Size and Methylation Index of Cell-Free and Cell-Surface-Bound DNA in Blood of Breast Cancer Patients in the Contest of Liquid Biopsy
Abstract
:1. Introduction
2. Results
2.1. Concentration and Size of cf DNA and csbDNA Fragments in Blood of HFs and Untreated BCPs
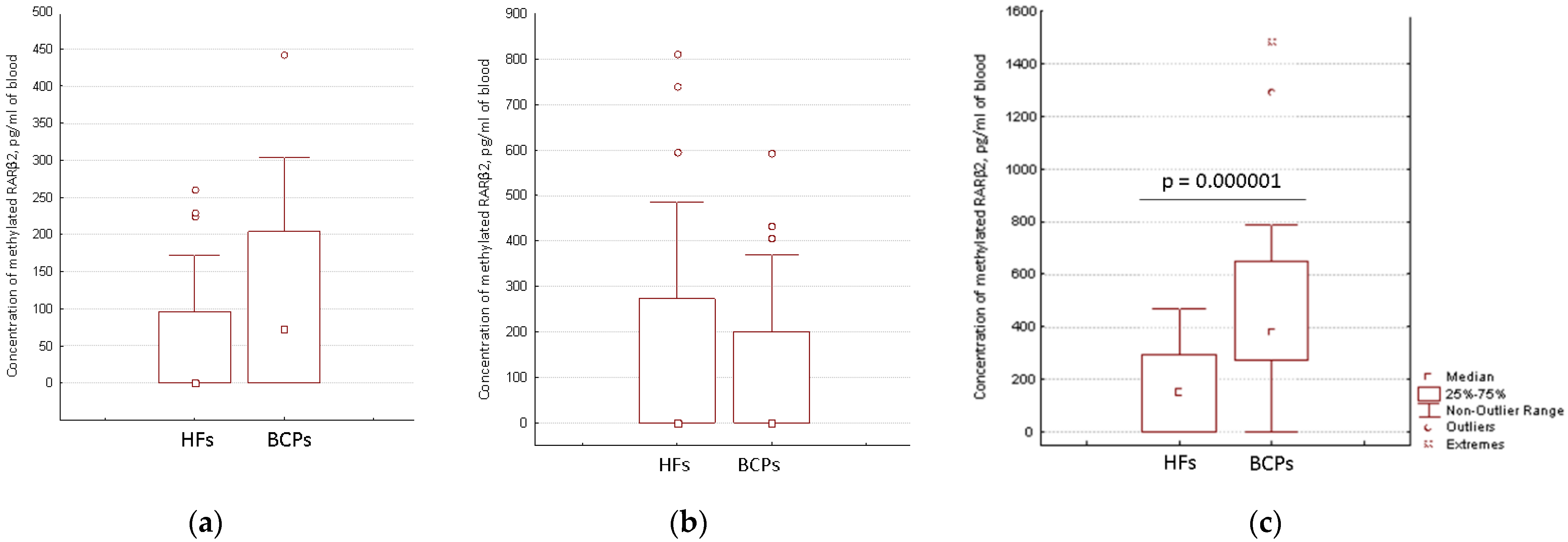
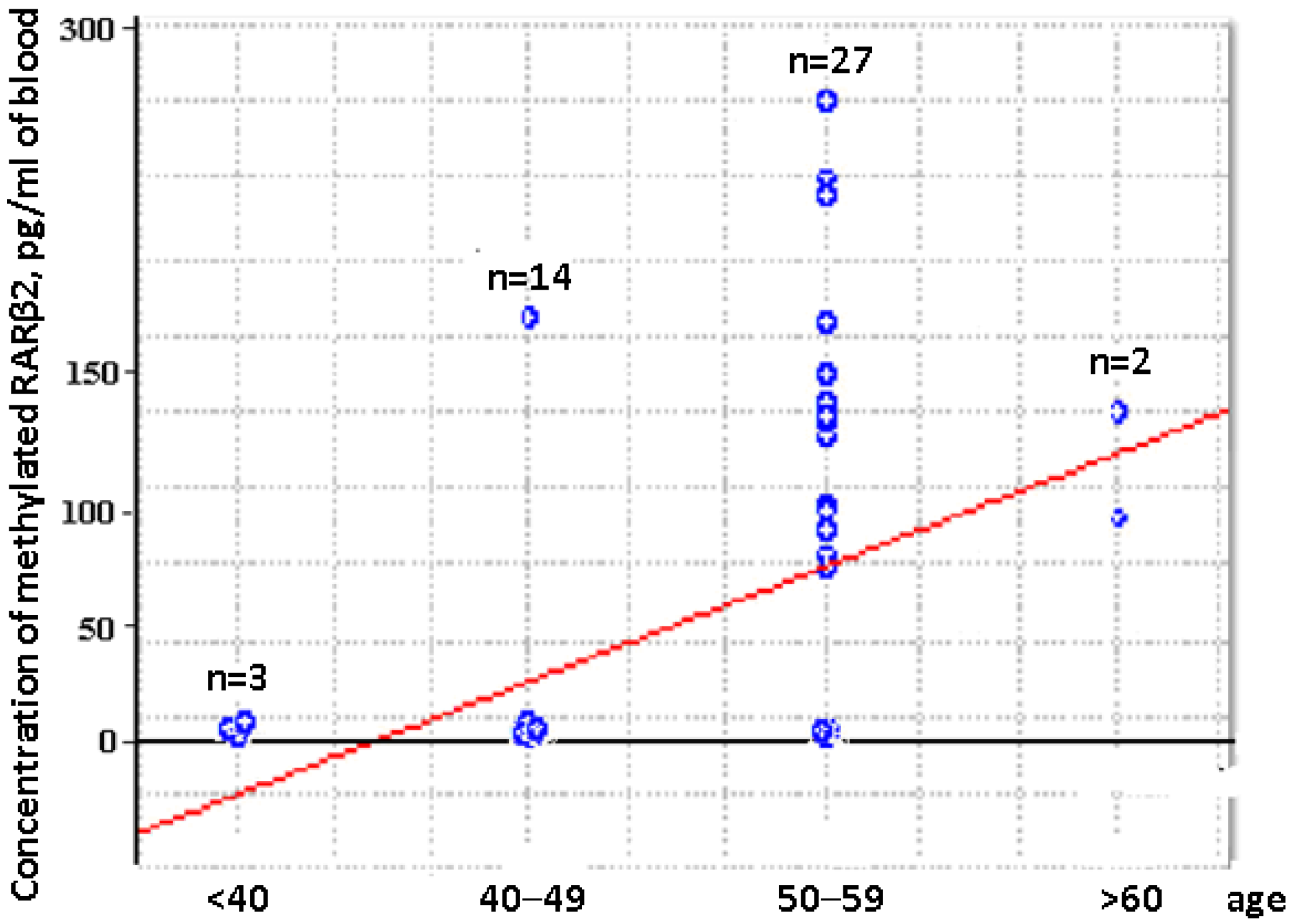
2.2. Distribution of Aberrantly Methylated and Unmethylated RARβ2 in Short DNA and Total DNA from Blood of HFs and BCPs
2.3. Concentration of Aberrantly Methylated RARβ2 in Blood of Untreated Luminal BCPs at I-II Stages of Disease
3. Discussion
4. Materials and Methods
4.1. Patients and Blood Treatment
4.2. CirDNA Isolation and Quantification of Short Fragments in Cell-Free and csb-DNA
4.3. Bisulfite Conversion and Methyl-Specific TaqMan PCR (MSP)
4.4. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Duque, G.; Manterola, C.; Otzen, T.; Arias, C.; Galindo, B.; Mora, M.; Guerrero, E.; García, N. Clinical utility of liquid biopsy in breast cancer: A systematic review. Clin. Genet. 2022, 101, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Palanca-Ballester, C.; Rodriguez-Casanova, A.; Torres, S.; Calabuig-Fariñas, S.; Exposito, F.; Serrano, D.; Redin, E.; Valencia, K.; Jantus-Lewintre, E.; Diaz-Lagares, A.; et al. Cancer Epigenetic Biomarkers in Liquid Biopsy for High Incidence Malignancies. Cancers 2021, 13, 3016. [Google Scholar] [CrossRef]
- Skvortsova, T.E.; Bryzgunova, O.E.; Lebedeva, A.O.; Mak, V.V.; Vlassov, V.V.; Laktionov, P.P. Methylated Cell-Free DNA In Vitro and In Vivo. In Circulating Nucleic Acids in Plasma and Serum; Gahan, P., Ed.; Springer: Dordrecht, Switzerland, 2010; Chapter 25; pp. 185–194. [Google Scholar]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, B.; Liebenberg, V.; Dietrich, D.; Schlegel, T.; Kneip, C.; Seegebarth, A.; Flemming, N.; Seemann, S.; Distler, J.; Lewin, J.; et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer 2010, 10, 600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rykov, S.V.; Filippova, E.A.; Loginov, V.I.; Braga, E.A. Gene Methylation in Circulating Cell-Free DNA from the Blood Plasma as Prognostic and Predictive Factor in Breast Cancer. Russ. J. Genet. 2021, 57, 1239–1252. [Google Scholar] [CrossRef]
