Analysis of Ser/Thr Kinase HASPIN-Interacting Proteins in the Spermatids
Abstract
1. Introduction
2. Results
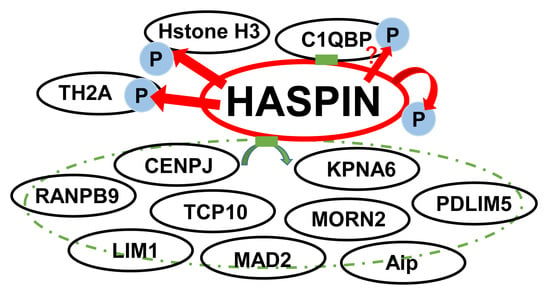
2.1. Screening of Proteins That Interact with HASPIN in a Mouse Testis cDNA Library Using Two-Hybrid System
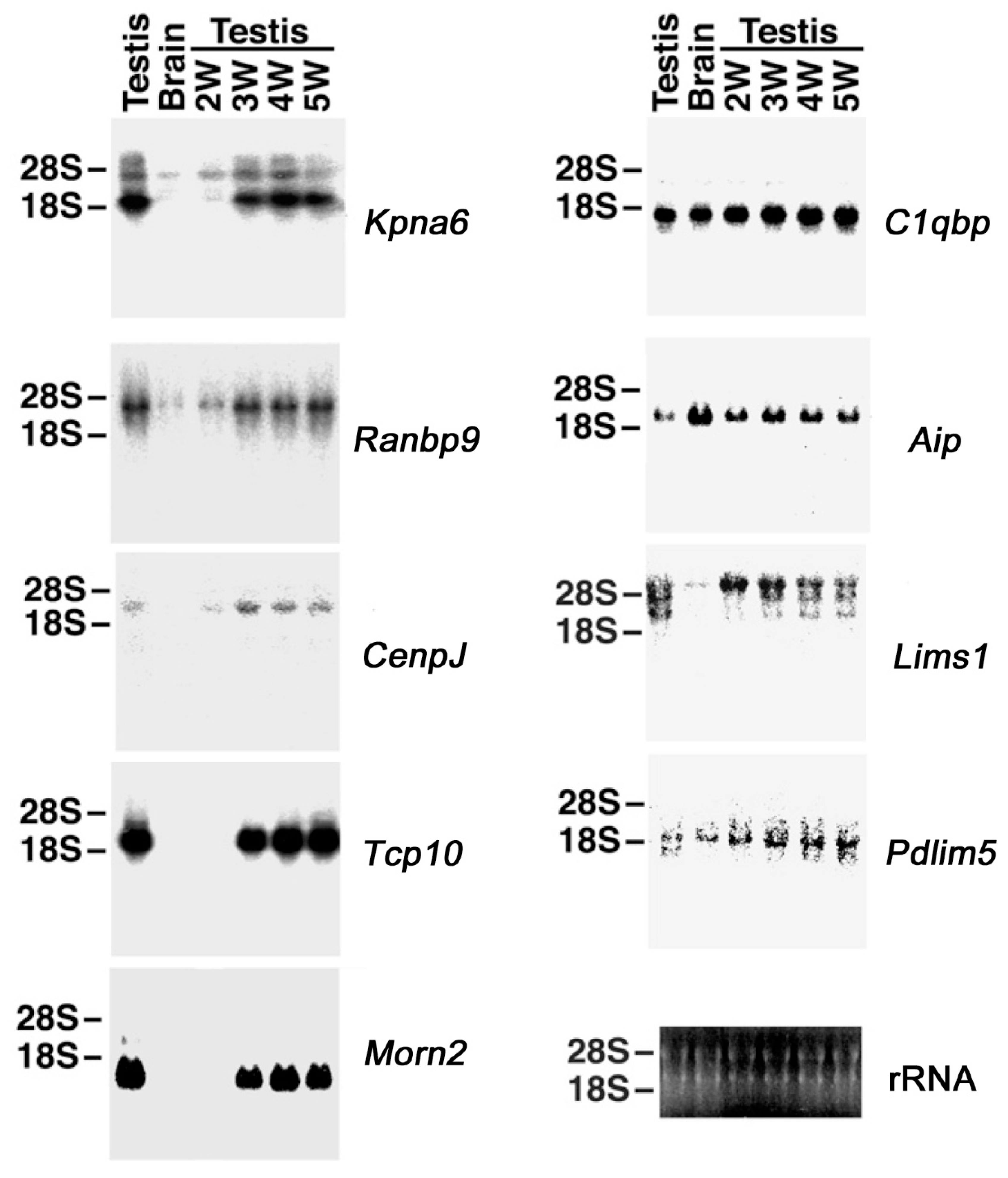
2.2. Northern Blotting of Genes Interacting with HASPIN
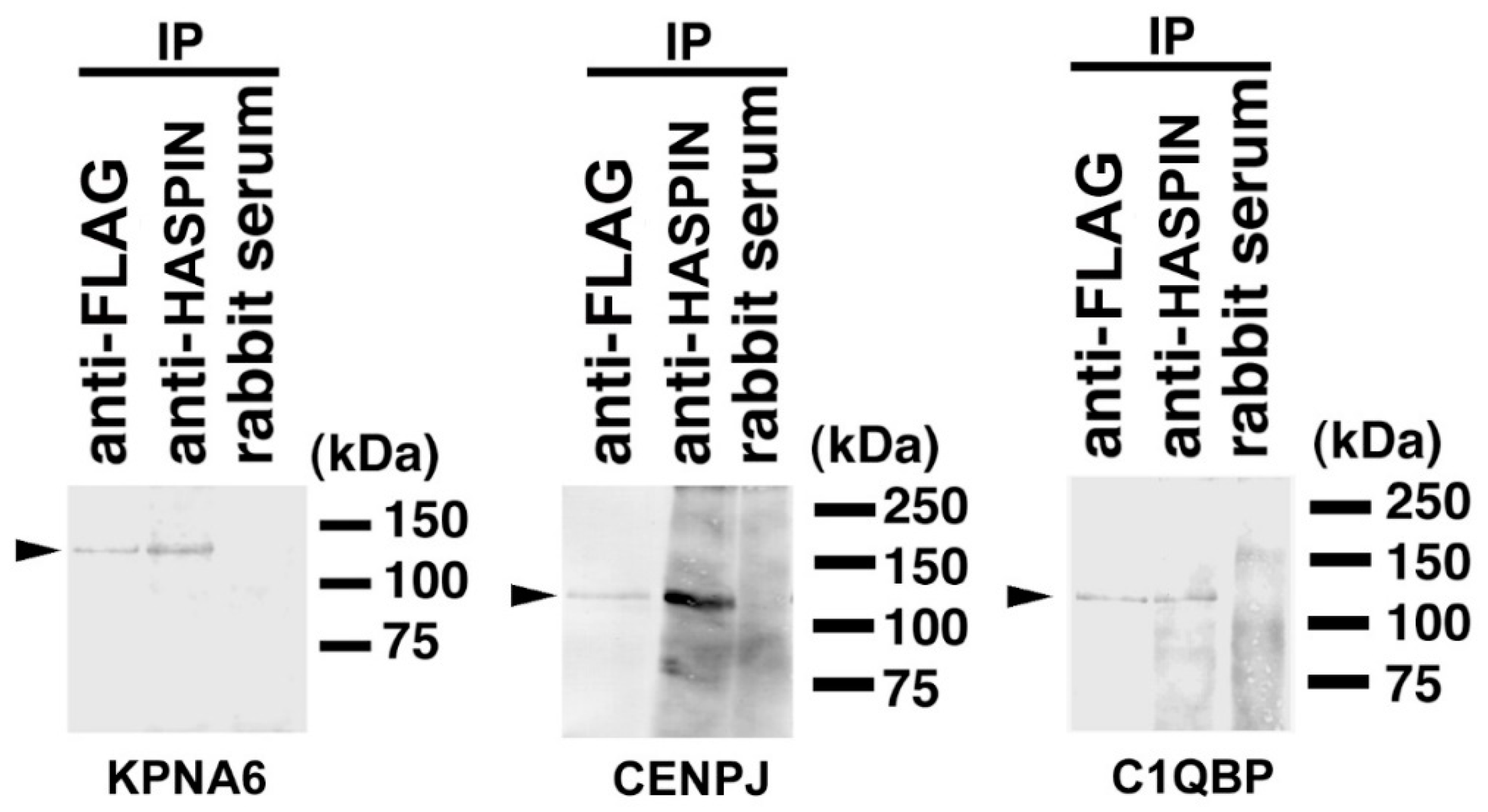
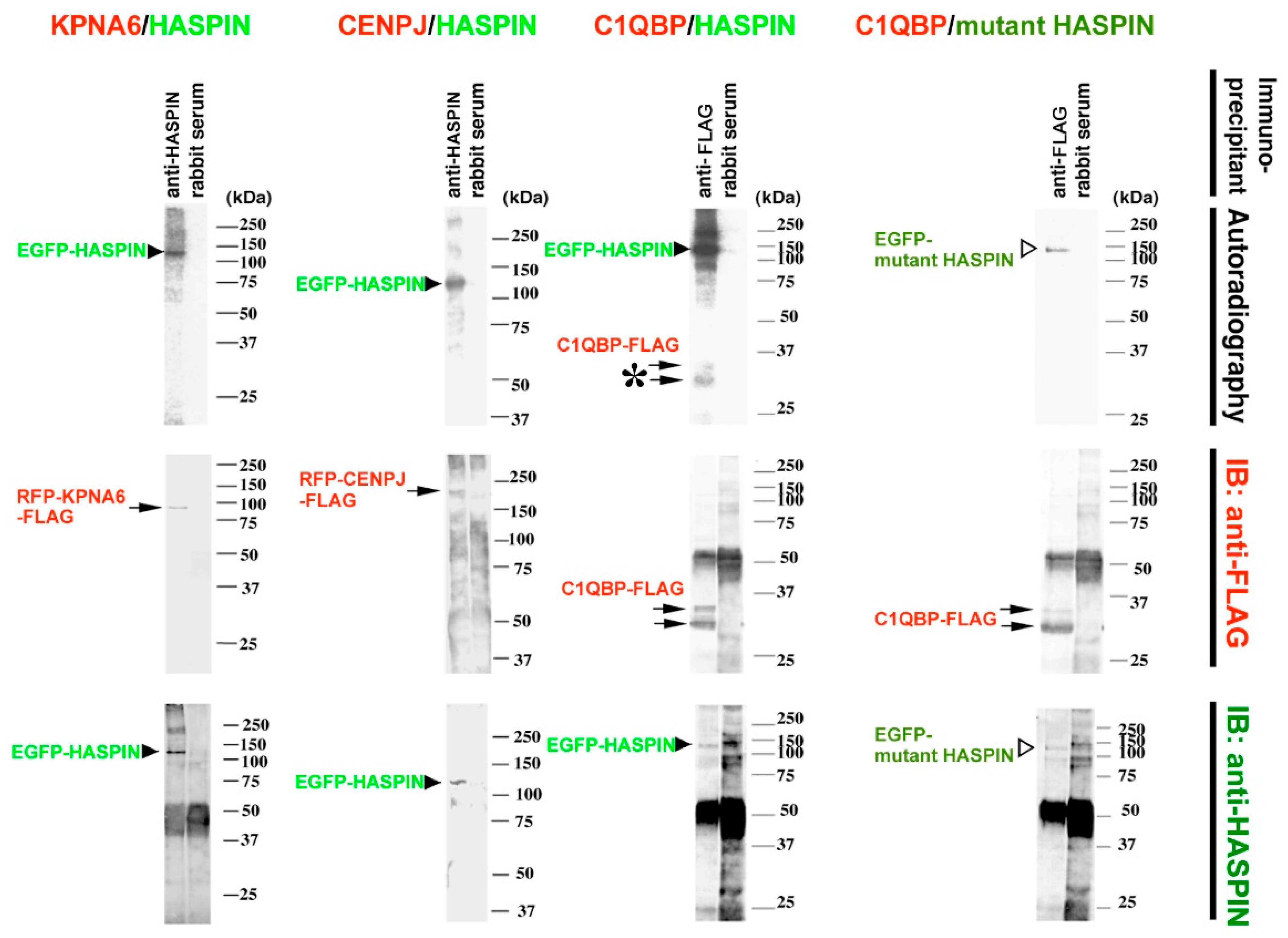
2.3. Binding Evaluation of HASPIN and Proteins Screened by Two-Hybrid System in Cultured Cells
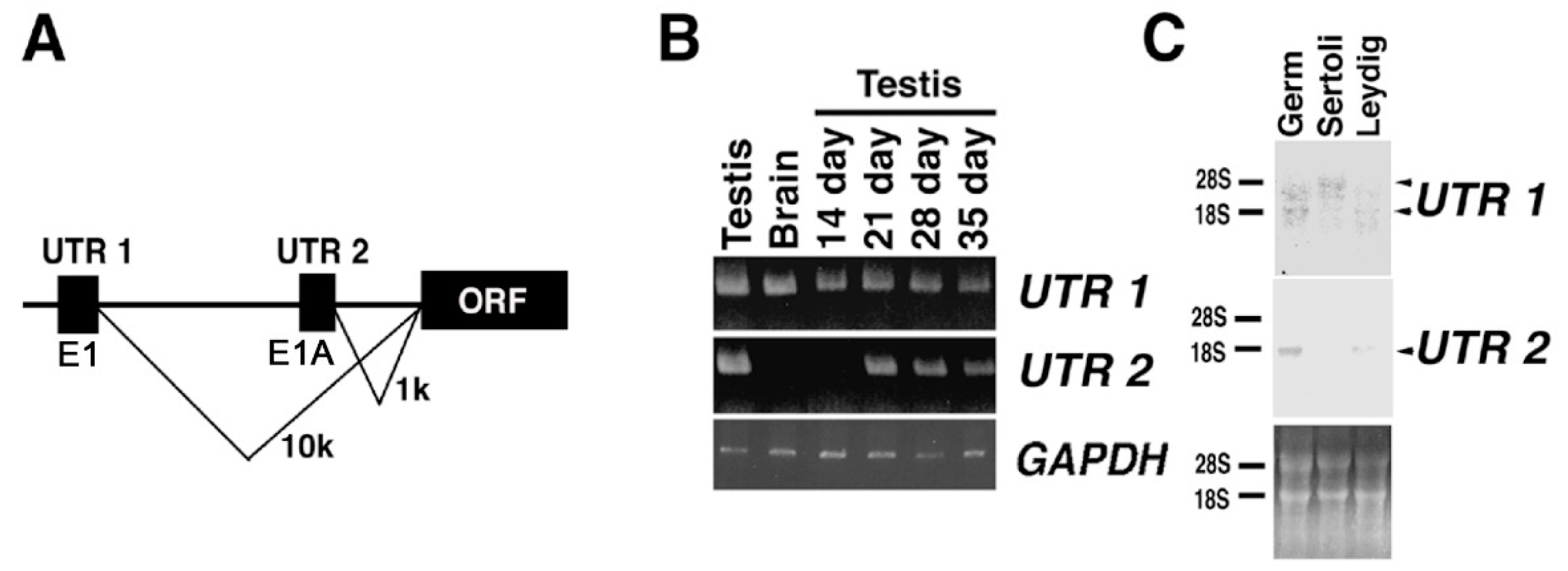
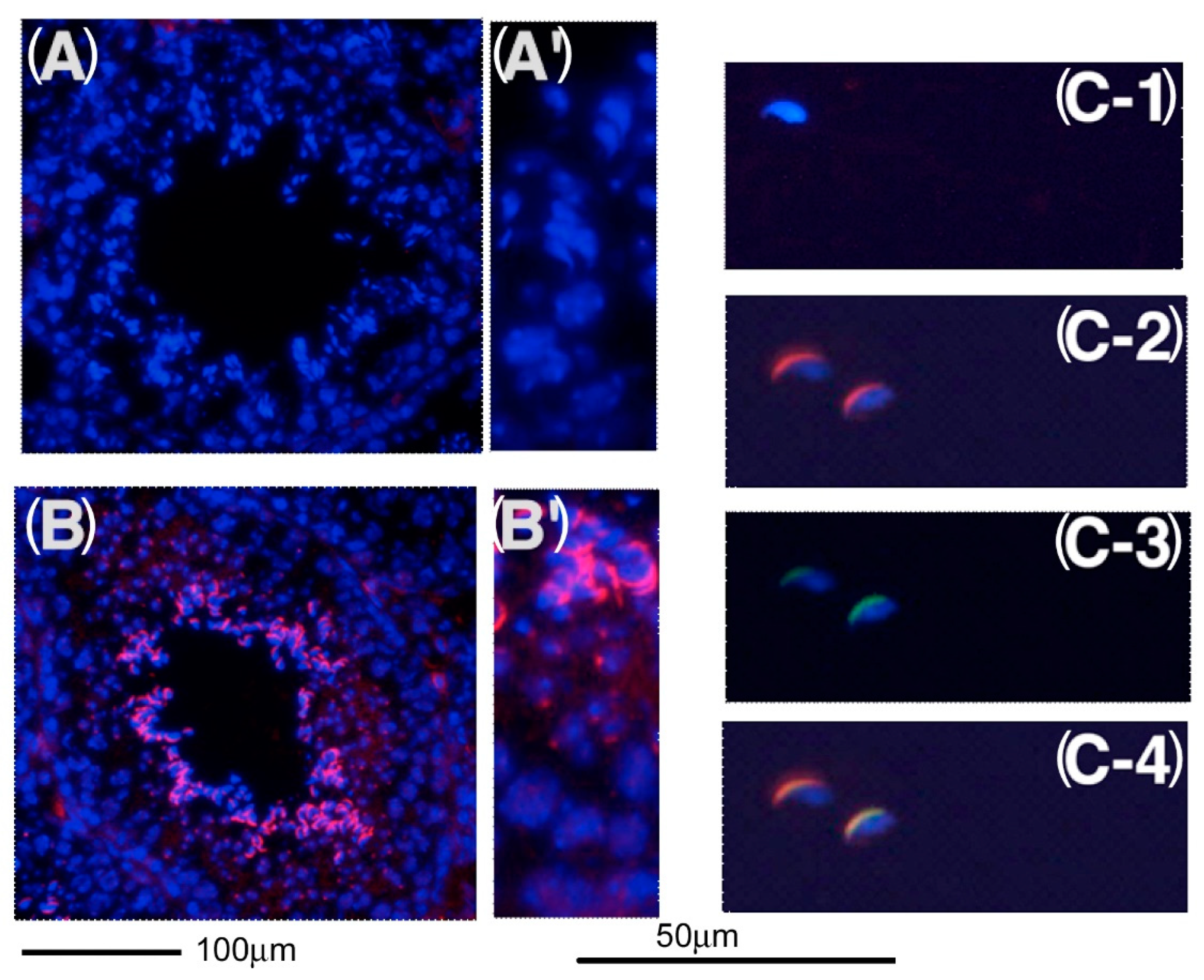
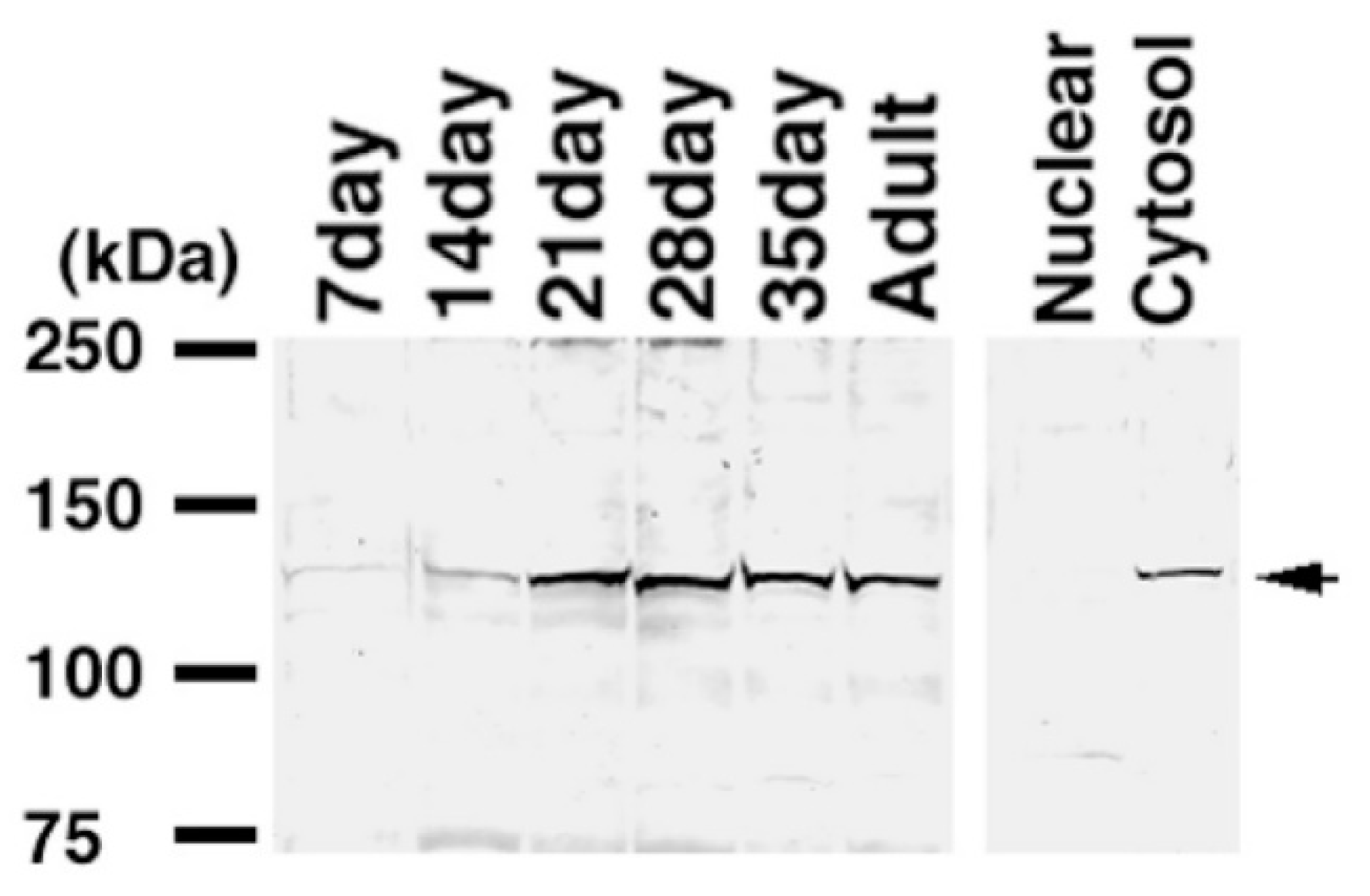
2.4. Expression of KPNA6/Importin α6 in the Testis
2.5. Expression of CENPJ/CPAP in the Testis
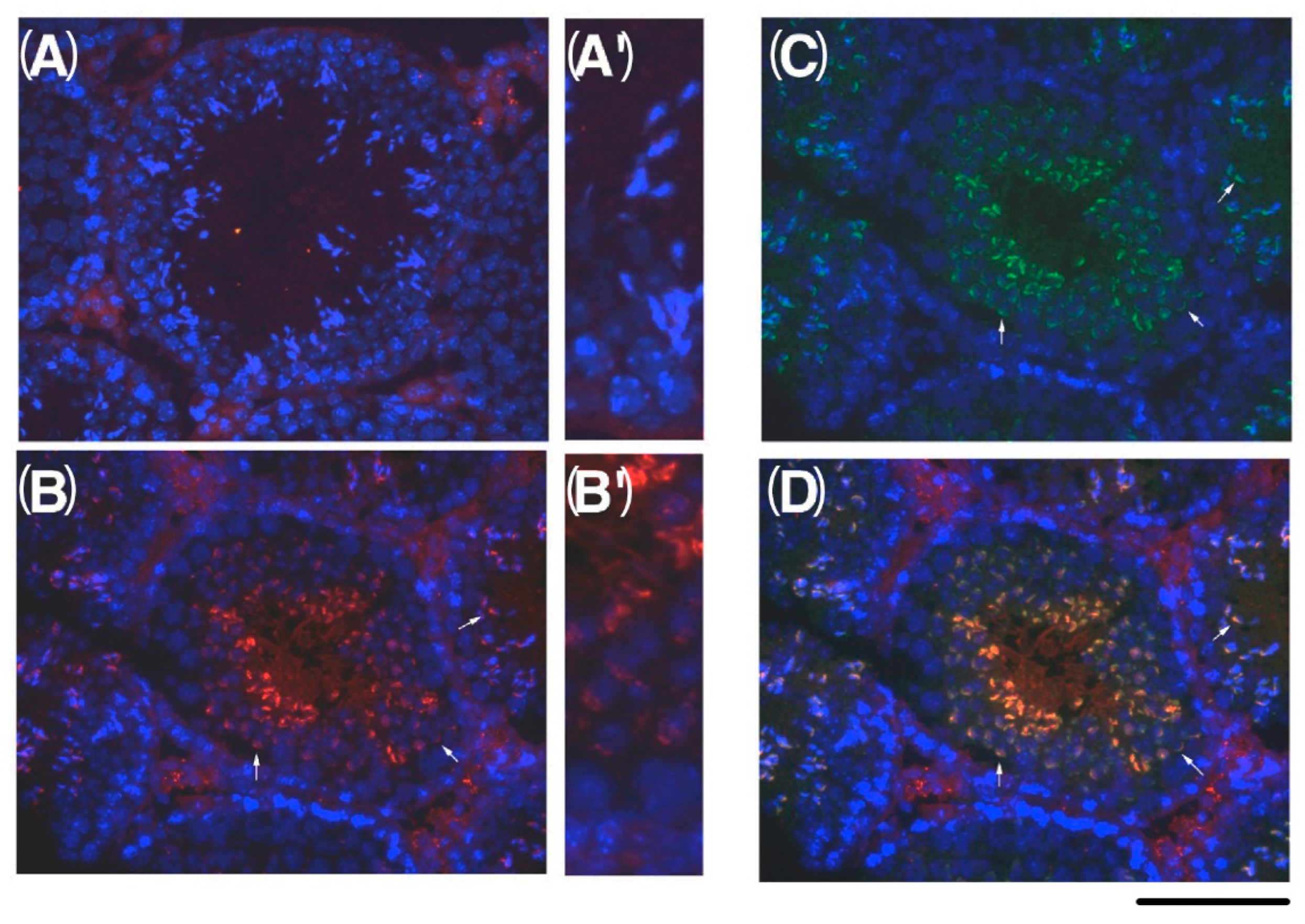
2.6. Expression of C1QBP/HABP1 in the Testis
2.7. Phosphorylation Target Protein of HASPIN
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Yeast Two-Hybrid System
4.3. Northern Blots and DNA Sequencing
4.4. RT-PCR
4.5. Western Blotting
4.6. Antibody Preparation
4.7. Expression Vectors
4.8. Transfection
4.9. Immunostaining
4.10. Immunoprecipitation
4.11. In Vitro Kinase Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tanaka, H.; Yoshimura, Y.; Nishina, Y.; Nozaki, M.; Nojima, H.; Nishimune, Y. Isolation and characterization of cDNA clones specifically expressed in testicular germ cells. FEBS Lett. 1994, 355, 4–10. [Google Scholar] [CrossRef]
- Tanaka, H.; Yoshimura, Y.; Nozaki, M.; Yomogida, K.; Tsuchida, J.; Tosaka, Y.; Habu, T.; Nakanishi, T.; Okada, M.; Nojima, H.; et al. Identification and characterization of a haploid germ cell-specific nuclear protein kinase (Haspin) in spermatid nuclei and its effects on somatic cells. J. Biol. Chem. 1999, 274, 17049–17057. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.M. The Haspin gene: Location in an intron of the integrin alphaE gene, associated transcription of an integrin alphaE-derived RNA and expression in diploid as well as haploid cells. Gene 2001, 267, 55–69. [Google Scholar] [CrossRef]
- Dai, J.; Sultan, S.; Taylor, S.S.; Higgins, J.M. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005, 19, 472–488. [Google Scholar] [CrossRef]
