Abstract
Ovarian cancer is a non-homogenous malignancy. High-grade serous carcinoma (HGSC) is the most common subtype, and its drug resistance mechanisms remain unclear. Despite the advantages of modern pharmacotherapy, high-grade ovarian cancer is associated with a poor prognosis and research into targeted therapies is in progress. The aim of the study was to assess the dominant energy substrate transport mechanism in ovarian cancer cells and to verify whether genomic aberrations could predict clinical outcomes using the Cancer Genome Atlas (TCGA) dataset. Total RNA was extracted from HGSC frozen tissues, and the expression of selected genes was compared to respective controls. GLUT1, FABPpm, MCT4 and SNAT1 genes were significantly overexpressed in carcinomas compared with controls, while expression of CD36/SR-B2, FATP1, FABP4, GLUT4, ASCT2 and LPL was decreased. No differences were found in FATP4, LAT1, MCT1 and FASN. The transcript content of mitochondrial genes such as PGC-1α, TFAM and COX4/1 was similar between groups, while the β-HAD level declined in ovarian cancer. Additionally, the MCT4 level was reduced and PGC-1α was elevated in cancer tissue from patients with ‘small’ primary tumor and omental invasion accompanied by ascites as compared to patients that exhibited greater tendencies to metastasize to lymph nodes with clear omentum. Based on TCGA, higher FABP4 and LPL and lower TFAM expression indicated poorer overall survival in patients with ovarian cancer. In conclusion, the presented data show that there is no exclusive energy substrate in HGSC. However, this study indicates the advantage of glucose and lactate transport over fatty acids, thereby suggesting potential therapeutic intervention targets to impede ovarian cancer growth.
1. Introduction
Ovarian cancer (OC) is the seventh overall and third gynecological most common cancer in women and is still one of the major causes of mortality with a five-year overall survival rate below 45% in advanced stages [,]. Most patients are diagnosed at advanced stages due to non-specific symptoms at the early stage, low efficiency of the diagnostic methods and a lack of screening tests to detect the cancer []. Although the effects of chemotherapy in OC are initially satisfactory, most OC cases exert major therapeutic implications during treatment and pose the threat of cancer recurrence after first-line treatment [].
OC is a non-homogenous malignancy which varies in genetic features, molecular morphology, clinical implications and therapy []. High-grade serous carcinoma (HGSC) is the most common subtype, accounting for over 70% of epithelial ovarian cancers (EOCs). The hallmark feature of HGSC is the diversity in the morphology of different cancer cells, depending on their intratumoral spatial localizations and the tumor microenvironment [], leading to widely diverse drug resistance mechanisms [].
The development of ovarian cancer requires an adequate energy supply, which is facilitated by the interactions between cancer cells and the tumor milieu [,]. The heterogeneous nature of OC manifests through the co-presence of cells highly dependent on glucose that shift their metabolism towards anaerobic processes even in the presence of oxygen, and ovarian cancer cells that rely on aerobic processes such as oxidative phosphorylation []. Glycolysis not only enables fast provision of ATP, but also generates intermediates that are further incorporated in the biosynthesis of amino acids and lipids. Free fatty acids play a crucial role in energy production, membrane synthesis in cancer cells and oncogenic signaling [], while their bioavailability is enhanced in a neoplastic environment []. As part of metabolic alterations, elevated rates of fatty acid synthesis due to increased expression of various lipogenic enzymes could account for cancer progression []. Moreover, if phospholipids with saturated fatty acids are abundantly expressed in cancer membranes, cancer cells are less prone to respond to chemotherapy, which also highlights the role of free fatty acids in cancer physiology []. Amino acids are indispensable for tumor growth as they provide nitrogen for purine and pyrimidine synthesis, serve as precursors in protein and glutathione biosynthesis, and are involved in a vast number of tumor signaling and gene expression pathways, e.g., extracellular signal-regulated protein kinase (ERK) or mammalian target of rapamycin (mTOR) cascades [,]. In particular, an increase in glutamine dependency in high-invasive OC preserves mitochondrial integrity by supporting reactive oxygen species (ROS) scavenging mechanisms and replenishing tricarboxylic acid cycles for energy generation []. Differences in predominantly used energy substrates occur even in patients with similar OC histopathological type, underlining the importance of metabolic adjustments in cancer progression []. According to The Cancer Genome Atlas, HGSC can be classified into immunoreactive, proliferative, differentiated, and mesenchymal cell types based on RNA sequencing and microarray data analysis []. Fatty acid, glucose and amino acid transporters contribute to the development, growth and metastasizing of OC, and thus become potential targets for energy metabolism-based molecular and novel cancer therapy in OC patients.
