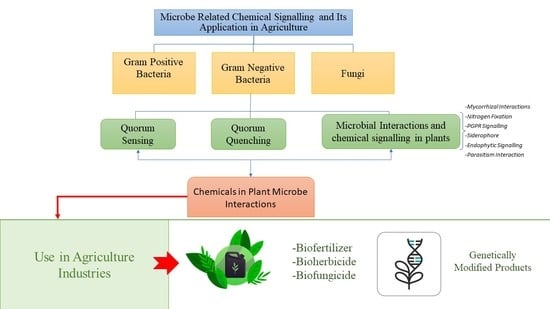
Microbe Related Chemical Signalling and Its Application in Agriculture
Abstract
:1. Introduction
2. Communication Mode between Microorganisms
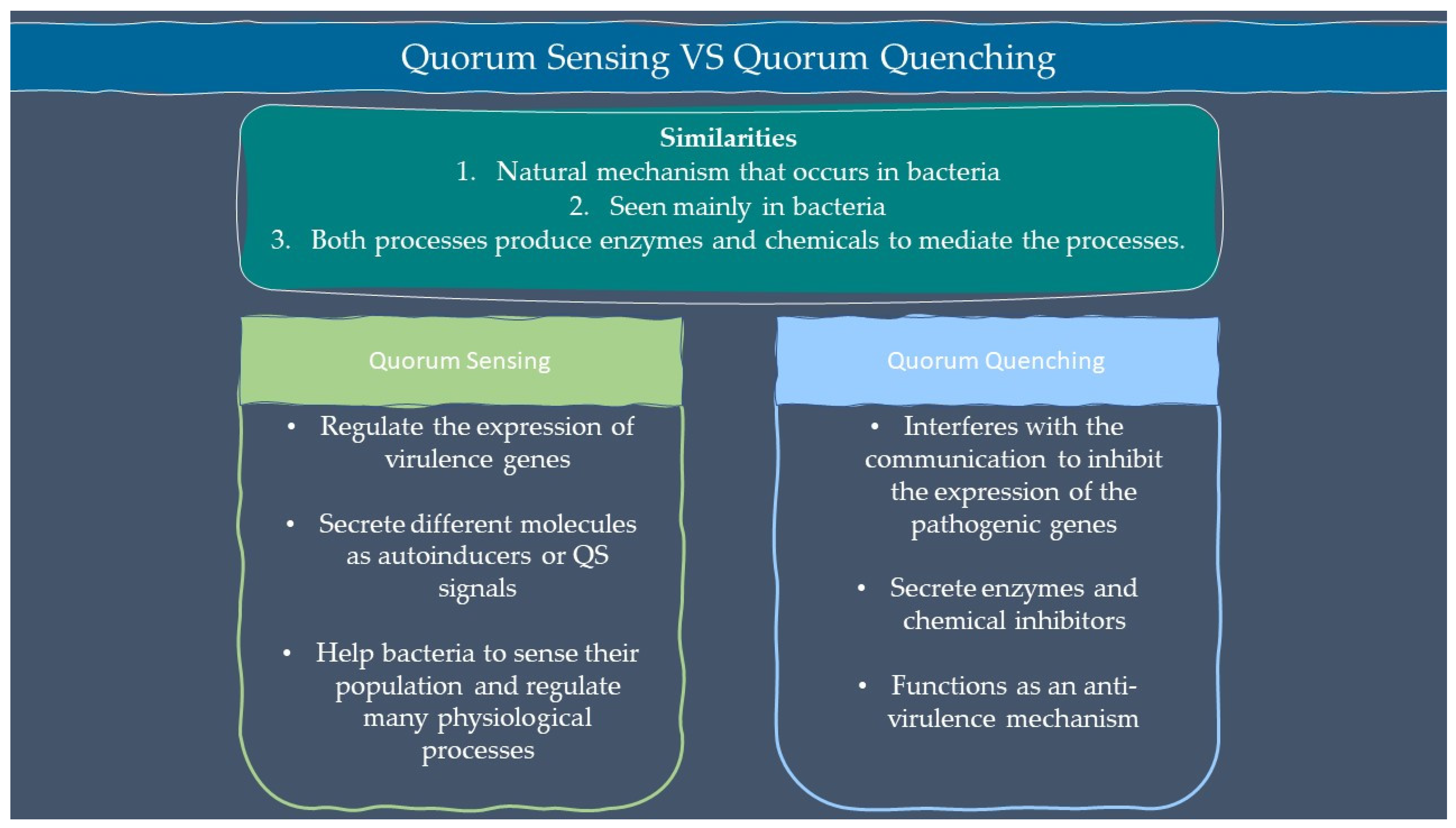
2.1. Quorum Sensing
2.2. Quorum Quenching
2.3. Chemical Signalling in Fungi
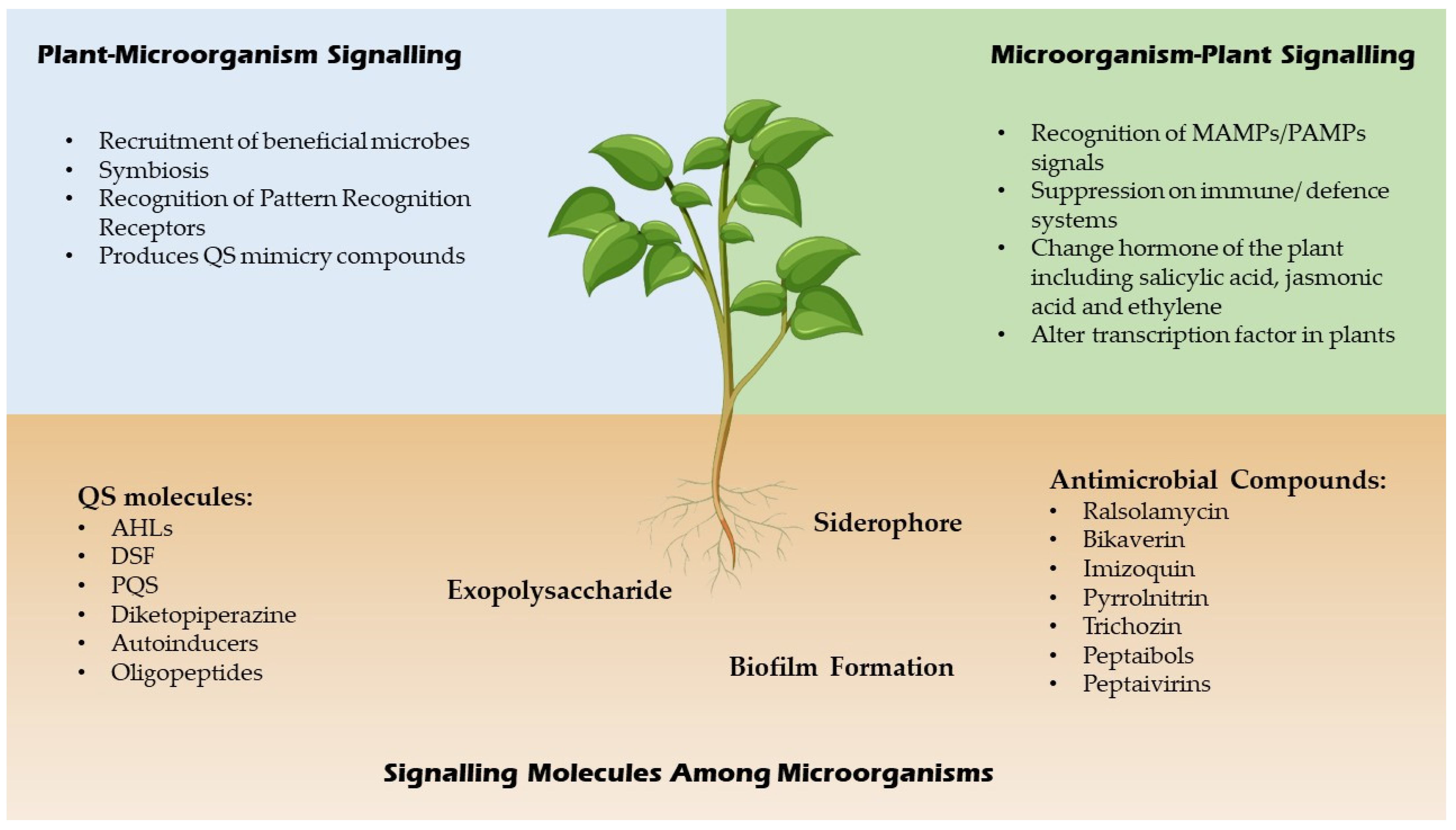
3. Microbial Interactions and Chemical Signalling in Plant
3.1. Mycorrhizal Interactions
3.2. Nitrogen Fixation
3.3. PGPR Signalling
3.4. Siderophore
3.5. Endophytic Signalling
3.6. Parasitism Interaction
3.6.1. Diffusible Signal Factor (DSF)
3.6.2. Exopolysaccharide (EPS)
3.6.3. Antimicrobial Compounds
4. Chemical Signals in Plant-Microbe/Pathogen Interactions
5. The Success of Microbial Chemicals in Improving Crop Yield and Growth
6. Genetically Modified Microbial Products in Agriculture
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hartmann, A.; Rothballer, M. Rhizotrophs: Plant Growth Promotion to Bioremediation; Springer: Berlin/Heidelberg, Germany, 2017; pp. 205–217. [Google Scholar]
- Fatima, K. Insights into Chemical Interaction between Plants and Microbes and its Potential Use in Soil Remediation. Biosci. Rev. 2019, 1, 39–45. [Google Scholar] [CrossRef]
- Guo, M.; Gamby, S.; Zheng, Y.; Sintim, H.O. Small Molecule Inhibitors of AI-2 Signaling in Bacteria: State-of-the-Art and Future Perspectives for Anti-Quorum Sensing Agents. Int. J. Mol. Sci. 2013, 14, 17694. [Google Scholar] [CrossRef] [PubMed]
- Babalola, O.O. Beneficial bacteria of agricultural importance. Biotechnol. Lett. 2010, 32, 1559–1570. [Google Scholar] [CrossRef] [PubMed]
- Tao, C.; Li, R.; Xiong, W.; Shen, Z.; Liu, S.; Wang, B.; Ruan, Y.; Geisen, S.; Shen, Q.; Kowalchuk, G.A. Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 2020, 8, 137. [Google Scholar] [CrossRef]
- Harding, D.P.; Raizada, M.N. Controlling weeds with fungi, bacteria and viruses: A review. Front. Plant Sci. 2015, 6, 659. [Google Scholar] [CrossRef] [PubMed]
- Wubs, E.R.J.; van der Putten, W.H.; Bosch, M.; Bezemer, T.M. Soil inoculation steers restoration of terrestrial ecosystems. Nat. Plants 2016, 2, 16107. [Google Scholar] [CrossRef]
- Boyette, C.D.; Bowling, A.J.; Vaughn, K.C.; Hoagland, R.E.; Stetina, K.C. Induction of infection in Sesbania exaltata by Colletotrichum gloeosporioides f. sp. aeschynomene formulated in an invert emulsion. World J. Microbiol. Biotechnol. 2010, 26, 951–956. [Google Scholar] [CrossRef]
- Dessaux, Y.; Grandclément, C.; Faure, D. Engineering the Rhizosphere. Trends Plant Sci. 2016, 21, 266–278. [Google Scholar] [CrossRef]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, T. Using Morphogenic Genes to Improve Recovery and Regeneration of Transgenic Plants. Plants 2019, 8, 38. [Google Scholar] [CrossRef]
- Calatrava-Morales, N.; McIntosh, M.; Soto, M.J. Regulation mediated by N-acyl homoserine lactone quorum sensing signals in the rhizobium-legume symbiosis. Genes 2018, 9, 263. [Google Scholar] [CrossRef]
- Rosier, A.; Bishnoi, U.; Lakshmanan, V.; Sherrier, D.J.; Bais, H.P. A perspective on inter-kingdom signaling in plant–beneficial microbe interactions. Plant Mol. Biol. 2016, 90, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Yeon, K.M. Quorum Sensing as Language of Chemical Signals, 1st ed.; Elsevier B.V.: Amsterdam, The Netherlands, 2018; Volume 81. [Google Scholar]
- Schenk, S.T.; Schikora, A. AHL-Priming functions via oxylipin and salicylic acid. Front. Plant Sci. 2015, 5, 784. [Google Scholar] [CrossRef]
- Saeki, E.K.; Kobayashi, R.K.T.; Nakazato, G. Quorum sensing system: Target to control the spread of bacterial infections. Microb. Pathog. 2020, 142, 104068. [Google Scholar] [CrossRef]
- Achari, G.A.; Ramesh, R. Recent Advances in Quorum Quenching of Plant Pathogenic Bacteria; Elsevier Inc.: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Ryan, R.P.; An, S.; Allan, J.H.; McCarthy, Y.; Dow, J.M. The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators. PLoS Pathog. 2015, 11, e1004986. [Google Scholar] [CrossRef] [PubMed]
- Ya’Ar Bar, S.; Dor, S.; Erov, M.; Afriat-Jurnou, L. Identification and Characterization of a New Quorum-Quenching N-acyl Homoserine Lactonase in the Plant Pathogen Erwinia amylovora. J. Agric. Food Chem. 2021, 69, 5652–5662. [Google Scholar] [CrossRef] [PubMed]
- Halloran, K.T.; Wang, Y.; Arora, K.; Chakravarthy, S.; Irving, T.C.; Bilsel, O.; Brooks, C.L.; Matthews, C.R. Frustration and folding of a TIM barrel protein. Proc. Natl. Acad. Sci. USA 2019, 116, 16378–16383. [Google Scholar] [CrossRef]
- Padder, S.A.; Prasad, R.; Shah, A.H. Quorum sensing: A less known mode of communication among fungi. Microbiol. Res. 2018, 210, 51–58. [Google Scholar] [CrossRef]
- Yin, X.-T.; Xu, L.-N.; Xu, L.; Fan, S.-S.; Liu, Z.-Y.; Zhang, X.-Y. Evaluation of the efficacy of endophytic Bacillus amyloliquefaciens against Botryosphaeria dothidea and other phytopathogenic microorganisms. Afr. J. Microbiol. Res. 2011, 5, 340–345. [Google Scholar]
- Landini, P.; Antoniani, D.; Burgess, J.G.; Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 2010, 86, 813–823. [Google Scholar] [CrossRef]
- Barriuso, J.; Hogan, D.A.; Keshavarz, T.; Martínez, M.J. Role of quorum sensing and chemical communication in fungal biotechnology and pathogenesis. FEMS Microbiol. Rev. 2018, 42, 627–638. [Google Scholar] [CrossRef]