- Wang, F.; Ulyanova, N.P.; van der Waal, M.S.; Patnaik, D.; Lens, S.M.; Higgins, J.M. A Positive Feedback Loop Involving Haspin and Aurora B Promotes CPC Accumulation at Centromeres in Mitosis. Curr. Biol. 2001, 21, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Hada, M.; Kim, J.; Inoue, E.; Fukuda, Y.; Tanaka, H.; Watanabe, Y.; Okada, Y. TH2A is phosphorylated at meiotic centromere by Haspin. Chromosoma 2017, 126, 769–780. [Google Scholar] [CrossRef]
- Higgins, J.M. Haspin-like proteins: A new family of evolutionarily conserved putative eukaryotic protein kinases. Protein Sci. 2001, 10, 1677–1684. [Google Scholar] [CrossRef]
- Kurihara, D.; Matsunaga, S.; Omura, T.; Higashiyama, T.; Fukui, K. Identification and characterization of plant Haspin kinase as a histone H3 threonine kinase. BMC Plant Biol. 2011, 11, 73. [Google Scholar] [CrossRef]
- Soupsana, K.; Karanika, E.; Kiosse, F.; Christogianni, A.; Sfikas, Y.; Topalis, P.; Batistatou, A.; Kanaki, Z.; Klinakis, A.; Politou, A.S.; et al. Distinct roles of haspin in stem cell division and male gametogenesis. Sci. Rep. 2021, 11, 19901. [Google Scholar] [CrossRef]
- Shimada, M.; Goshima, T.; Matsuo, H.; Johmura, Y.; Haruta, M.; Murata, K.; Tanaka, H.; Ikawa, M.; Nakanishi, K.; Nakanishi, M. Essential role of autoactivation circuitry on Aurora B-mediated H2AX-pS121 in mitosis. Nat. Commun. 2016, 7, 12059. [Google Scholar] [CrossRef]
- Huertas, D.; Soler, M.; Moreto, J.; Villanueva, A.; Martinez, A.; Vidal, A.; Charlton, M.; Moffat, D.; Patel, S.; McDermott, J.; et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene 2012, 31, 1408–1418. [Google Scholar] [CrossRef]
- Liu, N.; Qadri, F.; Busch, H.; Huegel, S.; Sihn, G.; Chuykin, I.; Hartmann, E.; Bader, M.; Rother, F. Kpna6 deficiency causes infertility in male mice by disrupting spermatogenesis. Development 2021, 148, dev198374. [Google Scholar] [CrossRef]
- Tsuji, L.; Takumi, T.; Imamoto, N.; Yoneda, Y. Identification of novel homologues of mouse importin alpha, the alpha subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 1997, 416, 30–34. [Google Scholar] [CrossRef]
- Tanaka, H.; Pereira, L.A.; Nozaki, M.; Tsuchida, J.; Sawada, K.; Mori, H.; Nishimune, Y. A germ cell-specific nuclear antigen recognized by a monoclonal antibody raised against mouse testicular germ cells. Int. J. Androl. 1997, 20, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Shibata, N.; Tsunekawa, N.; Okamoto-Ito, S.; Akasu, R.; Tokumasu, A.; Noce, T. Mouse RanBPM is a partner gene to a germline specific RNA helicase, mouse vasa homolog protein. Mol. Reprod. Dev. 2004, 67, 1–7. [Google Scholar] [CrossRef]
