A Comparative Study of the Plasma Chemokine Profile in COVID-19 Patients Infected with Different SARS-CoV-2 Variants
Abstract
:1. Introduction
2. Results
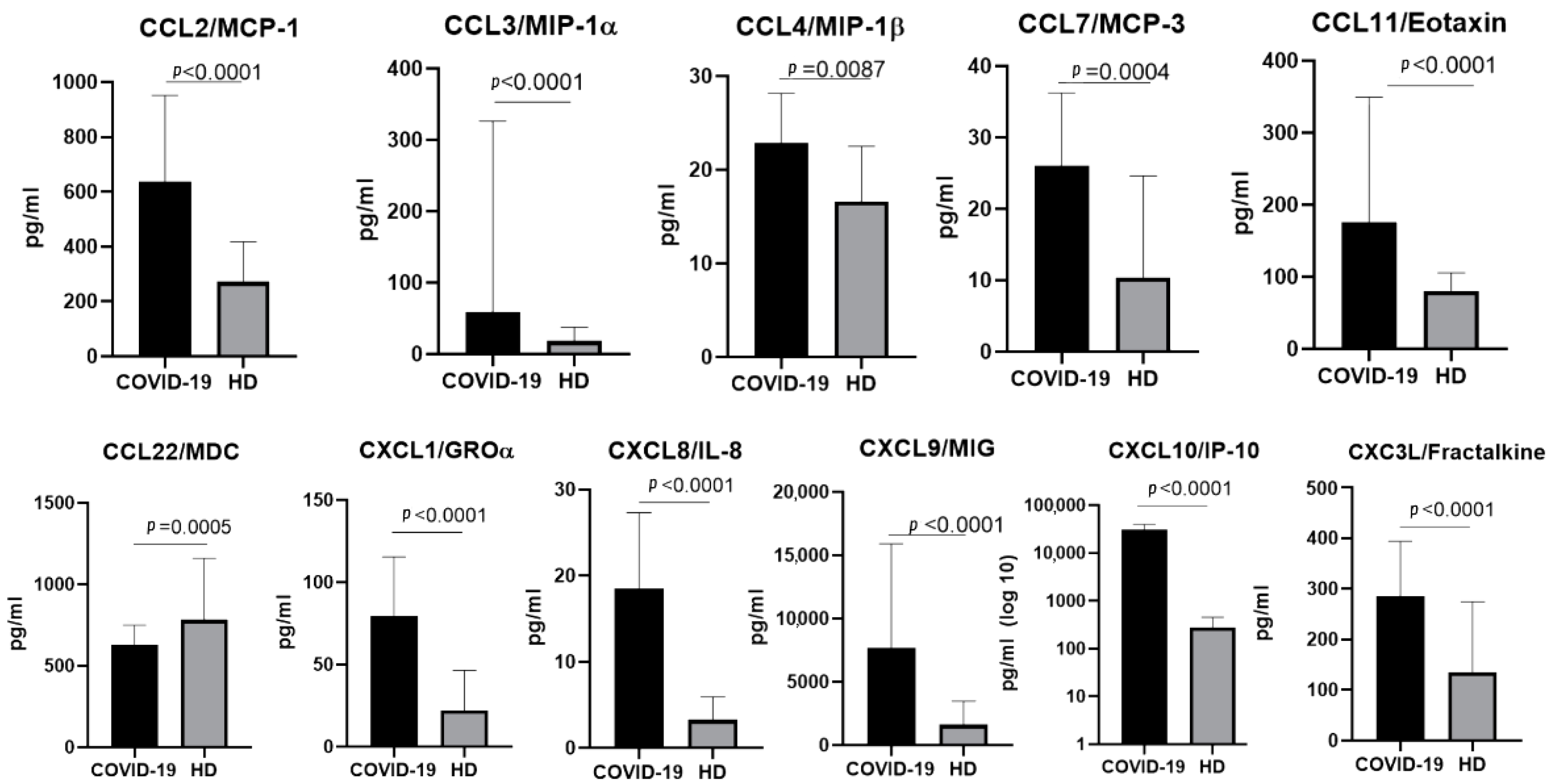
2.1. The Chemokine Profile in Patients Infected with the Ancestral Wuhan Variant
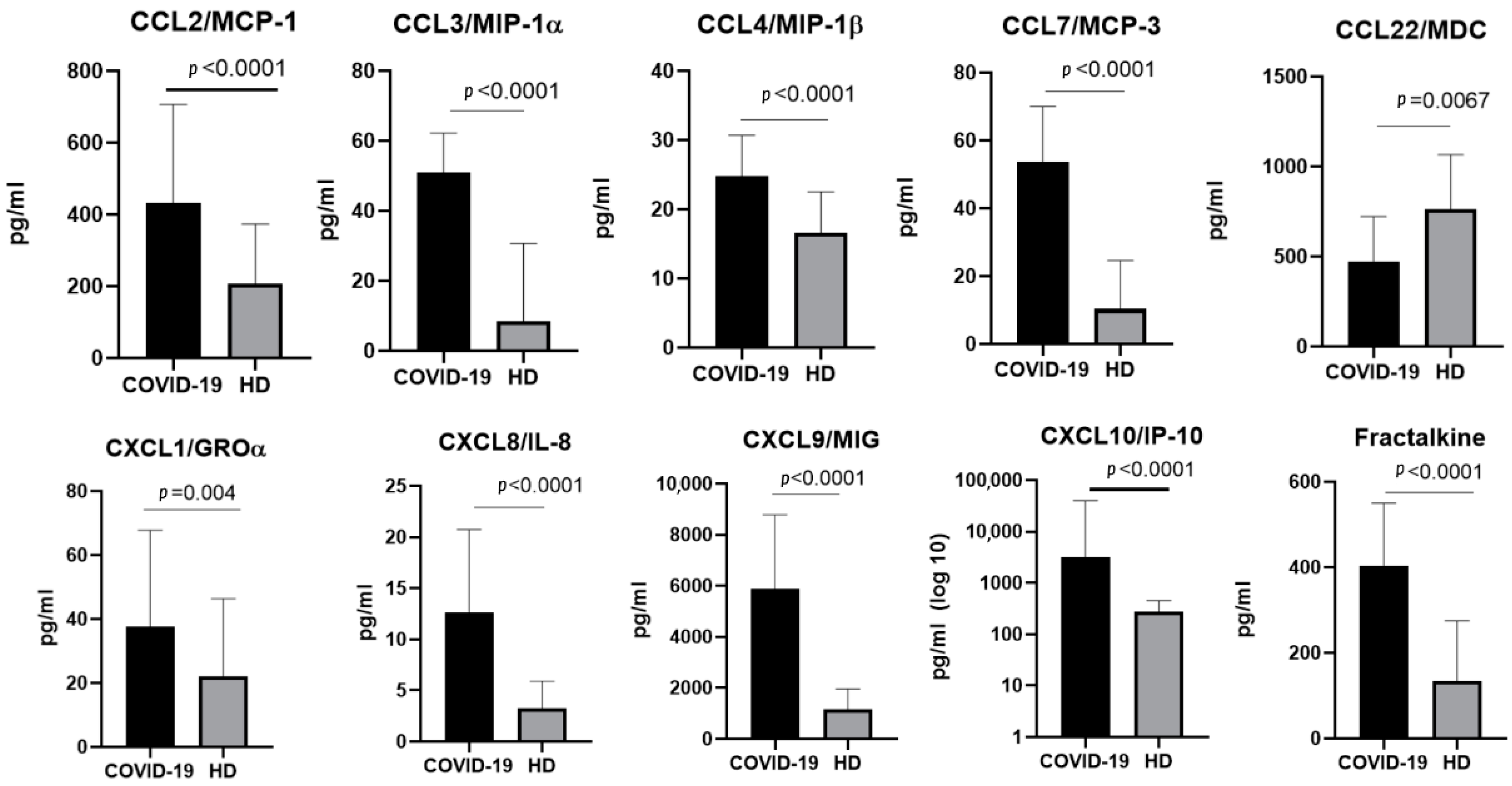
2.2. The Chemokine Profile in Patients Infected with the Alpha Variant
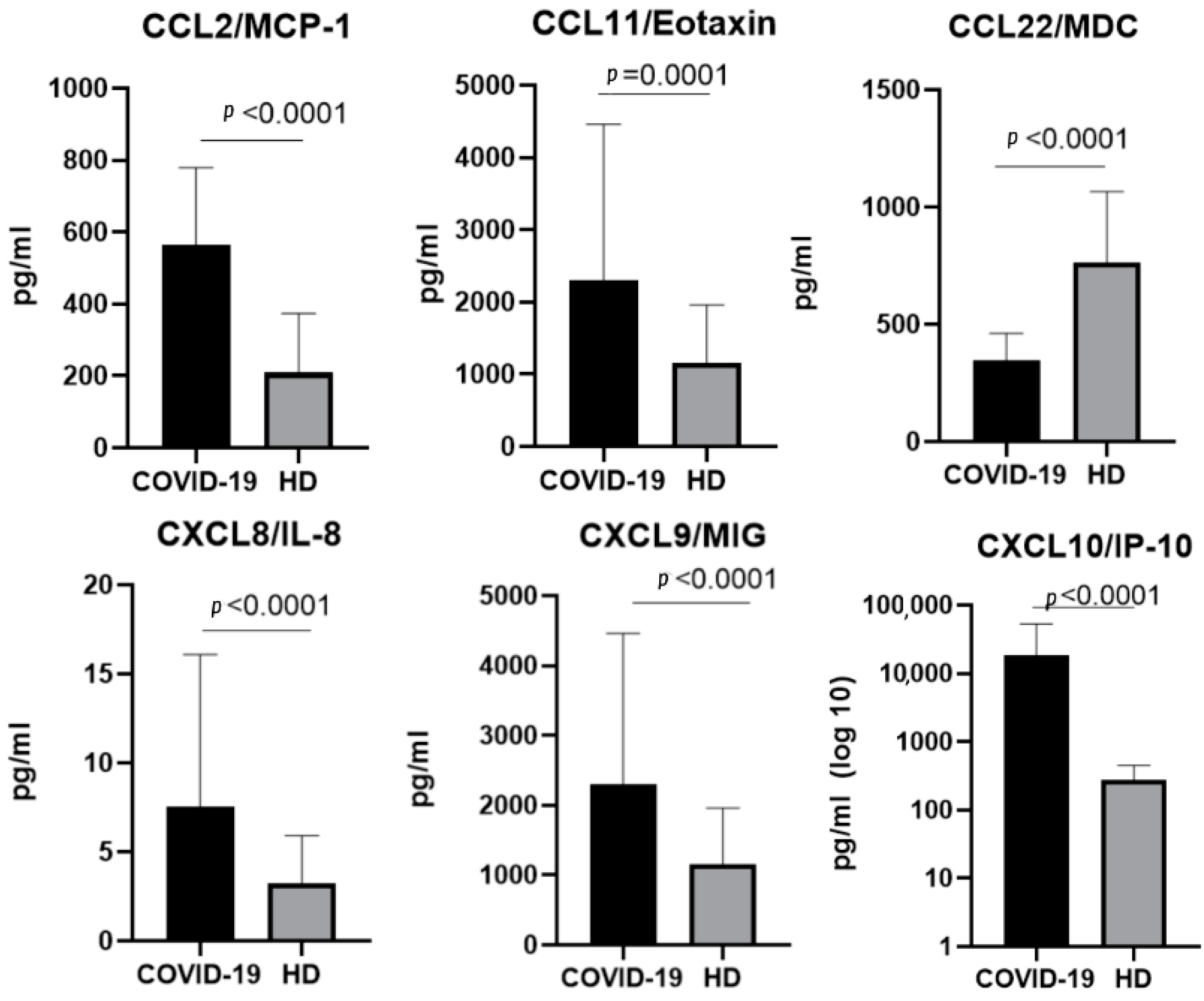
2.3. The Chemokine Profile in Patients Infected with the Delta Variant