- Cottier, F.; Mühlschlegel, F.A. Communication in Fungi. Int. J. Microbiol. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gorai, P.S.; Barman, S.; Gond, S.K.; Mandal, N.C. Chapter 28—Trichoderma; Amaresan, N., Senthil Kumar, M., Annapurna, K., Kumar, K., Sankaranarayanan, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 571–591. [Google Scholar]
- Macías-Rodríguez, L.; Contreras-Cornejo, H.A.; Adame-Garnica, S.G.; del-Val, E.; Larsen, J. The interactions of Trichoderma at multiple trophic levels: Inter-kingdom communication. Microbiol. Res. 2020, 240, 126552. [Google Scholar] [CrossRef] [PubMed]
- Speckbacher, V.; Zeilinger, S. Secondary metabolites of mycoparasitic fungi. In Secondary Metabolites—Sources and Applications; InTech: London, UK, 2018; pp. 37–55. [Google Scholar]
- Lutz, M.P.; Feichtinger, G.; Défago, G.; Duffy, B. Mycotoxigenic Fusarium and Deoxynivalenol Production Repress Chitinase Gene Expression in the Biocontrol Agent Trichoderma atroviride P1. Appl. Environ. Microbiol. 2003, 69, 3077–3084. [Google Scholar] [CrossRef]
- Montibus, M.; Ducos, C.; Bonnin-Verdal, M.-N.; Bormann, J.; Ponts, N.; Richard-Forget, F.; Barreau, C. The bZIP Transcription Factor Fgap1 Mediates Oxidative Stress Response and Trichothecene Biosynthesis but Not Virulence in Fusarium graminearum. PLoS ONE 2013, 8, e83377. [Google Scholar] [CrossRef]
- Ponts, N.; Pinson-Gadais, L.; Verdal-Bonnin, M.-N.; Barreau, C.; Richard-Forget, F. Accumulation of deoxynivalenol and its 15-acetylated form is significantly modulated by oxidative stress in liquid cultures of Fusarium graminearum. FEMS Microbiol. Lett. 2006, 258, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.B. Higher plant–lower plant interactions: Phytoalexins and phytotoxins. In Introduction to Ecological Biochemistry; Elsevier: Amsterdam, The Netherlands, 1993; pp. 264–297. [Google Scholar]
- Van Rij, E.T.; Girard, G.; Lugtenberg, B.J.J.; Bloemberg, G.V. Influence of fusaric acid on phenazine-1-carboxamide synthesis and gene expression of Pseudomonas chlororaphis strain PCL1391. Microbiology 2005, 151, 2805–2814. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, N.; Keller, N.P. Mycotoxins in Conversation with Bacteria and Fungi. Front. Microbiol. 2019, 10, 403. [Google Scholar] [CrossRef]
- Tian, Y.; Tan, Y.; Liu, N.; Yan, Z.; Liao, Y.; Chen, J.; de Saeger, S.; Yang, H.; Zhang, Q.; Wu, A. Detoxification of Deoxynivalenol via Glycosylation Represents Novel Insights on Antagonistic Activities of Trichoderma when Confronted with Fusarium graminearum. Toxins 2016, 8, 335. [Google Scholar] [CrossRef]
- López-Díaz, C.; Rahjoo, V.; Sulyok, M.; Ghionna, V.; Martín-Vicente, A.; Capilla, J.; di Pietro, A.; López-Berges, M.S. Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 2018, 19, 440–453. [Google Scholar] [CrossRef]
- Ruiz, J.A.; Bernar, E.M.; Jung, K. Production of Siderophores Increases Resistance to Fusaric Acid in Pseudomonas protegens Pf-5. PLoS ONE 2015, 10, e0117040. [Google Scholar] [CrossRef]
- Wongsuk, T.; Pumeesat, P.; Luplertlop, N. Fungal quorum sensing molecules: Role in fungal morphogenesis and pathogenicity. J. Basic Microbiol. 2016, 56, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ma, A.; Zhao, G.; Yun, J.; Liu, X.; Zhang, H.; Zhuang, G. Effect of Farnesol on Penicillium Decumbens’s Morphology and Cellulase Production. Bioresources 2011, 6, 3252–3259. [Google Scholar]
- Lei, X.; Deng, B.; Ruan, C.; Deng, L.; Zeng, K. Phenylethanol as a quorum sensing molecule to promote biofilm formation of the antagonistic yeast Debaryomyces nepalensis for the control of black spot rot on jujube. Postharvest Biol. Technol. 2022, 185, 111788. [Google Scholar] [CrossRef]
- Jurick, W.M.; Peng, H.; Beard, H.S.; Garrett, W.M.; Lichtner, F.J.; Luciano-Rosario, D.; Macarisin, O.; Liu, Y.; Peter, K.A.; Gaskins, V.L.; et al. Blistering1 modulates penicillium expansum virulence via vesicle-mediated protein secretion. Mol. Cell. Proteomics 2020, 19, 344–361. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, M.J.; Bartholomew, H.P.; Fonseca, J.M.; Gaskins, V.L.; Prusky, D.; Jurick, W.M. Delivering the goods: Fungal secretion modulates virulence during host–pathogen interactions. Fungal Biol. Rev. 2021, 36, 76–86. [Google Scholar] [CrossRef]
- Brodhun, F.; Feussner, I. Oxylipins in fungi. FEBS J. 2011, 278, 1047–1063. [Google Scholar] [CrossRef]
- Yashiroda, Y.; Yoshida, M. Intraspecies cell–cell communication in yeast. FEMS Yeast Res. 2019, 19, 71. [Google Scholar] [CrossRef]
- Schmidt, R.; Etalo, D.W.; de Jager, V.; Gerards, S.; Zweers, H.; de Boer, W.; Garbeva, P. Microbial Small Talk: Volatiles in Fungal-Bacterial Interactions. Front. Microbiol. 2016, 6, 1495. [Google Scholar] [CrossRef]
- Egbe, N.E.; Dornelles, T.O.; Paget, C.M.; Castelli, L.M.; Ashe, M.P. Farnesol inhibits translation to limit growth and filamentation in C. albicans and S. cerevisiae. Microb. Cell 2017, 4, 294. [Google Scholar] [CrossRef]
- Liu, P.; Luo, L.; Guo, J.; Liu, H.; Wang, B.; Deng, B.; Long, C.A.; Cheng, Y. Farnesol induces apoptosis and oxidative stress in the fungal pathogen Penicillium expansum. Mycologia 2010, 102, 311–318. [Google Scholar] [CrossRef]