- Panagopoulou, M.; Esteller, M.; Chatzaki, E. Circulating Cell-Free DNA in Breast Cancer: Searching for Hidden Information towards Precision Medicine. Cancers 2021, 13, 728. [Google Scholar] [CrossRef] [PubMed]
- Delmonico, L.; Alves, G.; Bines, J. Cell free DNA biology and its involvement in breast carcinogenesis. Adv. Clin. Chem. 2020, 97, 171–223. [Google Scholar]
- Wang, W.; Liang, M.; Ma, G.; Li, L.; Zhou, W.; Xia, T.; Xie, H.; Wang, S. Plasma cell-free DNA integrity plus circulating tumor cells: A potential biomarker of no distant metastasis breast cancer. Neoplasma 2017, 64, 611–618. [Google Scholar] [CrossRef]
- Udomruk, S.; Orrapin, S.; Pruksakorn, D.; Chaiyawat, P. Size distribution of cell-free DNA in oncology. Crit. Rev. Oncol. Hematol. 2021, 166, 103455. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445. [Google Scholar] [CrossRef] [Green Version]
- Boone, B.A.; Orlichenko, L.; Schapiro, N.E.; Loughran, P.; Gianfrate, G.C.; Ellis, J.T.; Zeh, H.J. The receptor for advanced glycation end products (RAGE) enhances autophagy and neutrophil extracellular traps in pancreatic cancer. Cancer Gene Ther. 2015, 22, 326–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamkovich, S.; Bryzgunova, O. Protease Activity and Cell-Free DNA in Blood Plasma of Healthy Donors and Breast Cancer Patients. J. Immunoass. Immunochem. 2016, 37, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Tamkovich, S.N.; Cherepanova, A.V.; Yarmoshchuk, S.V.; Permyakova, V.I.; Anykeeva, O.Y.; Laktionov, P.P. Redistribution of Free- and Cell-Surface-Bound DNA in Blood of Benign and Malignant Prostate Tumor Patients. Acta Nat. 2015, 7, 115–118. [Google Scholar] [CrossRef]
- Wu, Z.-S.; Wu, Q.; Yang, J.-H.; Wang, H.-Q.; Ding, X.-D.; Yang, F.; Xu, X.-C. Prognostic significance of MMP-9 and TIMP-1 serum and tissue expression in breast cancer. Int. J. Cancer 2008, 122, 2050–2056. [Google Scholar] [CrossRef] [PubMed]
