Blood Test for Breast Cancer Screening through the Detection of Tumor-Associated Circulating Transcripts
Abstract
:1. Introduction
2. Results
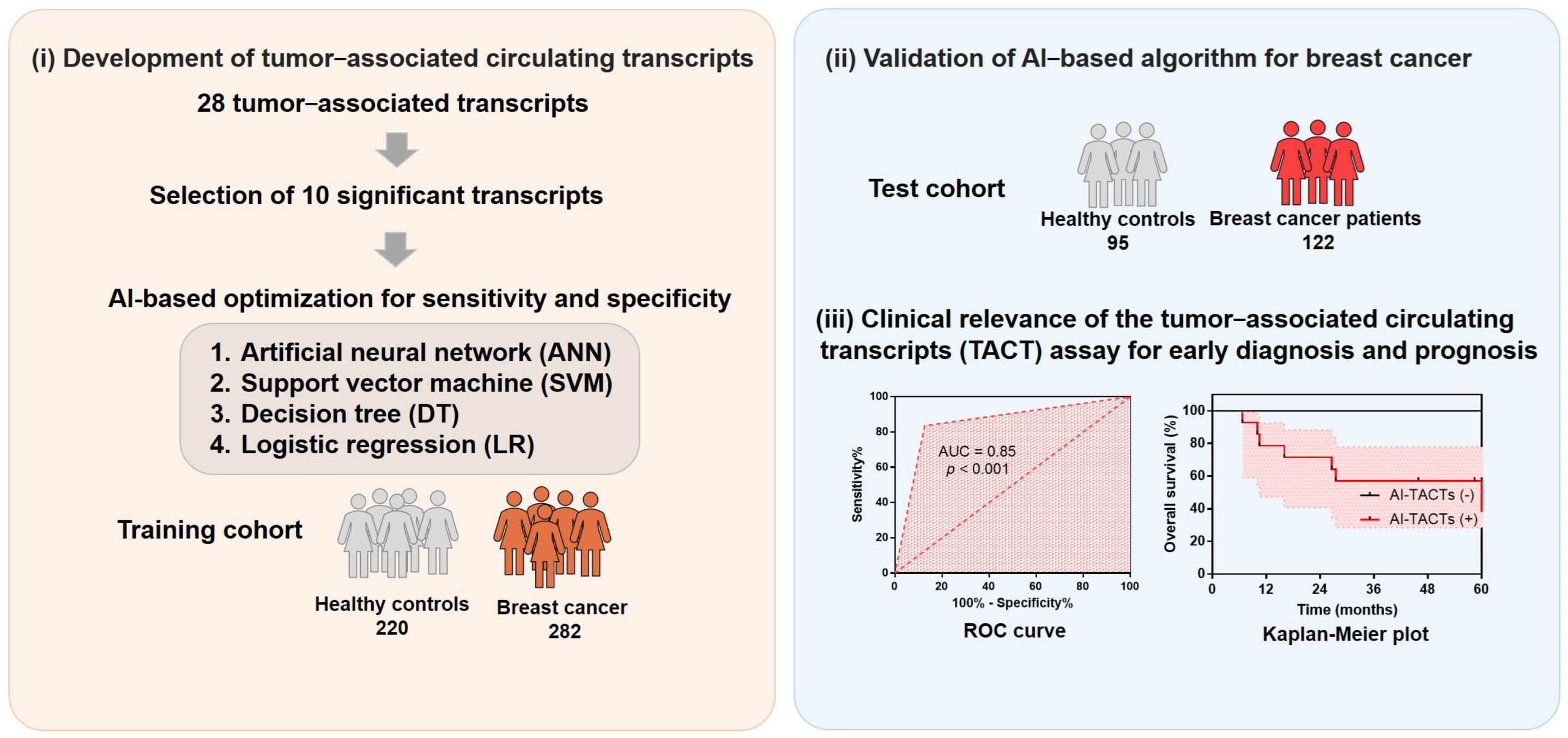
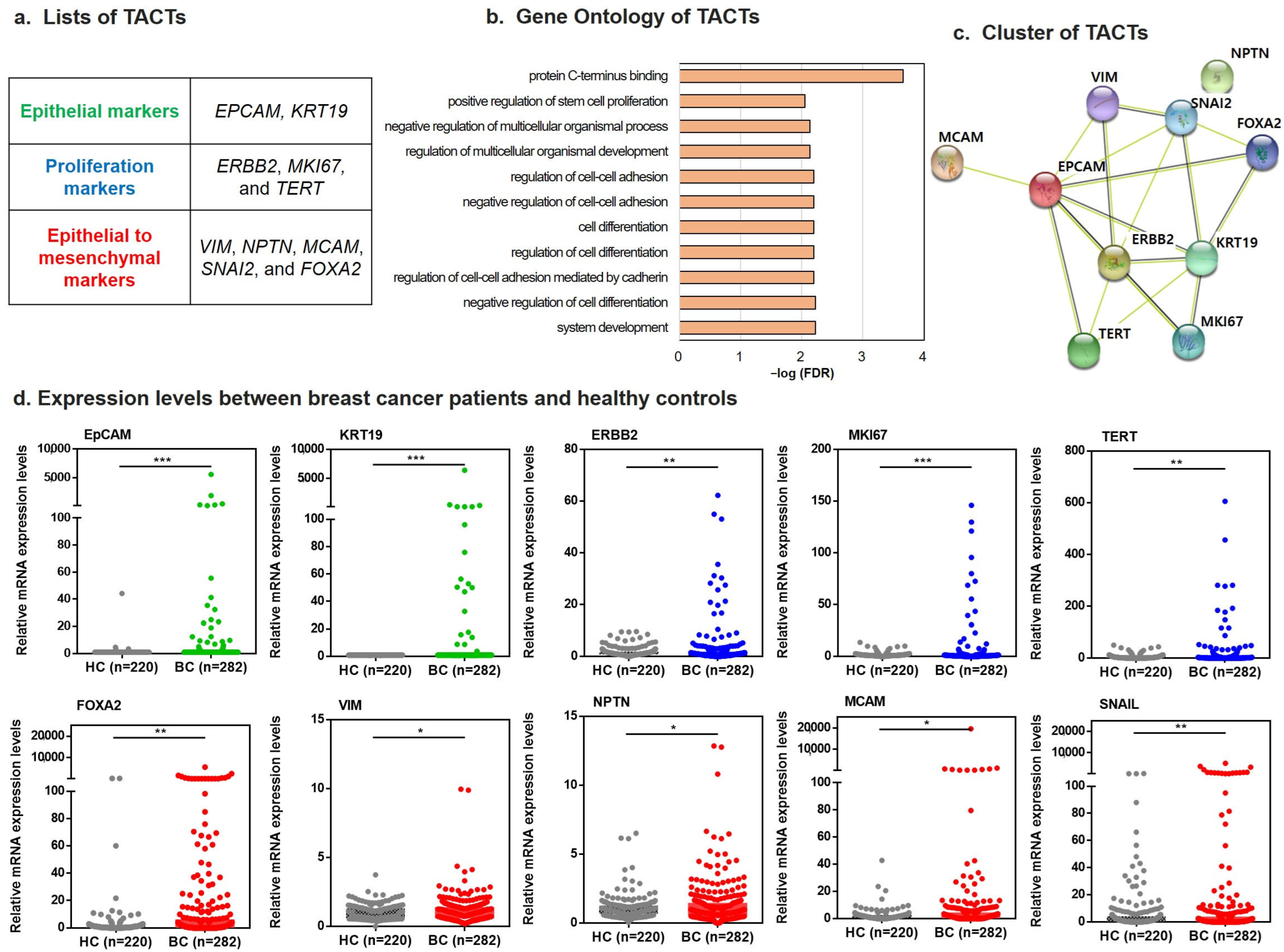
2.1. Development of TACTs for Breast Cancer Diagnosis
2.2. Assessment for Performance of TACTs in Patients with Breast Cancer versus Healthy Controls
2.3. Predictive Modeling of TACTs Using the Training Set and Test Set
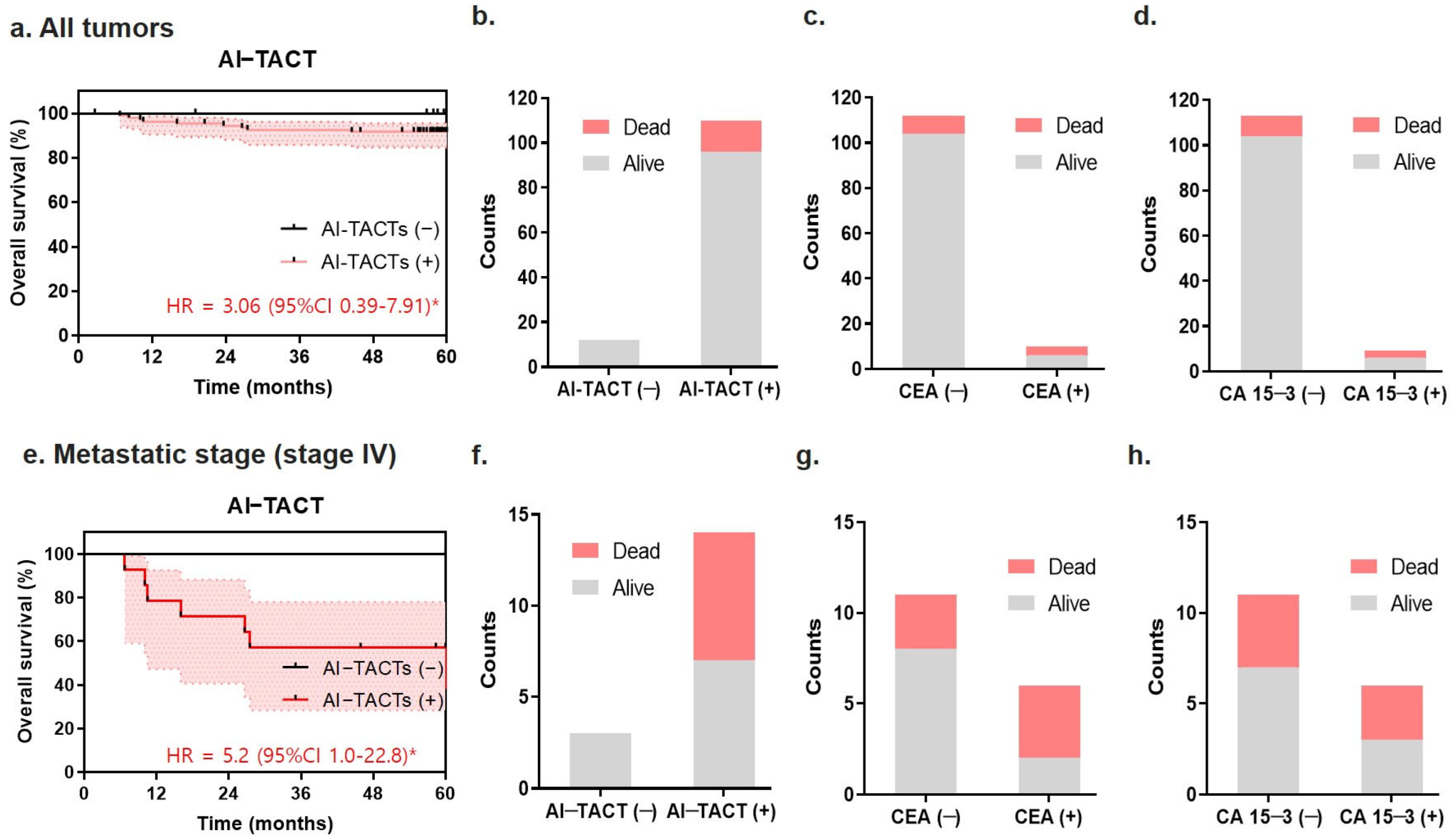
2.4. Diagnostic Performance of TACTs According to Breast Cancer Stage
2.5. Predictive Ability of the TACT Assay for Poor Prognosis in Metastatic Breast Cancers
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Selection and Blood Spiking Test of Tumor-Associated Circulating Transcripts
4.3. Patient Cohorts
4.4. Peripheral Blood from Patients and Healthy Controls
4.5. Reverse Transcription-Quantitative PCR Assay
4.6. Sensitivity and Specificity by Deep Learning Analysis
4.7. Bioinformatic Analysis of TACTs
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Kaiser, J. ‘Liquid biopsy’ for cancer promises early detection. Science 2018, 359, 259. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabieres, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Killock, D. Diagnosis: CancerSEEK and destroy—A blood test for early cancer detection. Nat. Rev. Clin. Oncol. 2018, 15, 133. [Google Scholar] [CrossRef]
- Liu, R.; Gao, Q.; Foltz, S.M.; Fowles, J.S.; Yao, L.; Wang, J.T.; Cao, S.; Sun, H.; Wendl, M.C.; Sethuraman, S.; et al. Co-evolution of tumor and immune cells during progression of multiple myeloma. Nat. Commun. 2021, 12, 2559. [Google Scholar] [CrossRef]
- Wang, H.Y.; Ahn, S.; Kim, S.; Park, S.; Park, S.; Han, H.; Sohn, J.H.; Kim, S.; Lee, H. Detection of circulating tumor cells in patients with breast cancer using the quantitative RT-PCR assay for monitoring of therapy efficacy. Exp. Mol. Pathol. 2014, 97, 445–452. [Google Scholar] [CrossRef]