- Kobayashi, D.Y.; Crouch, J.A. Bacterial/Fungal interactions: From pathogenes to mutualistic endosymbionts. Annu. Rev. Phytopathol. 2009, 47, 63–82. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, U.; Ouziad, F.; Marner, F.J.; Bothe, H. The bacterium Paenibacillus validus stimulates growth of the arbuscular mycorrhizal fungus Glomus intraradices up to the formation of fertile spores. FEMS Microbiol. Lett. 2006, 254, 258–267. [Google Scholar] [CrossRef]
- Lackner, G.; Moebius, N.; Partida-Martinez, L.P.; Boland, S.; Hertweck, C. Evolution of an endofungal Lifestyle: Deductions from the Burkholderia rhizoxinica Genome. BMC Genomics 2011, 12, 210. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; Ho, L.; Rees, C.A.; Hill, J.E.; Nodwell, J.R.; Elliot, M.A. Streptomyces exploration is triggered by fungal interactions and volatile signals. Elife 2017, 6, e21738. [Google Scholar] [CrossRef] [PubMed]
- Scherlach, K.; Hertweck, C. Mediators of mutualistic microbe–microbe interactions. Nat. Prod. Rep. 2018, 35, 303–308. [Google Scholar] [CrossRef]
- Nadarajah, K.; Abdul Rahman, N.S.N. Plant–microbe interaction: Aboveground to belowground, from the good to the bad. Int. J. Mol. Sci. 2021, 22, 10388. [Google Scholar] [CrossRef]
- Nadarajah, K.; Kumar, I.S. Molecular microbial biodiversity assessment in the mycorrhizosphere. In Mycorrhizosphere and Pedogenesis; Springer: Singapore, 2019; pp. 401–420. [Google Scholar]
- Earl, A.M.; Losick, R.; Kolter, R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 2008, 16, 269–275. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Zhu, H. Genetic and Molecular Mechanisms Underlying Symbiotic Specificity in Legume-Rhizobium Interactions. Front. Plant Sci. 2018, 9, 313. [Google Scholar] [CrossRef]
- Rasmann, S.; Turlings, T.C.J. Root signals that mediate mutualistic interactions in the rhizosphere. Curr. Opin. Plant Biol. 2016, 32, 62–68. [Google Scholar] [CrossRef]
- Santi, C.; Bogusz, D.; Franche, C. Biological nitrogen fixation in non-legume plants. Ann. Bot. 2013, 111, 743–767. [Google Scholar] [CrossRef]
- Nadarajah, K.K. Rhizosphere interactions: Life below ground. In Plant-Microbe Interaction: An Approach to Sustainable Agriculture; Springer: Singapore, 2016; pp. 3–23. [Google Scholar]
- González, J.E.; Marketon, M.M. Quorum Sensing in Nitrogen-Fixing Rhizobia. Microbiol. Mol. Biol. Rev. 2003, 67, 574–592. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.K.; Schmidt, M.A.; Hynes, M.F. Molecular Biology in the Improvement of Biological Nitrogen Fixation by Rhizobia and Extending the Scope to Cereals. Microorganisms 2021, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Gühl, K.; Holmer, R.; Xiao, T.T.; Shen, D.; Wardhani, T.A.K.; Geurts, R.; van Zeijl, A.; Kohlen, W. The Effect of Exogenous Nitrate on LCO Signalling, Cytokinin Accumulation, and Nodule Initiation in Medicago truncatula. Genes 2021, 12, 988. [Google Scholar] [CrossRef]
- Helman, Y.; Chernin, L. Silencing the mob: Disrupting quorum sensing as a means to fight plant disease. Mol. Plant Pathol. 2015, 16, 316–329. [Google Scholar] [CrossRef]
- Bukhat, S.; Imran, A.; Javaid, S.; Shahid, M.; Majeed, A.; Naqqash, T. Communication of plants with microbial world: Exploring the regulatory networks for PGPR mediated defense signaling. Microbiol. Res. 2020, 238, 126486. [Google Scholar] [CrossRef]
- Tsuge, K.; Inoue, S.; Ano, T.; Itaya, M.; Shoda, M. Horizontal Transfer of Iturin a Operon, itu, to Bacillus subtilis 168 and Conversion into an Iturin A Producer. Antimicrob. Agents Chemother. 2005, 49, 4641–4648. [Google Scholar] [CrossRef]
- Hashem, A.; Tabassum, B.; Fathi Abd_Allah, E. Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 2019, 26, 1291–1297. [Google Scholar] [CrossRef]
- Koumoutsi, A.; Chen, X.-H.; Vater, J.; Borriss, R. DegU and YczE Positively Regulate the Synthesis of Bacillomycin D by Bacillus amyloliquefaciens Strain FZB42. Appl. Environ. Microbiol. 2007, 73, 6953–6964. [Google Scholar] [CrossRef]
- Gong, A.-D.; Li, H.-P.; Yuan, Q.-S.; Song, X.-S.; Yao, W.; He, W.-J.; Zhang, J.-B.; Liao, Y.-C. Antagonistic Mechanism of Iturin A and Plipastatin A from Bacillus amyloliquefaciens S76-3 from Wheat Spikes against Fusarium graminearum. PLoS ONE 2015, 10, e0116871. [Google Scholar] [CrossRef]
- Romero, D.; Traxler, M.F.; López, D.; Kolter, R. Antibiotics as Signal Molecules. Chem. Rev. 2011, 111, 5492. [Google Scholar] [CrossRef] [PubMed]
- López, D.; Vlamakis, H.; Losick, R.; Kolter, R. Paracrine signaling in a bacterium. Genes Dev. 2009, 23, 1631–1638. [Google Scholar] [CrossRef] [PubMed]
- López, D.; Fischbach, M.A.; Chu, F.; Losick, R.; Kolter, R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2009, 106, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Rahman, F.B.; Sarkar, B.; Moni, R.; Rahman, M.S. Molecular Genetics of Surfactin and Its Effects on Different Sub-populations of Bacillus subtilis. Biotechnol. Rep. 2021, 32, e00686. [Google Scholar] [CrossRef]
- Zhi, Y.; Wu, Q.; Xu, Y. Genome and transcriptome analysis of surfactin biosynthesis in Bacillus amyloliquefaciens MT45. Sci. Rep. 2017, 7, 40976. [Google Scholar] [CrossRef]