- Fuksiewicz, M.; Kotowicz, B.; Rutkowski, A.; Kowalska, M. The matrix metalloproteinase-7 and pro-enzyme of metalloproteinase-1 as a potential marker for patients with rectal cancer without distant metastasis. Tumor Biol. 2015, 36, 3629–3635. [Google Scholar] [CrossRef]
- Ivanov, M.; Baranova, A.; Butler, T.; Spellman, P.; Mileyko, V. Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics 2015, 16 (Suppl. S13), S1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Zhang, R.; Li, J.; Zhang, R. Size profile of cell-free DNA: A beacon guiding the practice and innovation of clinical testing. Theranostics 2020, 10, 4737–4748. [Google Scholar] [CrossRef]
- Ponomaryova, A.A.; Rykova, E.Y.; Cherdyntseva, N.V.; Skvortsova, T.E.; Dobrodeev, A.Y.; Zav’yalov, A.A.; Bryzgalov, L.O.; Tuzikov, S.A.; Vlassov, V.V.; Laktionov, P.P. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer 2013, 81, 397–403. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBO-511 CAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Tanas, A.S.; Sigin, V.O.; Kalinkin, A.I.; Litviakov, N.V.; Slonimskaya, E.M.; Ibragimova, M.K.; Ignatova, E.O.; Simonova, O.A.; Kuznetsova, E.B.; Kekeeva, T.V.; et al. Genome-Wide Methylotyping Resolves Breast Cancer Epigenetic Heterogeneity and Suggests Novel Therapeutic Perspectives. Epigenomics 2019, 11, 605–617. [Google Scholar] [CrossRef] [Green Version]
- Skvortsova, T.E.; Rykova, E.Y.; Tamkovich, S.N.; Bryzgunova, O.E.; Starikov, A.V.; Kuznetsova, N.P.; Vlassov, V.V.; Laktionov, P.P. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Br. J. Cancer 2006, 94, 1492–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kougioumtsidou, N.; Vavoulidis, E.; Nasioutziki, M.; Symeonidou, M.; Pratilas, G.C.; Mareti, E.; Petousis, S.; Chatzikyriakidou, A.; Grimbizis, G.; Theodoridis, T.; et al. DNA Methylation Patterns of RAR-beta2 and RASSF1A Gene Promoters in FNAB Samples from Greek Population with Benign or Malignant Breast Lesions. Diagn. Cytopathol. 2021, 49, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Yari, K.; Rahimi, Z. Promoter Methylation Status of the Retinoic Acid Receptor-Beta 2 Gene in Breast Cancer Patients: A Case Control Study and Systematic Review. Breast Care 2019, 14, 117–123. [Google Scholar] [CrossRef]
