tool. For. Ecol. Manag. 2020, 473, 118322. [Google Scholar] [CrossRef]
- Kyre, B.R.; Rodrigues, T.B.; Rieske, L.K. RNA interference and validation of reference genes for gene expression analyses using qPCR in southern pine beetle, Dendroctonus frontalis. Sci. Rep. 2019, 9, 5640. [Google Scholar] [CrossRef]
- Pampolini, F.; Rodrigues, T.B.; Leelesh, R.S.; Kawashima, T.; Rieske, L.K. Confocal microscopy provides visual evidence and confirms the feasibility of dsRNA delivery to emerald ash borer through plant tissues. J. Pest Sci. 2020, 93, 1143–1153. [Google Scholar] [CrossRef]
- Bragg, Z.; Rieske, L.K. Feasibility of Systemically Applied dsRNAs for Pest-Specific RNAi-Induced Gene Silencing in White Oak. Front. Plant Sci. 2022, 13, 638. [Google Scholar] [CrossRef]
- Thatcher, R.C.; Connor, M.D. Identification and Biology of Southern Pine Bark Beetles. Integr. Pest Manag. Handb. 1985, 634, 1–14. [Google Scholar]
- Blanche, C.A.; Hodges, J.D.; Nebeker, T.E.; Moehring, D.M. Southern Pine Beetle: The Host Dimension; Mississippi Agriculture and Forestry, Experiment Station (MAFES): Mississippi State, MS, USA, 1983; Volume 917. [Google Scholar]
- Thatcher, R.C.; Searcy, J.L.; Coster, J.E.; Hertel, G.D. The Southern Pine Beetle; USDA, Expanded Southern Pine Beetle Research and Application Program, Forest Service, Science and Education Administration: Pineville, LA, USA, 1980; Volume Technical Bulletin 1631. [Google Scholar]
- De La Giroday, H.M.C.; Carroll, A.L.; Aukema, B.H. Breach of the northern Rocky Mountain geoclimatic barrier: Initiation of range expansion by the mountain pine beetle. J. Biogeogr. 2012, 39, 1112–1123. [Google Scholar] [CrossRef]
- Dodds, K.J.; Aoki, C.F.; Arango-Velez, A.; Cancelliere, J.; D’Amato, A.W.; Digirolomo, M.F.; Rabaglia, R.J. Expansion of Southern Pine Beetle into Northeastern Forests: Management and Impact of a Primary Bark Beetle in a New Region. J. For. 2018, 116, 178–191. [Google Scholar] [CrossRef]
- Kurz, W.A.; Dymond, C.C.; Stinson, G.; Rampley, G.J.; Neilson, E.T.; Carroll, A.L.; Ebata, T.; Safranyik, L. Mountain pine beetle and forest carbon feedback to climate change. Nature 2008, 452, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.S.; Obar, R.A.; Schroeder, C.C.; Austin, T.W.; Poodry, C.A.; Wadsworth, S.C.; Vallee, R.B. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature 1991, 351, 583–586. [Google Scholar] [CrossRef]
- Boltz, B.A.; Bongarten, B.C.; Teskey, R.O. Seasonal patterns of net photosynthesis of loblolly pine from diverse origins. Can. J. For. Res. 1986, 16, 1063–1068. [Google Scholar] [CrossRef]
- McGregor, W.H.D.; Kramer, P.J. Seasonal Trends in Rates of Photosynthesis and Respiration of Loblolly Pine and White Pine Seedlings. Am. J. Bot. 1963, 50, 760–765. [Google Scholar] [CrossRef]
- Nedlo, J.E.; Martin, T.A.; Vose, J.M.; Teskey, R.O. Growing season temperatures limit growth of loblolly pine (Pinus taeda L.) seedlings across a wide geographic transect. Trees 2009, 23, 751–759. [Google Scholar] [CrossRef]