- Korenblum, E.; de Araujo, L.V.; Guimarães, C.R.; de Souza, L.M.; Sassaki, G.; Abreu, F.; Nitschke, M.; Lins, U.; Freire, D.M.G.; Barreto-Bergter, E.; et al. Purification and characterization of a surfactin-like molecule produced by Bacillus sp. H2O-1 and its antagonistic effect against sulfate reducing bacteria. BMC Microbiol. 2012, 12, 252. [Google Scholar] [CrossRef]
- Farzand, A.; Moosa, A.; Zubair, M.; Khan, A.R.; Massawe, V.C.; Tahir, H.A.S.; Sheikh, T.M.M.; Ayaz, M.; Gao, X. Suppression of Sclerotinia sclerotiorum by the Induction of Systemic Resistance and Regulation of Antioxidant Pathways in Tomato Using Fengycin Produced by Bacillus amyloliquefaciens FZB42. Biomolecules 2019, 9, 613. [Google Scholar] [CrossRef]
- Istivan, T.S.; Coloe, P.J. Phospholipase A in Gram-negative bacteria and its role in pathogenesis. Microbiology 2006, 152, 1263–1274. [Google Scholar] [CrossRef]
- Kulimushi, P.Z.; Arias, A.A.; Franzil, L.; Steels, S.; Ongena, M. Stimulation of fengycin-type antifungal lipopeptides in Bacillus amyloliquefaciens in the presence of the maize fungal pathogen Rhizomucor variabilis. Front. Microbiol. 2017, 8, 850. [Google Scholar] [CrossRef]
- Sharma, G.; Dang, S.; Gupta, S.; Gabrani, R. Antibacterial Activity, Cytotoxicity, and the Mechanism of Action of Bacteriocin from Bacillus subtilis GAS101. Med. Princ. Pract. 2018, 27, 186–192. [Google Scholar] [CrossRef]
- Fernandez, M.; Godino, A.; Príncipe, A.; López Ramírez, V.; Quesada, J.M.; Rigo, V.; Espinosa-Urgel, M.; Morales, G.M.; Fischer, S. Characterization of the bacteriocins and the PrtR regulator in a plant-associated Pseudomonas strain. J. Biotechnol. 2020, 307, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Khalid, S.; Keller, N.P. Chemical signals driving bacterial–fungal interactions. Environ. Microbiol. 2021, 23, 1334–1347. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, A.; de Stradis, A.; Lo Cantore, P.; Iacobellis, N.S. Biocide effects of volatile organic compounds produced by potential biocontrol rhizobacteria on Sclerotinia sclerotiorum. Front. Microbiol. 2015, 6, 1056. [Google Scholar] [CrossRef] [PubMed]
- Janakiev, T.; Dimkić, I.; Unković, N.; Ljaljević Grbić, M.; Opsenica, D.; Gašić, U.; Stanković, S.; Berić, T. Phyllosphere Fungal Communities of Plum and Antifungal Activity of Indigenous Phenazine-Producing Pseudomonas synxantha Against Monilinia laxa. Front. Microbiol. 2019, 10, 2287. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Parmar, P.; Patel, B.; Goswami, D.; Saraf, M. Breaking bad: Better call gingerol for improving antibiotic susceptibility of Pseudomonas aeruginosa by inhibiting multiple quorum sensing pathways. Microbiol. Res. 2021, 252, 126863. [Google Scholar] [CrossRef]
- Hanson, L.E.; Howell, C.R. Elicitors of Plant Defense Responses from Biocontrol Strains of Trichoderma viren. Phytopathology 2004, 94, 171–176. [Google Scholar] [CrossRef]
- Reino, J.L.; Guerrero, R.F.; Hernández-Galán, R.; Collado, I.G. Secondary metabolites from species of the biocontrol agent Trichoderma. Phytochem. Rev. 2007, 7, 89–123. [Google Scholar] [CrossRef]
- Santoyo, G.; Urtis-Flores, C.A.; Loeza-Lara, P.D.; Orozco-Mosqueda, M.D.C.; Glick, B.R. Rhizosphere Colonization Determinants by Plant Growth-Promoting Rhizobacteria (PGPR). Biology 2021, 10, 475. [Google Scholar] [CrossRef]
- Esmaeilishirazifard, E.; Dariush, A.; Moschos, S.A.; Keshavarz, T. A novel antifungal property for the Bacillus licheniformis ComX pheromone and its possible role in inter-kingdom cross-talk. Appl. Microbiol. Biotechnol. 2018, 102, 5197–5208. [Google Scholar] [CrossRef]
- Islam, R.; Jeong, Y.T.; Lee, Y.S.; Song, C.H. Isolation and Identification of Antifungal Compounds from Bacillus subtilis C9 Inhibiting the Growth of Plant Pathogenic Fungi. Mycobiology 2018, 40, 59–66. [Google Scholar] [CrossRef]
- Nassimi, Z.; Taheri, P.; Tarighi, S. Farnesol altered morphogenesis and induced oxidative burst–related responses in Rhizoctonia solani AG1-IA. Mycologia 2019, 111, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Marketon, M.M.; Gronquist, M.R.; Eberhard, A.; González, J.E. Characterization of the Sinorhizobium meliloti sinR/sinI Locus and the Production of Novel N-Acyl Homoserine Lactones. J. Bacteriol. 2002, 184, 5686. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Fang, S.; MacDonald, J.; Xu, J.; Yuan, Z.-C. Isolation and characterization of Burkholderia cenocepacia CR318, a phosphate solubilizing bacterium promoting corn growth. Microbiol. Res. 2020, 233, 126395. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Yang, C.; Song, S.; Fu, S.; Sun, X.; Yang, L.; He, F.; Zhang, L.H.; Zhang, Y.; Deng, Y. A novel two-component system modulates quorum sensing and pathogenicity in Burkholderia cenocepacia. Mol. Microbiol. 2018, 108, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Xiang, W.; Cao, K.X.; Lu, X.; Yao, S.C.; Hung, D.; Huang, R.S.; Li, L.B. Characterization of volatile organic compounds emitted from endophytic burkholderia cenocepacia ETR-B22 by SPME-GC-MS and their inhibitory activity against various plant fungal pathogens. Molecules 2020, 25, 3765. [Google Scholar] [CrossRef]
- Elshafie, H.; Camele, I.; Racioppi, R.; Scrano, L.; Iacobellis, N.; Bufo, S. In Vitro Antifungal Activity of Burkholderia gladioli pv. agaricicola against Some Phytopathogenic Fungi. Int. J. Mol. Sci. 2012, 13, 16291–16302. [Google Scholar] [CrossRef]