- Ungerer, V.; Bronkhorst, A.J.; Holdenrieder, S. Preanalytical variables that affect the outcome of cell-free DNA measurements. Crit. Rev. Clin. Lab. Sci. 2020, 57, 484–507. [Google Scholar] [CrossRef] [PubMed]
- Tamkovich, S.N.; Laktionov, P.P. Cell-Surface-Bound Circulating DNA in the Blood: Biology and Clinical Application. IUBMB Life 2019, 71, 1201–1210. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.; Hesch, R.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Suzuki, N.; Kamataki, A.; Yamaki, J.; Homma, Y. Characterization of circulating DNA in healthy human plasma. Clin. Chim. Acta 2008, 387, 55–58. [Google Scholar] [CrossRef]
- Jiang, W.W.; Zahurak, M.; Goldenberg, D.; Milman, Y.; Park, H.L.; Westra, W.H.; Koch, W.; Sidransky, D.; Califano, J. Increased plasma DNA integrity index in head and neck cancer patients. Int. J. Cancer 2006, 119, 2673–2676. [Google Scholar] [CrossRef]
- Mouliere, F.; Robert, B.; Arnau Peyrotte, E.; Del Rio, M.; Ychou, M.; Molina, F.; Gongora, C.; Thierry, A.R. High fragmentation characterizes tumour-derived circulating DNA. PLoS ONE 2011, 6, e23418. [Google Scholar] [CrossRef]
- Kalinina, T.; Kononchuk, V.; Alekseenok, E.; Abdullin, G.; Sidorov, S.; Ovchinnikov, V.; Gulyaeva, L. Associations between the Levels of Estradiol-, Progesterone-, and Testosterone-Sensitive MiRNAs and Main Clinicopathologic Features of Breast Cancer. J. Pers. Med. 2022, 12, 4. [Google Scholar] [CrossRef]
- Tutanov, O.; Proskura, K.; Kamyshinsky, R.; Shtam, T.; Tsentalovich, Y.; Tamkovich, S. Proteomic Profiling of Plasma and Total Blood Exosomes in Breast Cancer: A Potential Role in Tumor Progression, Diagnosis, and Prognosis. Front. Oncol. 2020, 10, 580891. [Google Scholar] [CrossRef]