A substantial correlation between metabolic disturbances occurring in obesity and the development of human cancers was reported in both animal models and clinical trials [,,]. Obesity triggers a wide range of inflammatory effects, which induce a vast number of biological consequences, such as ROS and cytokine production and other mechanisms, by altering the insulin/IGF-1 axis or production of steroid hormone, hence promoting oncogenesis []. Although OC is not suspected to be an obesity-related cancer, some studies revealed a link between enhanced body mass index (BMI) and the likelihood of developing OC, although these findings were not confirmed by other studies [,,]. It is known that the biochemical interplay between adipocytes and OC cells might implicate invasive properties in OC and mediate carboplatin resistance. Moreover, advanced or recurrent OCs have the propensity to metastasize to adipose-rich tissue such as the omentum [].
A large body of evidence suggests that targeting energy substrate transport in poor prognostic patients with metabolically aggressive OC may be a plausible opportunity to enhance patients’ survival by introducing novel treatment schemes into conventional treatment. Therefore, the aim of this study was to assess metabolic and anthropometric factors in OC and their association with the level of expression of diverse energy substrate transporters in OC.
2. Results
2.1. Patient Characteristics
General characteristics of the control and study groups are presented in Table 1. The study group comprised 27 patients with HGSC, of which 23 patients were diagnosed with FIGO stage III–IV. Four patients carried the BRCA 1 or BRCA 2 tumor mutation. Interestingly, during surgical procedures (cytoreductive surgeries in HGSC) we observed two subgroups. Some patients presented a ‘small’ primary tumor and omental invasion accompanied by ascites (in our study group, seven patients). The other group had a greater tendency to metastasize to lymph nodes and omentum was clear (three patients). FIGO classification does not distinguish patients between these features. Based on in vivo research, OC cells have an affinity for fat tissue in the omentum. One of our hypotheses is that there might be molecular subgroups of HGSC that cause this heterogeneity (Table 2).

Table 1.
Study and control group characteristics. Values are presented as median and interquartile range.

Table 2.
Histopathological characteristics of study group.
2.2. Energy Substrate Transporters and Metabolism-Related Gene Expression
Among the tested fatty acid carriers, gene expression of cluster of differentiation 36/a scavenger receptor class B protein (CD36/SR-B2) and fatty acid transport protein 1 (FATP1) was lower in OC compared to controls (−37% and −62%, respectively). The mRNA level of membrane associated fatty acid binding protein (FABPpm) was upregulated (+89%), while FATP4 expression remained relatively constant between groups. The transcript content for cytosolic fatty acid binding proteins such as FABP4 was diminished in OC relative to control samples (−93%; Figure 1a–e and Figure S1). There were no significant changes in FASN expression that would indicate enhanced de novo synthesis of fatty acids in cancer cells. The level of lipoprotein lipase (LPL), required for hydrolytic release of fatty acids from triacylglycerols in circulating lipoprotein particles, lowered by 78% in ovarian cancer (Figures S1 and S2).