2.4. The Chemokine Profile in Patients Infected with the Omicron Variant
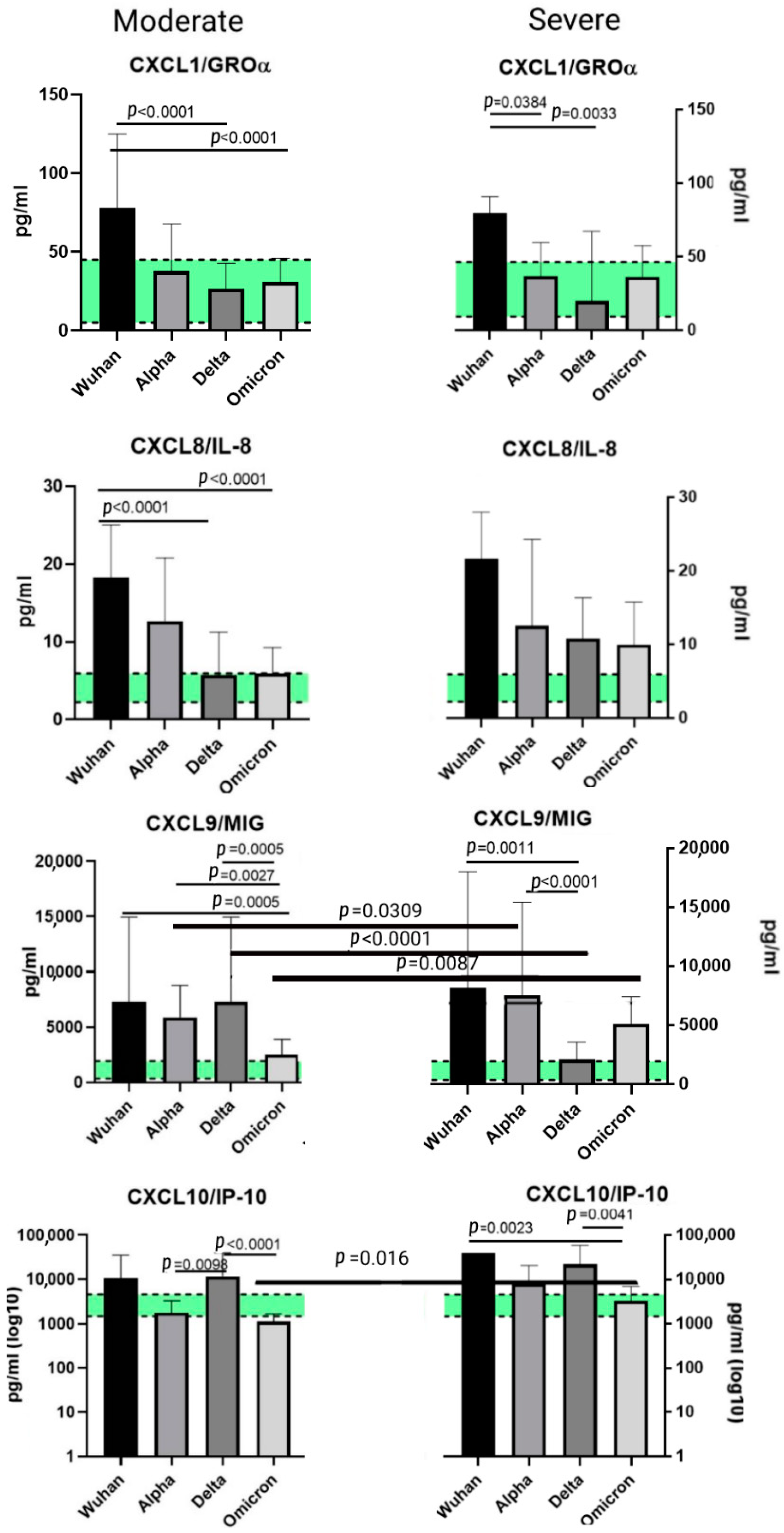
2.5. A Comparison of the Chemokines between the Variants
3. Discussion
3.1. Chemokines of the C-C Subfamily
3.1.1. CCL2/MCP-1
3.1.2. CCL3/MIP-1α and CCL4/MIP-1β
3.1.3. CCL7/MCP-3
3.1.4. CCL11/Eotaxin
3.1.5. CCL22/MDC
3.2. Chemokines of the C-X-C Subfamily
3.2.1. CXCL1/GROα
3.2.2. CXCL8/IL-8
3.2.3. CXCL9/MIG
3.2.4. CXCL10/IP-10
3.3. CX3C Family Chemokines: CX3CL1/Fractalkine
4. Materials and Methods
4.1. Samples
4.2. Patients
4.3. Genotyping of the SARS-CoV-2 Genetic Variants
4.4. Chemokine Detection
4.5. Statistical Analysis
5. Conclusions
- A statistically significant elevation in CCL2/MCP-1 concentrations in patients with COVID-19, independently from the variant. This tendency can be explained by the role MCP-1 plays in the attraction of monocytes to the sites of damaged vascular endothelium.
- A prominent increase in CXCL1/IP-10 levels in all the COVID-infected cohorts in comparison with healthy donors. IP-10 is often recognized as one of the markers of the cytokine storm; it is deeply involved in thrombosis and ARDS in patients with COVID-19.
- A statistically significant rise in CXCL8/IL-8 concentrations in patients with COVID-19, independently from the detected variant. This is a common finding in COVID-19 infection, and IL-8 is often recognized as a severity marker.