- Lin, R.K.; Su, C.M.; Lin, S.Y.; Thi Anh Thu, L.; Liew, P.L.; Chen, J.Y.; Tzeng, H.E.; Liu, Y.R.; Chang, T.H.; Lee, C.Y.; et al. Hypermethylation of TMEM240 predicts poor hormone therapy response and disease progression in breast cancer. Mol. Med. 2022, 28, 67. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Xiong, X. Promoter hypermethylation of RARβ2, DAPK, hMLH1, p14, and p15 is associated with progression of breast cancer: A PRISMA-compliant meta-analysis. Medicine 2018, 97, e13666. [Google Scholar] [CrossRef] [PubMed]
- Khodyrev, D.; Loginov, V.; Pronina, I.; Kazubskaya, T.; Garkavtseva, R.; Braga, E. Methylation of promoter region of RAR-β2 gene in renal cell, breast, and ovarian carcinomas. Russ. J. Genet. 2008, 44, 983–988. [Google Scholar] [CrossRef]
- Mirza, S.; Sharma, G.; Parshad, R.; Srivastava, A.; Gupta, S.D.; Ralhan, R. Clinical significance of Stratifin, ERα and PR promoter methylation in tumor and serum DNA in Indian breast cancer patients. Clin. Biochem. 2010, 43, 380–386. [Google Scholar] [CrossRef]
- Korshunova, Y.; Maloney, R.; Lakey, N.; Citek, R.; Bacher, B.; Budiman, A.; Ordway, J.; McCombie, W.; Leon, J.; Jeddeloh, J.; et al. Massively parallel bisulphite pyrosequencing reveals the molecular complexity of breast cancer-associated cytosine-methylation patterns obtained from tissue and serum DNA. Genome Res. 2008, 18, 19–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Sun, K.; Jiang, P.; Allen Chan, K.C.; Wong, J.; Cheng, Y.K.Y.; Liang, R.H.S.; Chan, W.; Ma, E.S.K.; Chan, S.L.; Cheng, S.H.; et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. USA 2015, 112, E5503–E5512. [Google Scholar] [CrossRef] [Green Version]
- Bryzgunova, O.E.; Konoshenko, M.Y.; Laktionov, P.P. Concentration of cell-free DNA in different tumor types. Expert Rev. Mol. Diagn. 2021, 21, 63–75. [Google Scholar] [CrossRef]
- Kolesnikova, E.V.; Tamkovich, S.N.; Bryzgunova, O.E.; Shelestyuk, P.I.; Permyakova, V.I.; Vlassov, V.V.; Tuzikov, A.S.; Laktionov, P.P.; Rykova, E.Y. Circulating DNA in the blood of gastric cancer patients. Ann. N. Y. Acad. Sci. 2008, 1137, 226–231. [Google Scholar] [CrossRef]