- Tenorio-Salgado, S.; Tinoco, R.; Vazquez-Duhalt, R.; Caballero-Mellado, J.; Perez-Rueda, E. Identification of volatile compounds produced by the bacterium Burkholderia tropica that inhibit the growth of fungal pathogens. Bioengineered 2013, 4, 236–243. [Google Scholar] [CrossRef]
- Deng, S. Phosphorus and selected metals and metalloids. In Principles and Applications of Soil Microbiology, 3rd ed.; Gentry, T.J., Fuhrmann, J.J., Zuberer, D.A.B.T.-P., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 523–555. [Google Scholar]
- Mitter, B.; Petric, A.; Shin, M.W.; Chain, P.S.G.; Hauberg-Lotte, L.; Reinhold-Hurek, B.; Nowak, J.; Sessitsch, A. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front. Plant Sci. 2013, 4, 120. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Living inside plants: Bacterial endophytes. Curr. Opin. Plant Biol. 2011, 14, 435–443. [Google Scholar] [CrossRef]
- Cornelis, P.; Bodilis, J. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ. Microbiol. Rep. 2009, 1, 256–262. [Google Scholar] [CrossRef]
- Joo, H.S.; Deyrup, S.T.; Shim, S.H. Endophyte-produced antimicrobials: A review of potential lead compounds with a focus on quorum-sensing disruptors. Phytochem. Rev. 2021, 20, 543–568. [Google Scholar] [CrossRef]
- Nair, D.N.; Padmavathy, S. Impact of endophytic microorganisms on plants, environment and humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef] [PubMed]
- Pinski, A.; Betekhtin, A.; Hupert-Kocurek, K.; Mur, L.A.J.; Hasterok, R. Defining the Genetic Basis of Plant–Endophytic Bacteria Interactions. Int. J. Mol. Sci. 2019, 20, 1947. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh Kumar, R.; Singh, R.P.; Mishra, P. Endophytes as emphatic communication barriers of quorum sensing in Gram-positive and Gram-negative bacteria—A review. Environ. Sustain. 2019, 2, 455–468. [Google Scholar] [CrossRef]
- Laue, H.; Schenk, A.; Li, H.; Lambertsen, L.; Neu, T.R.; Molin, S.; Ullrich, M.S. Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae. Microbiology 2006, 152, 2909–2918. [Google Scholar] [CrossRef]
- Heredia-Ponce, Z.; Gutiérrez-Barranquero, J.A.; Purtschert-Montenegro, G.; Eberl, L.; Cazorla, F.M.; de Vicente, A. Biological role of EPS from Pseudomonas syringae pv. syringae UMAF0158 extracellular matrix, focusing on a Psl-like polysaccharide. Npj Biofilms Microbiomes 2020, 6, 37. [Google Scholar] [CrossRef]
- Ishiga, T.; Ishiga, Y.; Betsuyaku, S.; Nomura, N. AlgU contributes to the virulence of Pseudomonas syringae pv. tomato DC3000 by regulating production of the phytotoxin coronatine. J. Gen. Plant Pathol. 2018, 84, 189–201. [Google Scholar] [CrossRef]
- Li, L.; Li, J.; Zhang, Y.; Wang, N. Diffusible signal factor (DSF)-mediated quorum sensing modulates expression of diverse traits in Xanthomonas citri and responses of citrus plants to promote disease. BMC Genomics 2019, 20, 55. [Google Scholar] [CrossRef]
- Ryan, R.P.; Dow, J.M. Communication with a growing family: Diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 2011, 19, 145–152. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, Y.; Chen, X.; Diab, A.A.; Ruan, L.; He, J.; Wang, H.; He, Y.-W. The Multiple DSF-family QS Signals are Synthesized from Carbohydrate and Branched-chain Amino Acids via the FAS Elongation Cycle. Sci. Rep. 2015, 5, 13294. [Google Scholar] [CrossRef]
- Poplawsky, A.R.; Chun, W. pigB determines a diffusible factor needed for extracellular polysaccharide slime and xanthomonadin production in Xanthomonas campestris pv. campestris. J. Bacteriol. 1997, 179, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Jacques, M.A.; Josi, K.; Darrasse, A.; Samson, R. Xanthomonas axonopodis pv. phaseoli var. fuscans is aggregated in stable biofilm population sizes in the phyllosphere of field-grown beans. Appl. Environ. Microbiol. 2005, 71, 2008–2015. [Google Scholar] [CrossRef] [PubMed]
- Aragón, W.; Reina-Pinto, J.J.; Serrano, M. The intimate talk between plants and microorganisms at the leaf surface. J. Exp. Bot. 2017, 68, 5339–5350. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Malamud, F.; Torres, P.S.; Roeschlin, R.; Rigano, L.A.; Enrique, R.; Bonomi, H.R.; Castagnaro, A.P.; Marano, M.R.; Vojnov, A.A. The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology 2011, 157, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Dow, J.M. Diffusible signal factor-dependent quorum sensing in pathogenic bacteria and its exploitation for disease control. J. Appl. Microbiol. 2017, 122, 2–11. [Google Scholar] [CrossRef]
- Ahuja, V.; Sangeetha, J.; Torvi, A.; Thangadurai, D.; Shettar, A.K.; David, M.; Thimmappa, S.C. Microbes and agricultural waste: A safe resource for the production of bionanomaterials. In Agri-Waste and Microbes for Production of Sustainable Nanomaterials; Elsevier: Amsterdam, The Netherlands, 2022; pp. 301–322. [Google Scholar]
- Bhattacharyya, C.; Roy, R.; Tribedi, P.; Ghosh, A.; Ghosh, A. Biofertilizers as substitute to commercial agrochemicals. In Agrochemicals Detection, Treatment and Remediation; Butterworth-Heinemann: Oxford, UK, 2020; pp. 263–290. [Google Scholar]