- A drop in the concentrations of CCL22/MDC in infected patients compared to healthy donors. In lung damage associated with other etiological factors (for instance, eosinophilic pneumonia or pulmonary hemorrhage), macrophage-derived chemokine was present in higher concentrations in plasma than it was in healthy donors. A decrease in the concentrations of this chemokine may be related to the life cycle of SARS-CoV-2. The severity of the disease is negatively correlated with the concentrations of MDC.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, X.-M.; Mannan, R.; Xiao, L.; Abdulfatah, E.; Qiao, Y.; Farver, C.; Myers, J.L.; Zelenka-Wang, S.; McMurry, L.; Su, F.; et al. Characterization of SARS-CoV-2 and host entry factors distribution in a COVID-19 autopsy series. Commun. Med. 2021, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Ozono, S.; Zhang, Y.; Ode, H.; Sano, K.; Tan, T.S.; Imai, K.; Miyoshi, K.; Kishigami, S.; Ueno, T.; Iwatani, Y.; et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021, 12, 848. [Google Scholar] [CrossRef] [PubMed]
- Samavati, L.; Uhal, B.D. ACE2, Much More Than Just a Receptor for SARS-COV-2. Front. Cell. Infect. Microbiol. 2020, 10, 317. [Google Scholar] [CrossRef] [PubMed]
- Attaway, A.H.; Scheraga, R.G.; Bhimraj, A.; Biehl, M.; Hatipoğlu, U. Severe COVID-19 pneumonia: Pathogenesis and clinical management. BMJ 2021, 372, n436. [Google Scholar] [CrossRef]
- D’Agnillo, F.; Walters, K.-A.; Xiao, Y.; Sheng, Z.-M.; Scherler, K.; Park, J.; Gygli, S.; Rosas, L.A.; Sadtler, K.; Kalish, H.; et al. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci. Transl. Med. 2021, 13, eabj7790. [Google Scholar] [CrossRef] [PubMed]
- Rendeiro, A.F.; Ravichandran, H.; Bram, Y.; Chandar, V.; Kim, J.; Meydan, C.; Park, J.; Foox, J.; Hether, T.; Warren, S.; et al. The spatial landscape of lung pathology during COVID-19 progression. Nature 2021, 593, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Szabo, P.A.; Dogra, P.; Gray, J.I.; Wells, S.B.; Connors, T.J.; Weisberg, S.P.; Krupska, I.; Matsumoto, R.; Poon, M.M.; Idzikowski, E.; et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity 2021, 54, 797–814.e6. [Google Scholar] [CrossRef]
- Suran, M. Autopsies Reveal Lung Damage Patterns From COVID-19. JAMA 2021, 326, 2463. [Google Scholar] [CrossRef] [PubMed]
- Upadhya, S.; Rehman, J.; Malik, A.B.; Chen, S. Mechanisms of lung injury induced by SARS-CoV-2 infection. Physiology 2022, 37, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Kommoss, F.K.; Schwab, C.; Tavernar, L.; Schreck, J.; Wagner, W.L.; Merle, U.; Jonigk, D.; Schirmacher, P.; Longerich, T. The Pathology of Severe COVID-19-Related Lung Damage. Dtsch. Arztebl. Int. 2020, 117, 500–506. [Google Scholar] [CrossRef]
- Stenmark, K.R.; Frid, M.G.; Gerasimovskaya, E.; Zhang, H.; McCarthy, M.K.; Thurman, J.M.; Morrison, T.E. Mechanisms of SARS-CoV-2-induced lung vascular disease: Potential role of complement. Pulm. Circ. 2021, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Matthay, M.A.; Leligdowicz, A.; Liu, K.D. Biological Mechanisms of COVID-19 Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2020, 202, 1489–1491. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 12 July 2022).