- Wang, H.Y.; Ahn, S.; Kim, S.; Park, S.; Jung, D.; Park, S.; Han, H.; Sohn, J.; Kim, S.; Lee, H. Detection of circulating tumor cell-specific markers in breast cancer patients using the quantitative RT-PCR assay. Int. J. Clin. Oncol. 2015, 20, 878–890. [Google Scholar] [CrossRef]
- Wang, X.M.; Zhang, Z.; Pan, L.H.; Cao, X.C.; Xiao, C. KRT19 and CEACAM5 mRNA-marked circulated tumor cells indicate unfavorable prognosis of breast cancer patients. Breast Cancer Res. Treat. 2019, 174, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Johannesdottir, S.A.; Horvath-Puho, E.; Ehrenstein, V.; Schmidt, M.; Pedersen, L.; Sorensen, H.T. Existing data sources for clinical epidemiology: The Danish National Database of Reimbursed Prescriptions. Clin. Epidemiol. 2012, 4, 303–313. [Google Scholar] [CrossRef]
- Behrendt, C.E.; Tumyan, L.; Gonser, L.; Shaw, S.L.; Vora, L.; Paz, I.B.; Ellenhorn, J.D.; Yim, J.H. Evaluation of expert criteria for preoperative magnetic resonance imaging of newly diagnosed breast cancer. Breast 2014, 23, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.; Fritsche, H.; Mennel, R.; Norton, L.; Ravdin, P.; Taube, S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. 2007, 25, 5287–5312. [Google Scholar] [CrossRef] [PubMed]
- Riehl, B.D.; Kim, E.; Bouzid, T.; Lim, J.Y. The Role of Microenvironmental Cues and Mechanical Loading Milieus in Breast Cancer Cell Progression and Metastasis. Front. Bioeng. Biotechnol. 2020, 8, 608526. [Google Scholar] [CrossRef] [PubMed]
- Bredemeier, M.; Edimiris, P.; Mach, P.; Kubista, M.; Sjöback, R.; Rohlova, E.; Kolostova, K.; Hauch, S.; Aktas, B.; Tewes, M.; et al. Gene Expression Signatures in Circulating Tumor Cells Correlate with Response to Therapy in Metastatic Breast Cancer. Clin. Chem. 2017, 63, 1585–1593. [Google Scholar] [CrossRef] [PubMed]