- Stone, W.; Wolfaardt, G. Measuring microbial metabolism in atypical environments. In Methods in Microbiology; Academic Press: Cambridge, MA, USA, 2018; Volume 45, pp. 123–144. [Google Scholar]
- Kambourova, M.; Oner, E.T.; Poli, A. Exopolysaccharides from prokaryotic microorganisms—Promising sources for white biotechnology processes. In Industrial Biorefineries & White Biotechnology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 523–554. [Google Scholar]
- Coutinho, T.A.; Wingfield, M.J. Ralstonia solanacearum and R. pseudosolanacearum on Eucalyptus: Opportunists or Primary Pathogens? Front. Plant Sci. 2017, 8, 761. [Google Scholar] [CrossRef]
- Kai, K. Bioorganic chemistry of signaling molecules in microbial communication. J. Pestic. Sci. 2019, 44, 200–207. [Google Scholar] [CrossRef]
- Meneses, C.H.S.G.; Rouws, L.F.M.; Simões-Araújo, J.L.; Vidal, M.S.; Baldani, J.I. Exopolysaccharide Production Is Required for Biofilm Formation and Plant Colonization by the Nitrogen-Fixing Endophyte Gluconacetobacter diazotrophicus. Mol. Plant-Microbe Interact. 2011, 24, 1448–1458. [Google Scholar] [CrossRef]
- Gupta, S.; Pandey, S. Exploiting the potential of plant growth-promoting rhizobacteria in legume production. In Abiotic Stress and Legumes; Elsevier: Amsterdam, The Netherlands, 2021; pp. 1–32. [Google Scholar]
- Zhou, J.; Lyu, Y.; Richlen, M.L.; Anderson, D.M.; Cai, Z. Quorum Sensing Is a Language of Chemical Signals and Plays an Ecological Role in Algal-Bacterial Interactions. Crit. Rev. Plant Sci. 2016, 35, 81–105. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2009, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Spraker, J.E.; Wiemann, P.; Baccile, J.A.; Venkatesh, N.; Schumacher, J.; Schroeder, F.C.; Sanchez, L.M.; Keller, N.P. Conserved Responses in a War of Small Molecules between a Plant-Pathogenic Bacterium and Fungi. MBio 2018, 9, e00820-18. [Google Scholar] [CrossRef] [PubMed]
- Khalid, S.; Baccile, J.A.; Spraker, J.E.; Tannous, J.; Imran, M.; Schroeder, F.C.; Keller, N.P. NRPS-Derived Isoquinolines and Lipopetides Mediate Antagonism between Plant Pathogenic Fungi and Bacteria. ACS Chem. Biol. 2018, 13, 171. [Google Scholar] [CrossRef] [PubMed]
- Van Pée, K.-H. Biosynthesis of halogenated alkaloids. In Alkaloids: Chemistry and Biology; Academic Press: Cambridge, MA, USA, 2012; Volume 71, pp. 167–210. [Google Scholar]
- Pawar, S.; Chaudhari, A.; Prabha, R.; Shukla, R.; Singh, D.P. Microbial pyrrolnitrin: Natural metabolite with immense practical utility. Biomolecules 2019, 9, 443. [Google Scholar] [CrossRef]
- Schuhegger, R.; Ihring, A.; Gantner, S.; Bahnweg, G.; Knappe, C.; Vogg, G.; Hutzler, P.; Schmid, M.; van Breusegem, F.; Eberl, L.; et al. Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant. Cell Environ. 2006, 29, 909–918. [Google Scholar] [CrossRef]
- Corral-Lugo, A.; Daddaoua, A.; Ortega, A.; Espinosa-Urgel, M.; Krell, T. Rosmarinic acid is a homoserine lactone mimic produced by plants that activates a bacterial quorum-sensing regulator. Sci. Signal. 2016, 9, ra1. [Google Scholar] [CrossRef]
- Petersen, M.; Abdullah, Y.; Benner, J.; Eberle, D.; Gehlen, K.; Hücherig, S.; Janiak, V.; Kim, K.H.; Sander, M.; Weitzel, C.; et al. Evolution of rosmarinic acid biosynthesis. Phytochemistry 2009, 70, 1663–1679. [Google Scholar] [CrossRef]
- Zhou, L.; Zheng, H.; Tang, Y.; Yu, W.; Gong, Q. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotechnol. Lett. 2012, 35, 631–637. [Google Scholar] [CrossRef]
- Nazzaro, F.; Fratianni, F.; d’Acierno, A.; de Feo, V.; Ayala-Zavala, F.J.; Gomes-Cruz, A.; Granato, D.; Coppola, R. Effect of polyphenols on microbial cell-cell communications. In Quorum Sensing; Academic Press: Cambridge, MA, USA, 2019; pp. 195–223. [Google Scholar]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rasamiravaka, T.; Stévigny, C.; Duez, P.; Rajaonson, S.; Diallo, B.; Mol, A.; Baucher, M.; El Jaziri, M. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in pseudomonas aeruginosa PAO1. Microbiology 2011, 157, 2120–2132. [Google Scholar] [CrossRef]
- Chagas, F.O.; Pessotti, R.D.C.; Caraballo-Rodríguez, A.M.; Pupo, M.T. Chemical signaling involved in plant–microbe interactions. Chem. Soc. Rev. 2018, 47, 1652–1704. [Google Scholar] [CrossRef]
- Nadarajah, K.; Abdul Hamid, N.W.; Abdul Rahman, N.S.N. SA-Mediated Regulation and Control of Abiotic Stress Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 5591. [Google Scholar] [CrossRef] [PubMed]
- Meena, V.S.; Meena, S.K.; Verma, J.P.; Kumar, A.; Aeron, A.; Mishra, P.K.; Bisht, J.K.; Pattanayak, A.; Naveed, M.; Dotaniya, M.L. Plant beneficial rhizospheric microorganism (PBRM) strategies to improve nutrients use efficiency: A review. Ecol. Eng. 2017, 107, 8–32. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef]
- Brooks, D.M.; Bender, C.L.; Kunkel, B.N. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 2005, 6, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Kremer, R.J. Bioherbicides and nanotechnology: Current status and future trends. In Nano-Biopesticides Today and Future Perspectives; Koul, O., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 353–366. [Google Scholar]
- National Research Council. Feasibility of Using Mycoherbicides for Controlling Illicit Drug Crops; National Academies Press: Washington, DC, USA, 2011.