- Gruss, O.J.; Carazo-Salas, R.E.; Schatz, C.A.; Guarguaglini, G.; Kast, J.; Wilm, K.; Le Bot, N.; Vernos, I.; Karsenti, E.; Mattaj, I.W. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 2001, 104, 83–93. [Google Scholar] [CrossRef]
- Nakamura, M.; Masuda, H.; Horii, J.; Kuma, K.; Yokoyama, N.; Ohba, T.; Nishitani, H.; Miyata, T.; Tanaka, M.; Nishimoto, T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J. Cell Biol. 1998, 143, 1041–1052. [Google Scholar] [CrossRef]
- Hung, L.Y.; Tang, C.J.; Tang, T.K. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol. Cell Biol. 2000, 20, 7813–7825. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.A.; Tanaka, H.; Nagata, Y.; Sawada, K.; Mori, H.; Chimelli, L.M.; Nishimune, Y. Identification and characterization of an antigen recognized by monoclonal antibody TRA 54 in mouse epididymis and vas deferens. Int. J. Androl. 1998, 21, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Batchu, R.B.; Datta, K. Purification, partial characterization of rat kidney hyaluronic acid binding protein and its localization on the cell surface. Eur. J. Cell Biol. 1991, 56, 58–67. [Google Scholar]
- Ghebrehiwet, B.; Lim, B.L.; Peerschke, E.I.; Willis, A.C.; Reid, K.B. Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular “heads” of C1q. J. Exp. Med. 1994, 179, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Petersen-Mahrt, S.K.; Estmer, C.; Ohrmalm, C.; Matthews, D.A.; Russell, W.C.; Akusjarvi, G. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 1999, 18, 1014–1024. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.M.; Deb, T.B.; Datta, K. Hyaluronic acid induced hyaluronic acid binding protein phosphorylation and inositol triphosphate formation in lymphocytes. Biochem. Mol. Biol. Int. 1996, 40, 327–337. [Google Scholar]
- Yu, L.; Loewenstein, P.M.; Zhang, Z.; Green, M. In vitro interaction of the human immunodeficiency virus type 1 Tat transactivator and the general transcription factor TFIIB with the cellular protein TAP. J. Virol. 1995, 69, 3017–3023. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.Q.; Eisen-Vandervelde, A.; Ray, S.; Hahn, Y.S. HCV core/gC1qR interaction arrests T cell cycle progression through stabilization of the cell cycle inhibitor p27Kip1. Virology 2003, 314, 271–282. [Google Scholar] [CrossRef]