- Van der Auwera, I.; Bovie, C.; Svensson, C.; Trinh, X.B.; Limame, R.; van Dam, P.; van Laere, S.J.; van Marck, E.A.; Dirix, L.Y.; Vermeulen, P.B. Quantitative methylation profiling in tumor and matched morphologically normal tissues from breast cancer patients. BMC Cancer 2010, 10, 97. [Google Scholar] [CrossRef] [Green Version]
- Bryzgunova, O.; Bondar, A.; Ruzankin, P.; Laktionov, P.; Tarasenko, A.; Kurilshikov, A.; Epifanov, R.; Zaripov, M.; Kabilov, M.; Laktionov, P. Locus-Specific Methylation of GSTP1, RNF219, and KIAA1539 Genes with Single Molecule Resolution in Cell-Free DNA from Healthy Donors and Prostate Tumor Patients: Application in Diagnostics. Cancers 2021, 13, 6234. [Google Scholar] [CrossRef] [PubMed]
- Kwabi-Addo, B.; Chung, W.; Shen, L.; Ittmann, M.; Wheeler, T.; Jelinek, J.; Issa, J.P. Age-related DNA methylation changes in normal human prostate tissues. Clin. Cancer Res. 2007, 13, 3796–3802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mc Auley, M.T. DNA methylation in genes associated with the evolution of ageing and disease: A critical review. Ageing Res. Rev. 2021, 72, 101488. [Google Scholar] [CrossRef] [PubMed]
- Tamkovich, S.N.; Tutanov, O.S.; Serdukov, D.S.; Belenikin, M.S.; Shlikht, A.G.; Kirushina, N.A.; Duzhak, T.G.; Voytsitskiy, V.E.; Tsentalovich, Y.P.; Tkachuk, V.A.; et al. Protein content of circulating nucleoprotein complexes. Adv. Exp. Med. Biol. 2016, 924, 133–136. [Google Scholar]
- Morozkin, E.S.; Loseva, E.M.; Morozov, I.V.; Kurilshikov, A.M.; Bondar, A.A.; Rykova, E.Y.; Rubtsov, N.B.; Vlassov, V.V.; Laktionov, P.P. A comparative study of cell-free apoptotic and genomic DNA using FISH and massive parallel sequencing. Expert Opin. Biol. Ther. 2012, 12, S141–S153. [Google Scholar] [CrossRef]
- Curigliano, G.; Burstein, H.J.; Winer, E.P.; Gnant, M.; Dubsky, P.; Loibl, S.; Colleoni, M.; Regan, M.M.; Piccart-Gebhart, M.; Senn, H.J.; et al. De-escalating and escalating treatments for early-stage breast cancer: The St. Gallen international expert consensus conference on the primary therapy of early breast cancer 2017. Ann. Oncol. 2017, 28, 1700–1712. [Google Scholar] [CrossRef]