- Chakraborty, A.; Ray, P. Mycoherbicides for the Noxious Meddlesome: Can Colletotrichum be a Budding Candidate? Front. Microbiol. 2021, 12, 2859. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shi, M.; Tian, Y.; Zhao, P.; Niu, Y.; Liao, M. Dirhamnolipid Produced by the Pathogenic Fungus Colletotrichum gloeosporioides BWH-1 and Its Herbicidal Activity. Molecules 2019, 24, 2969. [Google Scholar] [CrossRef] [PubMed]
- Boyette, C.D.; Hoagland, R.E.; Stetina, K.C. Extending the host range of the bioherbicidal fungus Colletotrichum gloeosporioides f. sp. aeschynomene. Biocontrol Sci. Technol. 2019, 29, 720–726. [Google Scholar] [CrossRef]
- Masi, M.; Zonno, M.C.; Cimmino, A.; Reveglia, P.; Berestetskiy, A.; Boari, A.; Vurro, M.; Evidente, A. On the metabolites produced by Colletotrichum gloeosporioides a fungus proposed for the Ambrosia artemisiifolia biocontrol; Spectroscopic data and absolute configuration assignment of colletochlorin A. Nat. Prod. Res. 2018, 32, 1537–1547. [Google Scholar] [CrossRef]
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial metabolites in nutrition, healthcare and agriculture. 3 Biotech 2017, 7, 15. [Google Scholar] [CrossRef]
- Asad, S.A. Mechanisms of action and biocontrol potential of Trichoderma against fungal plant diseases—A review. Ecol. Complex. 2022, 49, 100978. [Google Scholar] [CrossRef]
- Berrie, A.; Xu, X. Developing biopesticide-based programmes for managing powdery mildew in protected strawberries in the UK. Crop Prot. 2021, 149, 105766. [Google Scholar] [CrossRef]
- Pfordt, A.; Schiwek, S.; Karlovsky, P.; von Tiedemann, A. Trichoderma Afroharzianum Ear Rot—A New Disease on Maize in Europe. Front. Agron. 2020, 2, 11. [Google Scholar] [CrossRef]
- Khakimov, A.A.; Omonlikov, A.U.; Utaganov, S.B.U. Current status and prospects of the use of biofungicides against plant diseases. GSC Biol. Pharm. Sci. 2020, 13, 119–126. [Google Scholar] [CrossRef]
- Kumar, D.; Singh, M.K.; Singh, H.K.; Singh, K.N. Fungal biopesticides and their uses for control of insect pest and diseases. In Biofertilizers and Biopesticides in Sustainable Agriculture; Apple Academic Press: Palm Bay, FL, USA, 2020; pp. 43–70. [Google Scholar]
- Valantin-Morison, M.; Lasserre-Joulin, F.; Martinet, V.; Meiss, H.; Messéan, A.; Meynard, J.-M.; Paschalidou, F.; Perrin, B.; Rouabah, A. Integrating biocontrol into cropping system design. In Extended Biocontrol; Springer: Dordrecht, The Netherlands, 2022; pp. 233–244. [Google Scholar]
- Abbey, J.A.; Percival, D.; Abbey, L.; Asiedu, S.K.; Prithiviraj, B.; Schilder, A. Biofungicides as alternative to synthetic fungicide control of grey mould (Botrytis cinerea)—Prospects and challenges. Biocontrol Sci. Technol. 2019, 29, 207–228. [Google Scholar] [CrossRef]
- Korzeniewicz, R.; Baranowska, M.; Kwaśna, H.; Niedbała, G.; Behnke-Borowczyk, J. Communities of Fungi in Black Cherry Stumps and Effects of Herbicide. Plants 2020, 9, 1126. [Google Scholar] [CrossRef]
- Uludag, A.; Uremis, I.; Arslan, M. Biological weed control. In Non-Chemical Weed Control; Elsevier: Amsterdam, The Netherlands, 2018; pp. 115–132. [Google Scholar]
- Aneja, K.R.; Kumar, V.; Jiloha, P.; Kaur, M.; Sharma, C.; Surain, P.; Dhiman, R.; Aneja, A. Potential bioherbicides: Indian perspectives. In Biotechnology: Prospects and Applications; Springer: New Delhi, India, 2013; Volume 9788132216, pp. 197–215. [Google Scholar]
- U.S. Environmental Protection Agency. Notice Of Pesticide: X Registration SolviNix LC; U.S. Environmental Protection Agency: Washington, DC, USA, 2014.