- Meenakshi, J.; Goswami, S.K.; Datta, K. Constitutive expression of hyaluronan binding protein 1 (HABP1/p32/gC1qR) in normal fibroblast cells perturbs its growth characteristics and induces apoptosis. Biochem. Biophys. Res. Commun. 2003, 300, 686–693. [Google Scholar] [CrossRef]
- Ghosh, I.; Bharadwaj, A.; Datta, K. Reduction in the level of hyaluronan binding protein 1 (HABP1) is associated with loss of sperm motility. J. Reprod. Immunol. 2002, 53, 45–54. [Google Scholar] [CrossRef]
- Ghosh, I.; Datta, K. Sperm surface hyaluronan binding protein (HABP1) interacts with zona pellucida of water buffalo (Bubalus bubalis) through its clustered mannose residues. Mol. Reprod. Dev. 2003, 64, 235–244. [Google Scholar] [CrossRef]
- Grace, K.S.; Bronson, R.A.; Ghebrehiwet, B. Surface expression of complement receptor gC1q-R/p33 is increased on the plasma membrane of human spermatozoa after capacitation. Biol. Reprod. 2002, 66, 823–829. [Google Scholar] [CrossRef][Green Version]
- Ranganathan, S.; Ganguly, A.K.; Datta, K. Evidence for presence of hyaluronan binding protein on spermatozoa and its possible involvement in sperm function. Mol. Reprod. Dev. 1994, 38, 69–76. [Google Scholar] [CrossRef]
- Bharadwaj, A.; Ghosh, I.; Sengupta, A.; Cooper, T.G.; Weinbauer, G.F.; Brinkworth, M.H.; Nieschlag, E.; Datta, K. Stage-specific expression of proprotein form of hyaluronan binding protein 1 (HABP1) during spermatogenesis in rat. Mol. Reprod. Dev. 2002, 62, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Cechetto, J.D.; Sadacharan, S.K.; Berk, P.D.; Gupta, R.S. Immunogold localization of mitochondrial aspartate aminotransferase in mitochondria and on the cell surface in normal rat tissues. Histol. Histopathol. 2002, 17, 353–364. [Google Scholar] [PubMed]
- Tanaka, H.; Tsujimura, A. Pervasiveness of intronless genes expressed in haploid germ cell differentiation. Reprod. Med. Biol. 2021, 20, 255–259. [Google Scholar] [CrossRef]
- Takeshima, H.; Komazaki, S.; Nishi, M.; Iino, M.; Kangawa, K. Junctophilins: A novel family of junctional membrane complex proteins. Mol. Cell 2000, 6, 11–22. [Google Scholar]
- Dawid, I.B.; Breen, J.J.; Toyama, R. LIM domains: Multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998, 14, 156–162. [Google Scholar] [CrossRef]