- Babyshkina, N.; Vtorushin, S.; Dronova, T.; Patalyak, S.; Slonimskaya, E.; Kzhyshkowska, J.; Cherdyntseva, N.; Choynzonov, E. Impact of estrogen receptor α on the tamoxifen response and prognosis in luminal-A-like and luminal-B-like breast cancer. Clin. Exp. Med. 2019, 19, 547–556. [Google Scholar] [CrossRef]
- Hammond, M.E.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch. Pathol. Lab. Med. 2010, 134, e48–e72. [Google Scholar] [CrossRef]
- Lessey, B.A.; Palomino, W.A.; Apparao, K.B.; Young, S.L.; Lininger, R.A. Estrogen receptor-alpha (ER-alpha) and defects in uterine receptivity in women. Reprod. Biol. Endocrinol. 2006, 4 (Suppl. S1), S9. [Google Scholar] [CrossRef] [Green Version]
- Tamkovich, S.N.; Bryzgunova, O.E.; Rykova, E.Y.; Permyakova, V.I.; Vlassov, V.V.; Laktionov, P.P. Circulating nucleic acids in blood of healthy male and female donors. Clin. Chem. 2005, 51, 1317–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryzgunova, O.; Bondar, A.; Morozkin, E.; Mileyko, V.; Vlassov, V.; Laktionov, P. A reliable method to concentrate circulating DNA. Anal. Biochem. 2011, 408, 354–356. [Google Scholar] [CrossRef] [PubMed]

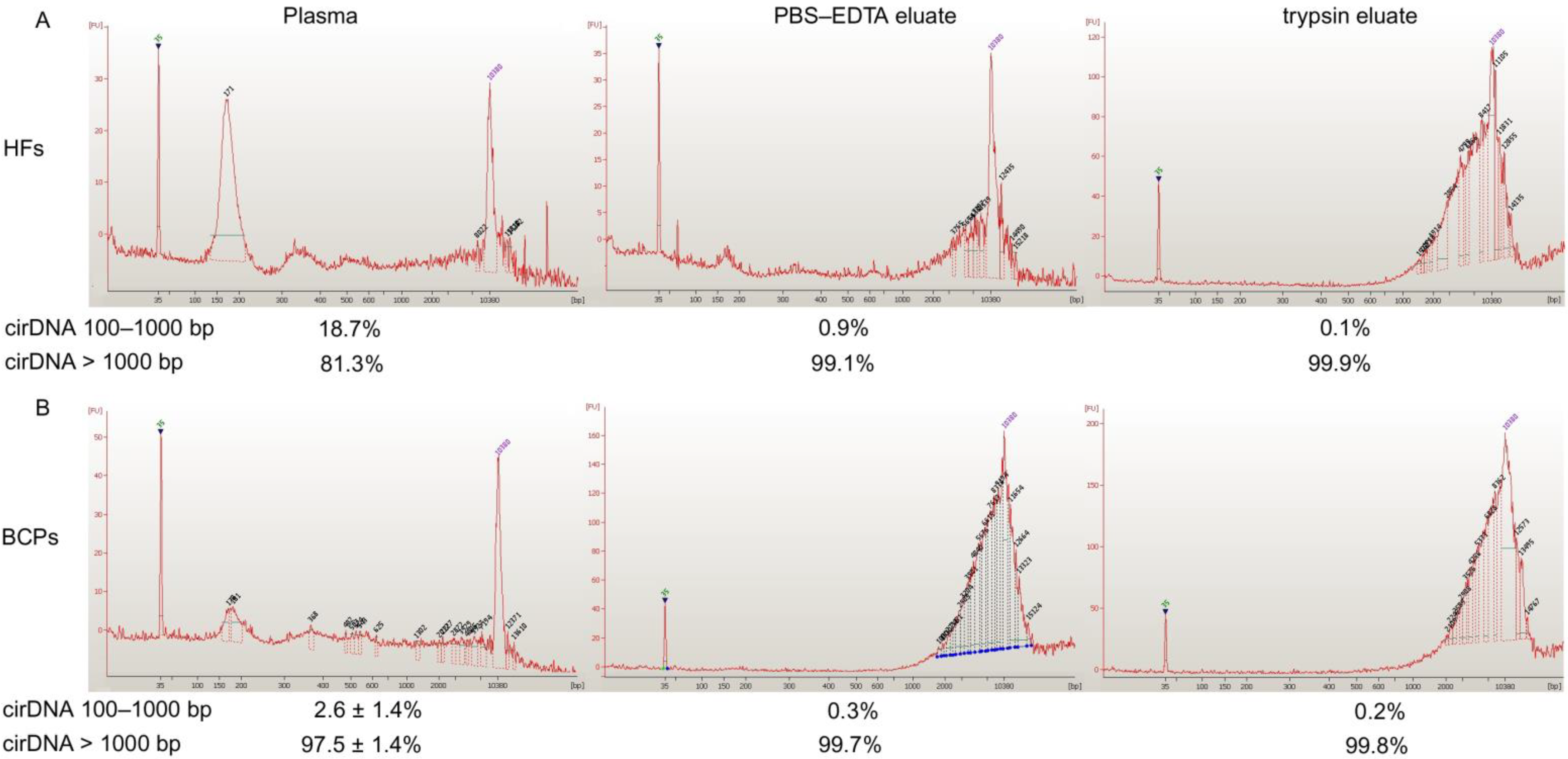
| Blood Fraction | cirDNA | HFs (n = 10) | BCP (n = 8) | p |
|---|---|---|---|---|
| Plasma | 100–1000 bp | ND | ND | |
| Total | 10 ± 3 | 33 ± 11 | <0.05 | |
| PBS-EDTA eluate | 100–1000 bp | ND | ND | |
| Total | 9 ± 4 | 15 ± 6 | ||
| Trypsin eluate | 100–1000 bp | ND | ND | |
| Total | 7 ± 4 | 61 ± 12 | <0.05 |
| Sample | Methylated RARβ2, pg/mL | Sensitivity | Specificity | ROC-Area |
|---|---|---|---|---|
| cfDNA | 70 | 52% | 65% | 0.711 |
| csbDNA | 330 | 74% | 63% | 0.752 |
| Total blood DNA (cfDNA + csbDNA) | 414 | 70% | 61% | 0.753 |
| N (%) | ||
|---|---|---|
| Tumor stage | T1 | 8 (31%) |
| T2 | 18 (69%) | |
| Lymph node status | N0 | 19 (73%) |
| N1 | 7 (21%) | |