- Abu-Dieyeh, M.H.; Bernier, J.; Watson, A.K. Sclerotinia minor avances fruiting and reduces germination in dandelion (Taraxacum officinale). Biocontrol Sci. Technol. 2007, 15, 815–825. [Google Scholar] [CrossRef]
- Kumar, V.; Singh, M.; Sehrawat, N.; Atri, N.; Singh, R.; Upadhyay, S.K.; Kumar, S.; Yadav, M. Mycoherbicide Control Strategy: Concept, Constraints, and Advancements. Biopestic. Int. 2021, 17, 29–40. [Google Scholar]
- Lizardi-Jiménez, M.A.; Hernández-Martínez, R. Solid state fermentation (SSF): Diversity of applications to valorize waste and biomass. 3 Biotech 2017, 7, 44. [Google Scholar] [CrossRef]
- Mousumi Das, M.; Aguilar, C.N.; Haridas, M.; Sabu, A. Production of bio-fungicide, Trichoderma harzianum CH1 under solid-state fermentation using coffee husk. Bioresour. Technol. Reports 2021, 15, 100708. [Google Scholar] [CrossRef]
- Zin, N.A.; Badaluddin, N.A. Biological functions of Trichoderma spp. for agriculture applications. Ann. Agric. Sci. 2020, 65, 168–178. [Google Scholar] [CrossRef]
- Cai, F.; Yu, G.; Wang, P.; Wei, Z.; Fu, L.; Shen, Q.; Chen, W. Harzianolide, a novel plant growth regulator and systemic resistance elicitor from Trichoderma harzianum. Plant Physiol. Biochem. 2013, 73, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Vinale, F.; Nigro, M.; Sivasithamparam, K.; Flematti, G.; Ghisalberti, E.L.; Ruocco, M.; Varlese, R.; Marra, R.; Lanzuise, S.; Eid, A.; et al. Harzianic acid: A novel siderophore from Trichoderma harzianum. FEMS Microbiol. Lett. 2013, 347, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Pirttilä, A.M.; Mohammad Parast Tabas, H.; Baruah, N.; Koskimäki, J.J. Biofertilizers and Biocontrol Agents for Agriculture: How to Identify and Develop New Potent Microbial Strains and Traits. Microorganisms 2021, 9, 817. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, I.V.; Sarrocco, S.; Malfatti, L.; Baroncelli, R.; Vannacci, G. CRISPR-Cas for Fungal Genome Editing: A New Tool for the Management of Plant Diseases. Front. Plant Sci. 2019, 10, 135. [Google Scholar] [CrossRef]
- Key, S.; Ma, J.K.C.; Drake, P.M.W. Genetically modified plants and human health. J. R. Soc. Med. 2008, 101, 290–298. [Google Scholar] [CrossRef]
- Abbas, M.S.T. Genetically engineered (Modified) crops (Bacillus thuringiensis crops) and the world controversy on their safety. Egypt. J. Biol. Pest Control 2018, 28, 52. [Google Scholar] [CrossRef]
- Schünemann, R.; Knaak, N.; Fiuza, L.M. Mode of Action and Specificity of Bacillus thuringiensis Toxins in the Control of Caterpillars and Stink Bugs in Soybean Culture. ISRN Microbiol. 2014, 2014, 35675. [Google Scholar] [CrossRef]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef]
- Bahariah, B.; Parveez, G.K.A.; Masani, M.Y.A.; Khalid, N. In silico Construction of phosphomannose isomerase (PMI) transformation vectors and evaluation of the effectiveness of vectors in tobacco (Nicotiana tabacum L.). Bioinformation 2012, 8, 151–157. [Google Scholar] [CrossRef]
- Tsuge, K.; Akiyama, T.; Shoda, M. Cloning, Sequencing, and Characterization of the Iturin a Operon. J. Bacteriol. 2001, 183, 6265–6273. [Google Scholar] [CrossRef]
- Dang, Y.; Zhao, F.; Liu, X.; Fan, X.; Huang, R.; Gao, W.; Wang, S.; Yang, C. Enhanced production of antifungal lipopeptide iturin A by Bacillus amyloliquefaciens LL3 through metabolic engineering and culture conditions optimization. Microb. Cell Fact. 2019, 18, 68. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Li, X.; Yu, H.; Yang, H.; Li, X.; Shen, Z. In situ enhancement of surfactin biosynthesis in Bacillus subtilis using novel artificial inducible promoters. Biotechnol. Bioeng. 2017, 114, 832–842. [Google Scholar] [CrossRef] [PubMed]
- An, S.-Q.; Potnis, N.; Dow, M.; Vorhölter, F.-J.; He, Y.-Q.; Becker, A.; Teper, D.; Li, Y.; Wang, N.; Bleris, L.; et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 2020, 44, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Tian, F.; Yang, F.; Chen, H.; Yuan, X.; Yang, C.-H.; Chen, Y.; Wang, Q.; He, C. Phosphodiesterase EdpX1 Promotes Xanthomonas oryzae pv. oryzae Virulence, Exopolysaccharide Production, and Biofilm Formation. Appl. Environ. Microbiol. 2018, 84, e01717-18. [Google Scholar] [CrossRef] [PubMed]
| Organism | Signal Molecule | Role | References |
|---|---|---|---|
| Fusarium | Fusaric acid |
| [35,36] |
| S. cerevisiae | Tryptophol |
| [37] |
| Farnesol |
| [38] | |
| Debaryomyces nepalensis | Phenylethanol |
| [39] |
| Penicillium spp. and Aspergillus spp. | Patulin |
| [40,41] |
| Oxylipins |
| [42,43] | |
| Penicillium sclerotiorum | Multicolic acid |
| [23] |
| Fusarium culmorum | Terpenes |
| [44] |
| Penicillium decumbens | Farnesol |
| [45] |
| Penicillium expansum | Farnesol |
| [46] |
| Fusarium graminearum | Farnesol |
| [46] |
| Organism Producing Quorum Sensing Molecule | Quorum Sensing Molecule Produced | Role of the Quorum Sensing Molecule | References |
|---|---|---|---|
| Arthrobacter agilis | Dimethylhexadecylamine |
| [86] |
| B. licheniformis | ComX pheromone |
| [87] |
| B. subtilis subsp. Subtilis C9 | Acetylbuanediol |
| [88] |
| P. aeruginosa | Rhamnolipids |
| [89] |
| P. fluorescens | 2,4-Diacetylphloroglucinol |
| [80] |
| Pseudomonas spp. | Dimethyl disulphide |
| [81] |
| Sinorhizobium meliloti | N-(tetrahydro-2-oxo-3-furanyl)-octanamide (C8-HL) |
| [90] |
| QS Mimicry Molecule | Plants | Affected Microbes | Role | References |
|---|---|---|---|---|
| Rosmarinic acid | Rosmarinus officinalis, Salvia officinalis, Thymus vulgaris, Melissa officinalis, Symphytum officinale, Aegiphila mollis, Ocimum basilicum | P. aeruginosa |
| [131,132] |
| Eugenol | Anethum graveolens, Syzygium aromaticum | Chromobacterium violaceum, P. aeruginosa |
| [133] |
| Curcumin | Curcuma longa | P. aeruginosa |
| [134] |
| Naringenin | Citrus sp., Ficus carica, Solanum lycopersicum | P. aeruginosa, C. violaceum |
| [134,135] |
| Type | Marketing Name | Active Ingredients | Target Pathogen, Diseases or Weeds | Mode of Action | References |
|---|---|---|---|---|---|
| Biofungicide | AQ10 Bio Fungicide | Spores of a naturally occurring Ampelomyces quisqualis strain AQ10 |
|
| [149] |
| Trichodex | Trichoderma harzi anum T39 |
|
| [150] | |
| Rootshield® WP | Trichoderma harzianum strain T-22 |
|
| [151] | |
| Binab T | Trichoderma harzianum and Trichoderma polysporum |
|
| [151] | |
| Primastop | Gliocladium catenulatum Strain J 1446 |
|
| [152] | |
| Contans WG | Coniothyrium minitans, strain CON/M/91-08 |
|
| [153] | |
| Biosave® | Pseudomonas syringae Strain ESC-11 |
|
| [154] | |
| Bioherbicide | Biochon | Chondrostereum purpureum |
|
| [155] |
| Dr. Biosedge | Puccinia canaliculata |
|
| [156,157] | |
| Solvinix | Tobacco mild green mosaic tobamovirus (TMGMV) |
|
| [158] | |
| Sarritor® | Sclerotinia minor IMI 344141 |
|
| [159,160] | |
| Organo-sol |
|
|
| [160] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdul Hamid, N.W.; Nadarajah, K. Microbe Related Chemical Signalling and Its Application in Agriculture. Int. J. Mol. Sci. 2022, 23, 8998. https://doi.org/10.3390/ijms23168998
Abdul Hamid NW, Nadarajah K. Microbe Related Chemical Signalling and Its Application in Agriculture. International Journal of Molecular Sciences. 2022; 23(16):8998. https://doi.org/10.3390/ijms23168998
Chicago/Turabian StyleAbdul Hamid, Nur Wahida, and Kalaivani Nadarajah. 2022. "Microbe Related Chemical Signalling and Its Application in Agriculture" International Journal of Molecular Sciences 23, no. 16: 8998. https://doi.org/10.3390/ijms23168998
APA StyleAbdul Hamid, N. W., & Nadarajah, K. (2022). Microbe Related Chemical Signalling and Its Application in Agriculture. International Journal of Molecular Sciences, 23(16), 8998. https://doi.org/10.3390/ijms23168998