- Lamb, J.R.; Tugendreich, S.; Hieter, P. Tetratrico peptide repeat interactions: To TPR or not to TPR? Trends Biochem. Sci. 1995, 20, 257–259. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. The HORMA domain: A common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 1998, 23, 284–286. [Google Scholar] [CrossRef]
- Peng, B.; Sutherland, D.K.; Sum, E.Y.; Olayioye, M.; Wittlin, S.; Tang, T.K.; Lindeman, G.J.; Visvader, J.E. CPAP is a novel stat5-interacting cofactor that augments stat5-mediated transcriptional activity. Mol. Endocrinol. 2002, 16, 2019–2033. [Google Scholar] [CrossRef]
- Ollendorff, V.; Donoghue, D.J. The serine/threonine phosphatase PP5 interacts withCDC16 and CDC27, two tetratricopeptide repeat-containing subunits of the anaphase-promoting complex. J. Biol. Chem. 1997, 272, 32011–32018. [Google Scholar] [CrossRef]
- Chen, J.; Fang, G. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 2001, 15, 1765–1770. [Google Scholar] [CrossRef]
- Wassmann, K.; Liberal, V.; Benezra, R. Mad2 phosphorylation regulates its association with Mad1 and the APC/C. EMBO J. 2003, 22, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ching, Y.P.; Chun, A.C.; Jin, D.Y. Nuclear localization of the cell cycle regulator CDH1 and its regulation by phosphorylation. J. Biol. Chem. 2003, 278, 12530–12536. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.; Chen, R.H. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat. Cell Biol. 2003, 5, 748–753. [Google Scholar] [CrossRef]
- Imamoto, N.; Shimamoto, T.; Takao, T.; Tachibana, T.; Kose, S.; Matsubae, M.; Sekimoto, T.; Shimonishi, Y.; Yoneda, Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 1995, 14, 3617–3626. [Google Scholar] [CrossRef]
- Gorlich, D.; Kostka, S.; Kraft, R.; Dingwall, C.; Laskey, R.A.; Hartmann, E.; Prehn, S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 1995, 5, 383–392. [Google Scholar] [CrossRef]
- Ho, H.C.; Tang, C.Y.; Suarez, S.S. Three-dimensional structure of the Golgi apparatus in mouse spermatids: A scanning electron microscopic study. Anat. Rec. 1999, 256, 189–194. [Google Scholar] [CrossRef]
- Moreno, R.D.; Ramalho-Santos, J.; Sutovsky, P.; Chan, E.K.; Schatten, G. Vesicular traffic and golgi apparatus dynamics during mammalian spermatogenesis: Implications for acrosome architecture. Biol. Reprod. 2000, 63, 89–98. [Google Scholar] [CrossRef]