- Danchin, A.; Timmis, K. SARS-CoV-2 variants: Relevance for symptom granularity, epidemiology, immunity (herd, vaccines), virus origin and containment? Environ. Microbiol. 2020, 22, 2001–2006. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Valdes, A.M.; Polidori, L.; Antonelli, M.; Penamakuri, S.; Nogal, A.; Louca, P.; May, A.; Figueiredo, J.C.; Hu, C.; et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: A prospective observational study from the ZOE COVID Study. Lancet 2022, 399, 1618–1624. [Google Scholar] [CrossRef]
- Abdullah, F.; Myers, J.; Basu, D.; Tintinger, G.; Ueckermann, V.; Mathebula, M.; Ramlall, R.; Spoor, S.; de Villiers, T.; Van der Walt, Z.; et al. Decreased severity of disease during the first global omicron variant COVID-19 outbreak in a large hospital in Tshwane, South Africa. Int. J. Infect. Dis. 2021, 116, 38–42. [Google Scholar] [CrossRef]
- Elizondo, V.; Harkins, G.W.; Mabvakure, B.; Smidt, S.; Zappile, P.; Marier, C.; Maurano, M.T.; Perez, V.; Mazza, N.; Beloso, C.; et al. SARS-CoV-2 genomic characterization and clinical manifestation of the COVID-19 outbreak in Uruguay. Emerg. Microbes Infect. 2021, 10, 51–65. [Google Scholar] [CrossRef]
- Nagy, Á.; Pongor, S.; Győrffy, B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int. J. Antimicrob. Agents 2020, 57, 106272. [Google Scholar] [CrossRef] [PubMed]
- Al Khatib, H.A.; Benslimane, F.M.; Elbashir, I.E.; Coyle, P.V.; Al Maslamani, M.A.; Al-Khal, A.; Al Thani, A.A.; Yassine, H.M. Within-Host Diversity of SARS-CoV-2 in COVID-19 Patients With Variable Disease Severities. Front. Cell. Infect. Microbiol. 2020, 10, 575613. [Google Scholar] [CrossRef]
- Hoang, V.-T.; Colson, P.; Levasseur, A.; Delerce, J.; Lagier, J.-C.; Parola, P.; Million, M.; Fournier, P.-E.; Raoult, D.; Gautret, P. Clinical outcomes in patients infected with different SARS-CoV-2 variants at one hospital during three phases of the COVID-19 epidemic in Marseille, France. Infect. Genet. Evol. 2021, 95, 105092. [Google Scholar] [CrossRef]
- Cao, C.; He, L.; Tian, Y.; Qin, Y.; Sun, H.; Ding, W.; Gui, L.; Wu, P. Molecular epidemiology analysis of early variants of SARS-CoV-2 reveals the potential impact of mutations P504L and Y541C (NSP13) in the clinical COVID-19 outcomes. Infect. Genet. Evol. 2021, 92, 104831. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Eaaswarkhanth, M.; Al Madhoun, A.; Al-Mulla, F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int. J. Infect. Dis. 2020, 96, 459–460. [Google Scholar] [CrossRef] [PubMed]
- Khalil, B.A.; Elemam, N.M.; Maghazachi, A.A. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J. 2021, 19, 976–988. [Google Scholar] [CrossRef] [PubMed]
- Jansen, L.; Tegomoh, B.; Lange, K.; Showalter, K.; Figliomeni, J.; Abdalhamid, B.; Iwen, P.C.; Fauver, J.; Buss, B.; Donahue, M. Investigation of a SARS-CoV-2 B.1.1.529 (Omicron) Variant Cluster—Nebraska, November–December 2021. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 1782–1784. [Google Scholar] [CrossRef]
- Bhavsar, I.; Miller, C.S.; Al-Sabbagh, M. Macrophage Inflammatory Protein-1 Alpha (MIP-1 alpha)/CCL3: As a Biomarker. Gen. Methods Biomark. Res. Appl. 2015, 2015, 223–249. [Google Scholar] [CrossRef]
- Menten, P.; Wuyts, A.; Van Damme, J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002, 13, 455–481. [Google Scholar] [CrossRef]
- Chen, L.; Wang, G.; Tan, J.; Cao, Y.; Long, X.; Luo, H.; Tang, Q.; Jiang, T.; Wang, W.; Zhou, J. Scoring cytokine storm by the levels of MCP-3 and IL-8 accurately distinguished COVID-19 patients with high mortality. Signal Transduct. Target. Ther. 2020, 5, 292. [Google Scholar] [CrossRef]