- Kocemba-Pilarczyk, K.A.; Dudzik, P.; Leśkiewicz, K. The relationship between expression of VIMENTIN and CD146 genes in breast cancer. Bio-Algorithms Med-Syst. 2021, 17, 1–7. [Google Scholar] [CrossRef]
- Nezamdoost, Z.; Saghebjoo, M.; Hoshyar, R.; Hedayati, M.; Keska, A. High-Intensity Training and Saffron: Effects on Breast Cancer-related Gene Expression. Med. Sci. Sports Exerc. 2020, 52, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, C.; Gao, W.; Chen, T.; Qian, T.; Hu, J.; Tan, Y. FOXA2 attenuates the epithelial to mesenchymal transition by regulating the transcription of E-cadherin and ZEB2 in human breast cancer. Cancer Lett. 2015, 361, 240–250. [Google Scholar] [CrossRef]
- Rodriguez-Pinto, D.; Sparkowski, J.; Keough, M.P.; Phoenix, K.N.; Vumbaca, F.; Han, D.K.; Gundelfinger, E.D.; Beesley, P.; Claffey, K.P. Identification of novel tumor antigens with patient-derived immune-selected antibodies. Cancer Immunol. Immunother. CII 2009, 58, 221–234. [Google Scholar] [CrossRef]
- Thierry, A.R. A Step Closer to Cancer Screening by Blood Test. Clin. Chem. 2018, 64, 1420–1422. [Google Scholar] [CrossRef] [PubMed]
- Watts, G. Liquid biopsy: Still early days for early detection. Lancet 2018, 391, 2593–2594. [Google Scholar] [CrossRef]
- Cayrefourcq, L.; Alix-Panabieres, C. Clinical relevance of liquid biopsy in breast cancer: Update in 2020. Expert Rev. Mol. Diagn. 2020, 20, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Labani-Motlagh, A.; Ashja-Mahdavi, M.; Loskog, A. The Tumor Microenvironment: A Milieu Hindering and Obstructing Antitumor Immune Responses. Front. Immunol. 2020, 11, 940. [Google Scholar] [CrossRef]