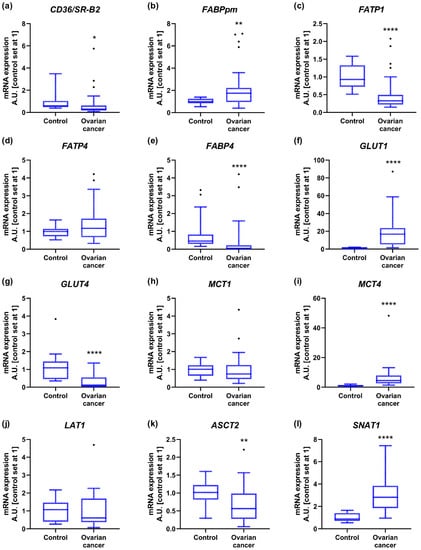
Figure 1.
Transcript levels of (a) cluster of differentiation 36/a scavenger receptor class B protein, (b) membrane associated fatty acid binding protein, (c) fatty acid transport protein 1, (d) fatty acid transport protein 4, (e) fatty acid binding protein 4, (f) glucose transporter 1, (g) glucose transporter 4, (h) monocarboxylate transporter 1, (i) monocarboxylate transporter 4, (j) Na+-independent neutral amino acid transporter, (k) Na+-dependent neutral amino acid transporter and (l) Na+-coupled neutral amino acid transporter 1 in ovarian control (n = 14) and cancer tissue (n = 27). Measurements were made in duplicate, and arithmetic means were used for subsequent investigation. Results are expressed in arbitrary units with control set as 1 and presented as median (min to max) value. Differences statistically significant at: * p < 0.05, ** p < 0.01, **** p < 0.0001.
The expression of glucose transport proteins was enhanced in the case of GLUT1 (+18-fold), but decreased for GLUT4 (−90%; Figure 1f,g and Figure S1). Regarding monocarboxylate transporters, increased transcript content was noticed for MCT4 (+4-fold), but not for MCT1 (Figure 1h,i and Figure S1). Importantly, the relative mRNA expression of GLUT1 and MCT4 in control ovarian samples was the lowest from all the tested energy substrate transporters, and increased to the highest extent during cancer development (Figure S1).
Furthermore, we demonstrated that the mRNA level of Na+-independent neutral amino acid transporter (LAT1) was comparable in both groups, while Na+-dependent neutral amino acid transporter expression declined in OC (ASCT2, −44%; Figure 1j,k and Figure S1). By contrast, the expression of Na+-coupled neutral amino acid transporter 1 (SNAT1) was markedly elevated in OC (+221%; Figure 1l and Figure S1).
Primary OC that developed metastasis was characterized by a lower expression of LPL (−59%) compared to tumors that had not spread to the lymph nodes. MCT4 level was markedly diminished (−57%) in patients with ‘small’ primary tumor and omental invasion accompanied by ascites compared to cancer tissue from patients with greater tendency to metastasize to lymph nodes with clear omentum. Regarding other genes, we did not notice significant differences in their levels with respect to the FIGO stage as well as the presence of lymphatic node invasion or ‘omental-cake’ (Table A1).
2.3. Mitochondrial Gene Expression
In the next step, we verified the expression of several mitochondrial genes and observed a significant decline in β-HAD level in OC (−60%) compared to controls. We did not notice considerable differences in the transcript content for PGC-1α, TFAM and COX4/1 in cancer tissue (Figure 2 and Figure S1). The expression of these genes did not differ with respect to the FIGO stage as well as the presence or absence of lymphatic node invasion or omentum ‘omental-cake’. Nonetheless, PGC-1α level was enhanced (+14-fold) in patients with ‘small’ primary tumor and omental invasion accompanied by ascites, compared to cancer tissue from patients characterized by a greater tendency to metastasize to lymph nodes with clear omentum (Table A1).