- South, D. Increasing Pine Survival and Early Growth by Planting Morphologically Improved Seedlings; Auburn University: Auburn, AL, USA, 1998; Volume 1. [Google Scholar]
- Mota-Sanchez, D.; Cregg, B.M.; Mccullough, D.G.; Poland, T.M.; Hollingworth, R.M. Distribution of trunk-injected 14C-imidacloprid in ash trees and effects on emerald ash borer (Coleoptera: Buprestidae) adults. Crop Prot. 2009, 28, 655–661. [Google Scholar] [CrossRef]
- Nix, K.; Lambdin, P.; Grant, J.; Coots, C.; Merten, P. Concentration Levels of Imidacloprid and Dinotefuran in Five Tissue Types of Black Walnut, Juglans nigra. Forests 2013, 4, 887–897. [Google Scholar] [CrossRef]
- Wu, C.; Dong, F.; Chen, X.; Zhang, T.; Mei, X.; Ning, J.; She, D. Spatial and temporal distribution, degradation, and metabolism of three neonicotinoid insecticides on different parts, especially pests’ target feeding parts of apple tree. Pest Manag. Sci. 2020, 76, 2190–2197. [Google Scholar] [CrossRef]
- Werner, R.A.; Lyon, D.L. Systemic Activity of Bidrin in Loblolly Pine Seedlings; Southeastern Forest Experiment Station, U.S. Department of Agriculture, Forest Service: Wahshington, DC, USA, 1970.
- Kaldis, A.; Berbati, M.; Melita, O.; Reppa, C.; Holeva, M.; Otten, P.; Voloudakis, A. Exogenously applied dsRNA molecules deriving from the Zucchini yellow mosaic virus (ZYMV) genome move systemically and protect cucurbits against ZYMV. Mol. Plant Pathol. 2018, 19, 883–895. [Google Scholar] [CrossRef]
- Dunoyer, P.; Melnyk, C.; Molnar, A.; Slotkin, R.K. Plant Mobile Small RNAs. Cold Spring Harb. Perspect. Biol. 2013, 5, a017897. [Google Scholar] [CrossRef]
- Fukudome, A.; Fukuhara, T. Plant dicer-like proteins: Double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant Res. 2016, 130, 33–44. [Google Scholar] [CrossRef]
- Rego-Machado, C.M.; Nakasu, E.Y.T.; Silva, J.M.F.; Lucinda, N.; Nagata, T.; Inoue-Nagata, A.K. siRNA biogenesis and advances in topically applied dsRNA for controlling virus infections in tomato plants. Sci. Rep. 2020, 10, 22277. [Google Scholar] [CrossRef]
- Dolgosheina, E.V.; Morin, R.D.; Aksay, G.; Sahinalp, S.C.; Magrini, V.; Mardis, E.R.; Mattsson, J.; Unrau, P.J. Conifers have a unique small RNA silencing signature. RNA 2008, 14, 1508–1515. [Google Scholar] [CrossRef]
- Gonzalez-Ibeas, D.; Martinez-Garcia, P.J.; Famula, R.A.; Delfino-Mix, A.; Stevens, K.A.; Loopstra, C.A.; Langley, C.H.; Neale, D.B.; Wegrzyn, J.L. Assessing the Gene Content of the Megagenome: Sugar Pine (Pinus lambertiana). G3 Genes Genomes Genet. 2016, 6, 3787–3802. [Google Scholar] [CrossRef][Green Version]
- Drew, A.P.; Ledig, F.T. Episodic Growth and Relative Shoot: Root Balance in Loblolly Pine Seedlings. Ann. Bot. 1980, 45, 143–148. [Google Scholar] [CrossRef]
- Kamthan, A.; Chaudhuri, A.; Kamthan, M.; Datta, A. Small RNAs in plants: Recent development and application for crop improvement. Front. Plant Sci. 2015, 6, 208. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Li, S.; Zhu, K.Y.; Ma, E.; Zhang, J. Characterization of a midgut-specific chitin synthase gene (LmCHS2) responsible for biosynthesis of chitin of peritrophic matrix in Locusta migratoria. Insect Biochem. Mol. Biol. 2012, 42, 902–910. [Google Scholar] [CrossRef]