- Ramalho-Santos, J.; Moreno, R.D.; Wessel, G.W.; Chan, E.K.; Schatten, G. Membrane trafficking machinery components associated with the mammalian acrosome during spermiogenesis. Exp. Cell Res. 2001, 267, 45–60. [Google Scholar] [CrossRef]
- Jasper, D.K. ER-annulate lamellar associations of mucosal epithelial cells of the rat jejunum. Cell Tissue Res. 1976, 175, 417–420. [Google Scholar] [CrossRef]
- Wischnitzer, S. The annulate lamellae. Int. Rev. Cytol. 1970, 27, 65–100. [Google Scholar]
- Cordes, V.C.; Rackwitz, H.R.; Reidenbach, S. Mediators of nuclear protein import target karyophilic proteins to pore complexes of cytoplasmic annulate lamellae. Exp. Cell Res. 1997, 237, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.; Russell, L.; Bundman, D.; Freund, M. Presence of microfilaments and tubular structures in boar spermatozoa after chemically inducing the acrosome reaction. Biol. Reprod. 1978, 19, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Stambaugh, R.; Smith, M. Tubulin and microtubule-like structures in mammalian acrosomes. J. Exp. Zool. 1978, 203, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef]
- Biggiogera, M.; Fakan, S.; Leser, G.; Martin, T.M.; Gordon, J. Immunoelectron microscopical visualization of ribonucleoproteins in the chromatoid body of mouse spermatids. Mol. Reprod. Dev. 1990, 26, 150–158. [Google Scholar] [CrossRef]
- Figueroa, J.; Burzio, L.O. Polysome-like structures in the chromatoid body of rat spermatids. Cell Tissue Res. 1998, 291, 575–579. [Google Scholar] [CrossRef]
- Ventela, S.; Toppari, J.; Parvinen, M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: Mechanisms of haploid gene product sharing. Mol. Biol. Cell 2003, 14, 2768–2780. [Google Scholar] [CrossRef]
- Bullard, D.C.; Schimenti, J.C. Molecular structure of Tcp-10 genes from the t complex responder locus. Mamm. Genome 1991, 1, 228–234. [Google Scholar] [CrossRef]
- Lyon, M. Transmission ratio distortion in mouse thaplotypes is due to multiple distorter genes acting on a responder locus. Cell 1984, 37, 621–628. [Google Scholar] [CrossRef]
- Herrmann, B.G.; Koschorz, B.; Werts, K.; McLaughlin, K.J.; Kispert, A. A protein kinase encoded by the t complex responder gene causes non-mendelian inheritance. Nature 1999, 402, 141–146. [Google Scholar] [CrossRef]