- Zhang, J.; Pan, S.; Jian, C.; Hao, L.; Dong, J.; Sun, Q.; Jin, H.; Han, X. Immunostimulatory Properties of Chemotherapy in Breast Cancer: From Immunogenic Modulation Mechanisms to Clinical Practice. Front. Immunol. 2021, 12, 819405. [Google Scholar] [CrossRef]
- Verigos, J.; Magklara, A. Revealing the Complexity of Breast Cancer by Next Generation Sequencing. Cancers 2015, 7, 2183–2200. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef]
- Ruan, G.T.; Zhu, L.C.; Gong, Y.Z.; Liao, X.W.; Wang, X.K.; Liao, C.; Wang, S.; Yan, L.; Xie, H.L.; Zhou, X.; et al. The diagnosis and prognosis values of WNT mRNA expression in colon adenocarcinoma. J. Cell Biochem. 2020, 121, 3145–3161. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, H. Meta-analysis of diagnostic accuracy of magnetic resonance imaging and mammography for breast cancer. J. Cancer Res. Ther. 2017, 13, 862–868. [Google Scholar] [CrossRef]
- Fass, L. Imaging and cancer: A review. Mol. Oncol. 2008, 2, 115–152. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Nguyen, Q.T. Molecular imaging for cancer diagnosis and surgery. Adv. Drug Deliv. Rev. 2014, 66, 90–100. [Google Scholar] [CrossRef]
- Wang, W.; Liu, M.; Wang, J.; Tian, R.; Dong, J.; Liu, Q.; Zhao, X.; Wang, Y. Analysis of the discriminative methods for diagnosis of benign and malignant solitary pulmonary nodules based on serum markers. Oncol. Res. Treat. 2014, 37, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Sun, X.; He, Y.; Liu, C.; Liu, H. Elevated Levels of Serum Tumor Markers CEA and CA15-3 Are Prognostic Parameters for Different Molecular Subtypes of Breast Cancer. PLoS ONE 2015, 10, e0133830. [Google Scholar] [CrossRef] [PubMed]