| Distant metastasis | M0 | 25 (96%) |
| M1 | 1 (4%) | |
| Molecular subtypes | Luminal A | 13 (50%) |
| Luminal B | 13 (50%) | |
| Histological type | Invasive ductal carcinoma | 26 (100%) |
| Gene | Sequence (5′-3′) | |
|---|---|---|
| LINE1 | Forward | TTCAACAAGAAGAGCTAACTATCC |
| Reverse | TTGTAGGTCACTCAGGACTTGC | |
| Probe | [5,6]-TAMRA-TGCACCCAATACAGGAGCACCCAGATTCA-BHQ2 | |
| RARβ2 GenBank X56849.1 926–1116, 161 b.p., dependent from methylation | Forward | AGG ATTGGGATGTCGAGAACGC |
| Reverse | CTCGACCAATCCAACCGAAACG | |
| RARβ2 GenBank X56849.1 931–1116, 186 bp, undependent from methylation | Forward | TTGTTTGAGGATTGGGATG |
| Reverse | TACCAT TTTCCAAACTTACTC | |
| RARβ2 GenBank X56849.1, 924–1117, 194 b.p., wild type | Forward | ATGCGAGCTGTTTGAGGACT |
| Reverse | TTACCATTTTCCAGGCTTGC | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamkovich, S.; Tupikin, A.; Kozyakov, A.; Laktionov, P. Size and Methylation Index of Cell-Free and Cell-Surface-Bound DNA in Blood of Breast Cancer Patients in the Contest of Liquid Biopsy. Int. J. Mol. Sci. 2022, 23, 8919. https://doi.org/10.3390/ijms23168919
Tamkovich S, Tupikin A, Kozyakov A, Laktionov P. Size and Methylation Index of Cell-Free and Cell-Surface-Bound DNA in Blood of Breast Cancer Patients in the Contest of Liquid Biopsy. International Journal of Molecular Sciences. 2022; 23(16):8919. https://doi.org/10.3390/ijms23168919
Chicago/Turabian StyleTamkovich, Svetlana, Alexey Tupikin, Anton Kozyakov, and Pavel Laktionov. 2022. "Size and Methylation Index of Cell-Free and Cell-Surface-Bound DNA in Blood of Breast Cancer Patients in the Contest of Liquid Biopsy" International Journal of Molecular Sciences 23, no. 16: 8919. https://doi.org/10.3390/ijms23168919
APA StyleTamkovich, S., Tupikin, A., Kozyakov, A., & Laktionov, P. (2022). Size and Methylation Index of Cell-Free and Cell-Surface-Bound DNA in Blood of Breast Cancer Patients in the Contest of Liquid Biopsy. International Journal of Molecular Sciences, 23(16), 8919. https://doi.org/10.3390/ijms23168919