- Chi, Y.; Ge, Y.; Wu, B.; Zhang, W.; Wu, T.; Wen, T.; Liu, J.; Guo, X.; Huang, C.; Jiao, Y.; et al. Serum Cytokine and Chemokine Profile in Relation to the Severity of Coronavirus Disease 2019 in China. J. Infect. Dis. 2020, 222, 746–754. [Google Scholar] [CrossRef]
- Mantovani, A.; A Gray, P.; Van Damme, J.; Sozzani, S. Macrophage-derived chemokine (MDC). J. Leukoc. Biol. 2000, 68, 400–404. [Google Scholar]
- Manabe, K.; Nishioka, Y.; Kishi, J.; Inayama, M.; Aono, Y.; Nakamura, Y.; Ogushi, F.; Bando, H.; Tani, K.; Sone, S. Elevation of macrophage-derived chemokine in eosinophilic pneumonia: A role of alveolar macrophages. J. Med Investig. 2005, 52, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.; Sutton, J.M.; Belizaire, R.M.; Friend, L.A.; Schuster, R.M.; Johannigman, T.A.; Miller, S.G.; Lentsch, A.B.; Pritts, T.A. Macrophage-Derived Chemokine (CCL22) Is a Novel Mediator of Lung Inflammation Following Hemorrhage and Resuscitation. Shock 2014, 42, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Tufa, A.; Gebremariam, T.H.; Manyazewal, T.; Getinet, T.; Webb, D.-L.; Hellström, P.M.; Genet, S. Cytokine and chemokine profile in patients hospitalized with COVID-19: A comparative study. bioRxiv 2022. [Google Scholar] [CrossRef]
- Becker, S.; Quay, J.; Koren, H.S.; Haskill, J.S. Constitutive and stimulated MCP-1, GRO alpha, beta, and gamma expression in human airway epithelium and bronchoalveolar macrophages. Am. J. Physiol. Cell. Mol. Physiol. 1994, 266 Pt 1, L278–L286. [Google Scholar] [CrossRef]
- Ma, K.; Yang, L.; Shen, R.; Kong, B.; Chen, W.; Liang, J.; Tang, G.; Zhang, B. Th17 cells regulate the production of CXCL1 in breast cancer. Int. Immunopharmacol. 2018, 56, 320–329. [Google Scholar] [CrossRef]
- Gudowska-Sawczuk, M.; Mroczko, B. What Is Currently Known about the Role of CXCL10 in SARS-CoV-2 Infection? Int. J. Mol. Sci. 2022, 23, 3673. [Google Scholar] [CrossRef]
- Arsentieva, N.A.; Liubimova, N.E.; Batsunov, O.K.; Korobova, Z.R.; Stanevich, O.V.; Lebedeva, A.A.; Vorobyov, E.A.; Vorobyova, S.V.; Kulikov, A.N.; Lioznov, D.A.; et al. Plasma cytokines in patients with COVID-19 during acute phase of the disease and following complete recovery. Med. Immunol. 2021, 23, 311–326. (In Russian) [Google Scholar] [CrossRef]
- McElvaney, O.J.; McEvoy, N.L.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Ni Choileain, O.; Clarke, J.; O’Connor, E.; Hogan, G.; et al. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care Med. 2020, 202, 812–821. [Google Scholar] [CrossRef]
- Li, L.; Li, J.; Gao, M.; Fan, H.; Wang, Y.; Xu, X.; Chen, C.; Liu, J.; Kim, J.; Aliyari, R.; et al. Interleukin-8 as a Biomarker for Disease Prognosis of Coronavirus Disease-2019 Patients. Front. Immunol. 2021, 11, 602395. [Google Scholar] [CrossRef]
- Callahan, V.; Hawks, S.; Crawford, M.; Lehman, C.; Morrison, H.; Ivester, H.; Akhrymuk, I.; Boghdeh, N.; Flor, R.; Finkielstein, C.; et al. The Pro-Inflammatory Chemokines CXCL9, CXCL10 and CXCL11 Are Upregulated Following SARS-CoV-2 Infection in an AKT-Dependent Manner. Viruses 2021, 13, 1062. [Google Scholar] [CrossRef]
- Tincati, C.; Cannizzo, E.S.; Giacomelli, M.; Badolato, R.; Monforte, A.D.; Marchetti, G. Heightened Circulating Interferon-Inducible Chemokines, and Activated Pro-Cytolytic Th1-Cell Phenotype Features Covid-19 Aggravation in the Second Week of Illness. Front. Immunol. 2020, 11, 580987. [Google Scholar] [CrossRef] [PubMed]