- Bone, B.; Wiberg, M.K.; Szabo, B.K.; Szakos, A.; Danielsson, R. Comparison of 99mTc-sestamibi scintimammography and dynamic MR imaging as adjuncts to mammography in the diagnosis of breast cancer. Acta Radiol. 2003, 44, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Malur, S.; Wurdinger, S.; Moritz, A.; Michels, W.; Schneider, A. Comparison of written reports of mammography, sonography and magnetic resonance mammography for preoperative evaluation of breast lesions, with special emphasis on magnetic resonance mammography. Breast Cancer Res. 2001, 3, 55–60. [Google Scholar] [CrossRef]
- Jemal, A.; Center, M.M.; DeSantis, C.; Ward, E.M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1893–1907. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.Z.; Garcia, E.; Glubrecht, D.D.; Poon, H.Y.; Mackey, J.R.; Godbout, R. CRABP1 is associated with a poor prognosis in breast cancer: Adding to the complexity of breast cancer cell response to retinoic acid. Mol. Cancer 2015, 14, 129. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar]
- Sui, S.; An, X.; Xu, C.; Li, Z.; Hua, Y.; Huang, G.; Sui, S.; Long, Q.; Sui, Y.; Xiong, Y.; et al. An immune cell infiltration-based immune score model predicts prognosis and chemotherapy effects in breast cancer. Theranostics 2020, 10, 11938–11949. [Google Scholar] [CrossRef]
- Kasangian, A.A.; Gherardi, G.; Biagioli, E.; Torri, V.; Moretti, A.; Bernardin, E.; Cordovana, A.; Farina, G.; Bramati, A.; Piva, S.; et al. The prognostic role of tumor size in early breast cancer in the era of molecular biology. PLoS ONE 2017, 12, e0189127. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kato, F.; Yamashita, H.; Baba, M.; Cui, Y.; Li, R.; Oyama-Manabe, N.; Shirato, H. Automatic Estimation of Volumetric Breast Density Using Artificial Neural Network-Based Calibration of Full-Field Digital Mammography: Feasibility on Japanese Women with and Without Breast Cancer. J. Digit. Imaging 2017, 30, 215–227. [Google Scholar] [CrossRef]