- Rao, C.M.; Deb, T.B.; Gupta, S.; Datta, K. Regulation of cellular phosphorylation of hyaluronan binding protein and its role in the formation of second messenger. Biochim. Biophys Acta 1997, 1336, 387–393. [Google Scholar] [CrossRef]
- Patrat, C.; Serres, C.; Jouannet, P. The acrosome reaction in human spermatozoa. Biol. Cell 2000, 92, 255–266. [Google Scholar] [CrossRef]
- Walensky, L.D.; Snyder, S.H. Inositol 1,4,5-trisphosphate receptors selectively localized to the acrosomes of mammalian sperm. J. Cell Biol. 1995, 130, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Fukami, K.; Nakao, K.; Inoue, T.; Kataoka, Y.; Kurokawa, M.; Fissore, R.A.; Nakamura, K.; Katsuki, M.; Mikoshiba, K.; Yoshida, N.; et al. Requirement of phospholipase Cdelta4 for the zona pellucida-induced acrosome reaction. Science 2001, 292, 920–923. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Wada, M.; Park, J. HASPIN kinase inhibitor CHR-6494 suppresses intestinal polyp development, cachexia, and hypogonadism in Apcmin/+ mice. Eur. J. Cancer Prev. 2020, 29, 482–485. [Google Scholar] [CrossRef]
- Saha, P.; Datta, K. Multi-functional, multicompartmental hyaluronan-binding protein 1 (HABP1/p32/gC1qR): Implication in cancer progression and metastasis. Oncotarget 2018, 9, 10784–10807. [Google Scholar] [CrossRef]
- Simos, G.; Georgatos, S.D. The lamin B receptor-associated protein p34 shares sequence homology and antigenic determinants with the splicing factor 2-associated protein p32. FEBS Lett. 1994, 346, 225–228. [Google Scholar] [PubMed]
- Koga, M.; Tanaka, H.; Yomogida, K.; Nozaki, M.; Tsuchida, J.; Ohta, H.; Nakamura, Y.; Masai, K.; Yoshimura, Y.; Yamanaka, M.; et al. Isolation and characterization of a haploid germ cell-specific novel complementary deoxyribonucleic acid; testis-specific homologue of succinyl CoA:3-Oxo acid CoA transferase. Biol. Reprod. 2000, 63, 1601–1609. [Google Scholar] [CrossRef][Green Version]
- Muta, T.; Kang, D.; Kitajima, S.; Fujiwara, T.; Hamasaki, N. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 1997, 272, 24363–24370. [Google Scholar] [CrossRef]
- Fukuda, E.; Tanaka, H.; Yamaguchi, K.; Takasaka, M.; Kawamura, Y.; Okuda, H.; Isotani, A.; Ikawa, M.; Shapiro, V.S.; Tsuchida, J.; et al. Identification and characterization of the antigen recognized by the germ cell mAb TRA98 using a human comprehensive wet protein array. Genes Cells 2021, 26, 180–189. [Google Scholar] [CrossRef]