- Quirch, M.; Lee, J.; Rehman, S. Hazards of the Cytokine Storm and Cytokine-Targeted Therapy in Patients With COVID-19: Review. J. Med. Internet Res. 2020, 22, e20193. [Google Scholar] [CrossRef] [PubMed]
- Pum, A.; Ennemoser, M.; Adage, T.; Kungl, A.J. Cytokines and Chemokines in SARS-CoV-2 Infections—Therapeutic Strategies Targeting Cytokine Storm. Biomolecules 2021, 11, 91. [Google Scholar] [CrossRef]
- Bazan, J.F.; Bacon, K.B.; Hardiman, G.; Wang, W.; Soo, K.; Rossi, D.; Greaves, D.R.; Zlotnik, A.; Schall, T.J. A new class of membrane-bound chemokine with a CX3C motif. Nature 1997, 385, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Fuentes, S.; Valdés, V.J.; Espinosa, B.; Gorocica-Rosete, P.; Salgado-Aguayo, A. Could SARS-CoV-2 blocking of ACE2 in endothelial cells result in upregulation of CX3CL1, promoting thrombosis in COVID-19 patients? Med. Hypotheses 2021, 151, 110570. [Google Scholar] [CrossRef]



| Variant | Wuhan (n = 56) | Alpha (n = 95) | Delta (n = 78) | Omicron (n = 57) | Healthy Donors (n = 51) |
|---|---|---|---|---|---|
| Age, years (Me; Q25, Q75) | 66 (32; 75) | 72 (28; 84) | 71 (45; 93) | 69 (32; 91) | 49 (24; 69) |
| Gender distribution | 50% female (n = 28); 50% male (n = 28) | 52.7% female (n = 50); 47.3% male (n = 45) | 64.1% female (n = 50); 35.9% male (n = 28) | 61.4% female (n = 35); 38.6% male (n = 22) | 58.8% female (n = 30), 41.2% male (n = 21) |
| Variant | % of Patients, Diagnosed with ‘Mild’ Infection | % of Patients, Diagnosed with ‘Moderate’ Infection | % of Patients, Diagnosed with ‘Severe’ Infection |
|---|---|---|---|
| Ancestral Wuhan | 43.5% (n = 30) | 46.4% (n = 32) | 10.1% (n = 7) |
| Alpha | 0 | 26.6% (n = 25) | 73.6% (n = 70) |
| Delta | 0 | 34.6% (n = 27) | 65.4% (n = 51) |
| Omicron | 0 | 75.4% (n = 43) | 24.6% (n = 14) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korobova, Z.R.; Arsentieva, N.A.; Liubimova, N.E.; Dedkov, V.G.; Gladkikh, A.S.; Sharova, A.A.; Chernykh, E.I.; Kashchenko, V.A.; Ratnikov, V.A.; Gorelov, V.P.; et al. A Comparative Study of the Plasma Chemokine Profile in COVID-19 Patients Infected with Different SARS-CoV-2 Variants. Int. J. Mol. Sci. 2022, 23, 9058. https://doi.org/10.3390/ijms23169058
Korobova ZR, Arsentieva NA, Liubimova NE, Dedkov VG, Gladkikh AS, Sharova AA, Chernykh EI, Kashchenko VA, Ratnikov VA, Gorelov VP, et al. A Comparative Study of the Plasma Chemokine Profile in COVID-19 Patients Infected with Different SARS-CoV-2 Variants. International Journal of Molecular Sciences. 2022; 23(16):9058. https://doi.org/10.3390/ijms23169058
Chicago/Turabian StyleKorobova, Zoia R., Natalia A. Arsentieva, Natalia E. Liubimova, Vladimir G. Dedkov, Anna S. Gladkikh, Alena A. Sharova, Ekaterina I. Chernykh, Victor A. Kashchenko, Vyacheslav A. Ratnikov, Victor P. Gorelov, and et al. 2022. "A Comparative Study of the Plasma Chemokine Profile in COVID-19 Patients Infected with Different SARS-CoV-2 Variants" International Journal of Molecular Sciences 23, no. 16: 9058. https://doi.org/10.3390/ijms23169058
APA StyleKorobova, Z. R., Arsentieva, N. A., Liubimova, N. E., Dedkov, V. G., Gladkikh, A. S., Sharova, A. A., Chernykh, E. I., Kashchenko, V. A., Ratnikov, V. A., Gorelov, V. P., Stanevich, O. V., Kulikov, A. N., Pevtsov, D. E., & Totolian, A. A. (2022). A Comparative Study of the Plasma Chemokine Profile in COVID-19 Patients Infected with Different SARS-CoV-2 Variants. International Journal of Molecular Sciences, 23(16), 9058. https://doi.org/10.3390/ijms23169058