- Nair, T.M. Building and Interpreting Artificial Neural Network Models for Biological Systems; Cartwright, H., Ed.; Artificial Neural Networks; Humana: New York, NY, USA, 2021; pp. 185–194. [Google Scholar]
- Bernal, J.; Kushibar, K.; Asfaw, D.S.; Valverde, S.; Oliver, A.; Martí, R.; Lladó, X. Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: A review. Artif. Intell. Med. 2019, 95, 64–81. [Google Scholar] [CrossRef]
- Sanchez-Cauce, R.; Perez-Martin, J.; Luque, M. Multi-input convolutional neural network for breast cancer detection using thermal images and clinical data. Comput. Methods Programs Biomed. 2021, 204, 106045. [Google Scholar] [CrossRef]
- Zhao, Y.; Pan, Z.; Namburi, S.; Pattison, A.; Posner, A.; Balachander, S.; Paisie, C.A.; Reddi, H.V.; Rueter, J.; Gill, A.J.; et al. CUP-AI-Dx: A tool for inferring cancer tissue of origin and molecular subtype using RNA gene-expression data and artificial intelligence. EBioMedicine 2020, 61, 103030. [Google Scholar] [CrossRef]

| Cohorts | Training Cohort, n (%) | Test Cohort, n (%) | ||
|---|---|---|---|---|
| Variable | Healthy Control (n = 220) | Breast Cancer (n = 282) | Healthy Control (n = 95) | Breast Cancer (n = 122) |
| Age at diagnosis | ||||
| <50 years | 174 (79.1) | 157 (55.7) | 66 (69.5) | 61 (50) |
| ≥50 years | 46 (20.9) | 125 (44.3) | 29 (30.5) | 61 (50) |
| TNM stage | ||||
| I | 126 (44.7) | 47 (38.6) | ||
| II | 66 (23.4) | 32 (26.2) | ||
| III | 14 (5.0) | 10 (8.2) | ||
| IV | 24 (8.5) | 17 (13.9) | ||
| Unknown | 52 (18.4) | 16 (13.1) | ||
| Therapy | ||||
| Adjuvant | 210 (74.5) | 92 (75.4) | ||
| Neoadjuvant | 48 (17.0) | 13 (10.7) | ||
| Metastasis | 24 (8.5) | 17 (13.9) | ||
| Subtypes | ||||
| Luminal A | 165 (58.5) | 65 (53.3) | ||
| Luminal B | 30 (10.6) | 21 (17.2) | ||
| HER-2 | 38 (13.5) | 18 (14.8) | ||
| Triple-negative | 45 (16.0) | 18 (14.8) | ||