- Nunes, F.M.F.; Aleixo, A.C.; Barchuk, A.R.; Bomtorin, A.D.; Grozinger, C.M.; Simões, Z.L.P. Non-Target Effects of Green Fluorescent Protein (GFP)-Derived Double-Stranded RNA (dsRNA-GFP) Used in Honey Bee RNA Interference (RNAi) Assays. Insects 2013, 4, 90–103. [Google Scholar] [CrossRef]
- Sim, S.; Ramirez, J.L.; Dimopoulos, G. Dengue Virus Infection of the Aedes aegypti Salivary Gland and Chemosensory Apparatus Induces Genes that Modulate Infection and Blood-Feeding Behavior. PLoS Pathog. 2012, 8, e1002631. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A Simple and Efficient Method for Isolating RNA from Pine Trees. Plant Mol. Biol. Report. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Palle, S.; Seeve, C.; Eckert, A.; Wegrzyn, J.; Neale, D.; Loopstra, C. Association of loblolly pine xylem development gene expression with single-nucleotide polymorphisms. Tree Physiol. 2013, 33, 763–774. [Google Scholar] [CrossRef]
- Chao, K.-H.; Barton, K.; Palmer, S.; Lanfear, R. sangeranalyseR: Simple and Interactive Processing of Sanger Sequencing Data in R. Genome Biol. Evol. 2021, 13, evab028. [Google Scholar] [CrossRef]



| Sequence 1 | Length | Sequence 2 | Length | Matches | Errors | Total | Match |
|---|---|---|---|---|---|---|---|
| Treatment-SHI | 390 | Recovered-SHI | 380 | 346 | 1 | 347 | 99.71% |
| Treatment-GFP | 267 | Recovered-GFP | 248 | 209 | 0 | 209 | 100.0% |
| Control-18s-SHI | 251 | Control-18s-GFP | 258 | 214 | 1 | 215 | 99.53% |
| Amplicon Name | Description |
|---|---|
| Treatment-SHI | PCR product used as the template to make dsSHI |
| Recovered-SHI | PCR product amplified using dsSHI primers on root tissue treated with dsSHI |
| Control-18s-SHI | PCR product amplified using 18s primers on root tissue treated with dsSHI |
| Treatment-GFP | PCR product used as template to make dsGFP |
| Recovered-GFP | PCR product amplified using dsGFP primers on root tissue treated with dsGFP |
| Control-18s-GFP | PCR product amplified using 18s primers on root tissue treated with dsGFP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bragg, Z.; Rieske, L.K. Spatial Distribution and Retention in Loblolly Pine Seedlings of Exogenous dsRNAs Applied through Roots. Int. J. Mol. Sci. 2022, 23, 9167. https://doi.org/10.3390/ijms23169167
Bragg Z, Rieske LK. Spatial Distribution and Retention in Loblolly Pine Seedlings of Exogenous dsRNAs Applied through Roots. International Journal of Molecular Sciences. 2022; 23(16):9167. https://doi.org/10.3390/ijms23169167
Chicago/Turabian StyleBragg, Zachary, and Lynne K. Rieske. 2022. "Spatial Distribution and Retention in Loblolly Pine Seedlings of Exogenous dsRNAs Applied through Roots" International Journal of Molecular Sciences 23, no. 16: 9167. https://doi.org/10.3390/ijms23169167
APA StyleBragg, Z., & Rieske, L. K. (2022). Spatial Distribution and Retention in Loblolly Pine Seedlings of Exogenous dsRNAs Applied through Roots. International Journal of Molecular Sciences, 23(16), 9167. https://doi.org/10.3390/ijms23169167