| Registrated Gene Name (NCBI Accetion Number) | Domain in Cloned Gene | Number Of Clones Obtained |
|---|---|---|
| CenpJ/CPAP (NP_001014996) | Glycine repeat | 5 |
| Kpna6/importin alpha 6 (NP_032494.3) | Whole | 2 |
| Morn2 (NP_001347369) | MORN motif | 2 |
| C1qbp/HABP1 (NP_031599.2) | Whole | 1 |
| Ranbp9/RanBPM (NP_001391577.1) | SPRY domain | 1 |
| Tcp10/t-complex protein 10 (X58170) | Glycine repeat | 1 |
| Aip (NP_001263213.1) | Tetratricopeptide repeat | 1 |
| Lims1 (NP_001180232.1) | LIM domain | 1 |
| Pdlim5 (NP_001177781.1) | LIM domain | 1 |
| Mad2l2 (NP_001292349.1) | HORMA domain | 1 |
| Name | Reference |
|---|---|
| anti-KPNA6-rabbit IgG | this study |
| anti-CENPJ-rabbit IgG | this study |
| anti-C1QBP rabbit IgG | Muta et al. [69] |
| anti-HASPIN rabbit serum | Tanaka et al. [2] |
| Integrin β1 antibody | SantaCruz Biotechnology (SC-8978) |
| Anti-FLAG antibody | Sigma-Aldrich (F3165) |
| monoclonal antibody TRA98 | Fukuda et al. [14,70] |
| monoclonal antibody TRA54 | Pereira et al. [17] |
| rabibit anti-ACTB antibody | Cell Signaling (4967) |
| anti-rabbit IgG HRP-linked whole Ab (from donkey) | Amersham Biosciences (NA934V) |
| rabbit anti-rat IgG HRP-secondly antibody | DAKO (P0450) |
| donkey anti-rabbit IgG, biotinylated species specific antibody | Amersham Biosciences (SAB3700865) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maeda, N.; Tsuchida, J.; Nishimune, Y.; Tanaka, H. Analysis of Ser/Thr Kinase HASPIN-Interacting Proteins in the Spermatids. Int. J. Mol. Sci. 2022, 23, 9060. https://doi.org/10.3390/ijms23169060
Maeda N, Tsuchida J, Nishimune Y, Tanaka H. Analysis of Ser/Thr Kinase HASPIN-Interacting Proteins in the Spermatids. International Journal of Molecular Sciences. 2022; 23(16):9060. https://doi.org/10.3390/ijms23169060
Chicago/Turabian StyleMaeda, Naoko, Junji Tsuchida, Yoshitake Nishimune, and Hiromitsu Tanaka. 2022. "Analysis of Ser/Thr Kinase HASPIN-Interacting Proteins in the Spermatids" International Journal of Molecular Sciences 23, no. 16: 9060. https://doi.org/10.3390/ijms23169060
APA StyleMaeda, N., Tsuchida, J., Nishimune, Y., & Tanaka, H. (2022). Analysis of Ser/Thr Kinase HASPIN-Interacting Proteins in the Spermatids. International Journal of Molecular Sciences, 23(16), 9060. https://doi.org/10.3390/ijms23169060