| Unknown | 4 (1.4) | 0 (0) | ||
| CEA | ||||
| Positive | 26 (9.2) | 10 (8.2) | ||
| Negative | 256 (90.8) | 112 (91.8) | ||
| CA15-3 | ||||
| Positive | 19 (6.7) | 9 (7.4) | ||
| Negative | 263 (93.3) | 113 (92.6) | ||
| Survival status | ||||
| Alive | 242 (85.8) | 110 (90.2) | ||
| Dead | 40 (14.2) | 12 (9.8) | ||
| Mammography (category) | ||||
| <4 | 19 (6.7) | 8 (6.6) | ||
| ≥4 | 263 (93.3) | 104 (85.2) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Ahn, S.; Kim, J.Y.; Kim, J.; Han, H.J.; Hwang, D.; Park, J.; Park, H.S.; Park, S.; Kim, G.M.; et al. Blood Test for Breast Cancer Screening through the Detection of Tumor-Associated Circulating Transcripts. Int. J. Mol. Sci. 2022, 23, 9140. https://doi.org/10.3390/ijms23169140
Park S, Ahn S, Kim JY, Kim J, Han HJ, Hwang D, Park J, Park HS, Park S, Kim GM, et al. Blood Test for Breast Cancer Screening through the Detection of Tumor-Associated Circulating Transcripts. International Journal of Molecular Sciences. 2022; 23(16):9140. https://doi.org/10.3390/ijms23169140
Chicago/Turabian StylePark, Sunyoung, Sungwoo Ahn, Jee Ye Kim, Jungho Kim, Hyun Ju Han, Dasom Hwang, Jungmin Park, Hyung Seok Park, Seho Park, Gun Min Kim, and et al. 2022. "Blood Test for Breast Cancer Screening through the Detection of Tumor-Associated Circulating Transcripts" International Journal of Molecular Sciences 23, no. 16: 9140. https://doi.org/10.3390/ijms23169140
APA StylePark, S., Ahn, S., Kim, J. Y., Kim, J., Han, H. J., Hwang, D., Park, J., Park, H. S., Park, S., Kim, G. M., Sohn, J., Jeong, J., Song, Y. U., Lee, H., & Kim, S. I. (2022). Blood Test for Breast Cancer Screening through the Detection of Tumor-Associated Circulating Transcripts. International Journal of Molecular Sciences, 23(16), 9140. https://doi.org/10.3390/ijms23169140