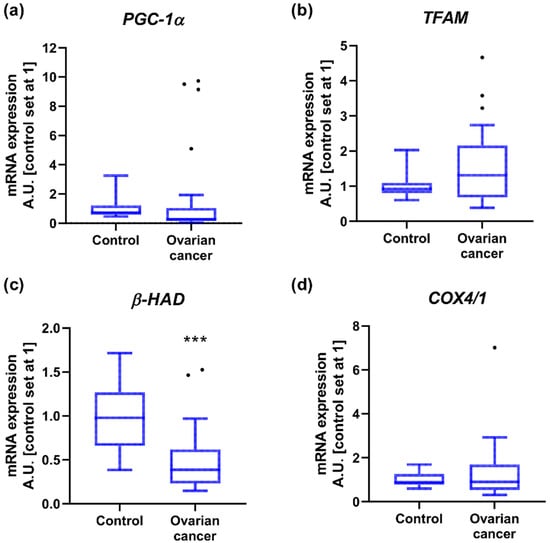
Figure 2.
Transcript levels of (a) peroxisome proliferator-activated receptor gamma co-activator 1α, (b) mitochondrial transcription factor A, (c) acetyl-CoA acyltransferase and (d) cytochrome c oxidase subunit 4 isoform 1 in ovarian control (n = 14) and cancer tissue (n = 27). Measurements were made in duplicate and arithmetic means were used for subsequent investigation. Results are expressed in arbitrary units with control set as 1 and presented as median (min to max) value. Differences statistically significant at *** p < 0.001.
2.4. Associations of Gene Expression with BMI in Patients with Ovarian Cancer
Overweightness, defined as a BMI greater than 25 kg/m2, was not related to changes in transcript content in the studied genes. Obesity (BMI > 30 kg/m2), however, was associated with higher expression of FABPpm, PGC-1α and FASN in OC compared to tissue samples obtained from non-obese patients (Table 3).

Table 3.
Log2-fold changes in gene expression in ovarian cancer tissue obtained from overweight (n = 9) or obese patients (n = 10) compared to lean individuals (n = 8).
2.5. Correlations
A correlation analysis among the values of expression of energy substrate transporters in control samples revealed negative associations between FABP4 and GLUT1 (p = 0.001, r = −0.807) as well as LAT1 and SNAT1 (p = 0.037, r = −0.569). Positive relationships involved FABP4 with GLUT4 (p = 0.02, r = 0.622), FATP4 and MCT1 (p = 0.0003, r = 0.846), FATP4 and MCT4 (p = 0.005, r = 0.723), MCT1 and GLUT1 (p = 0.013, r = 0.657), and MCT4 and ASCT2 (p = 0.03, r = 0.587). In cancer samples, we observed significant correlations between FABP4 and FABPpm (p = 0.009, r = 0.490), FATP4 and MCT1 (p = 0.009, r = 0.489), FATP4 and FABPpm (p = 0.0004, r = 0.636), and SNAT1 and GLUT1 (p = 0.005, r = 0.527) (Figure 3).
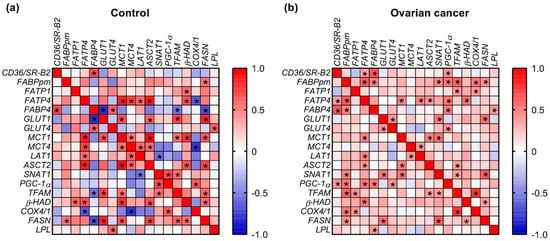
Figure 3.
Heat map of correlations between the expression of energy substrate transporters and mitochondrial genes in (a) ovarian control and (b) cancer tissue. The relationships between the analyzed parameters were assayed via Spearman correlation coefficient. Correlations that were statistically significant (p < 0.05) are indicated with an asterisk.
The positive associations that were present in both control and cancer samples included relationships between FABP4 and CD36/SR-B2 (control: p = 0.008, r = 0.692; cancer: p = 0.0003, r = 0.647), FATP4 and LAT1 (control: p = 0.037, r = 0.569; cancer: p = 0.012, r = 0.478), FATP4 and ASCT2 (control: p = 0.0001, r = 0.868; cancer: p = 0.03, r = 0.418), MCT1 and ASCT2 (control: p = 0.0003, r = 0.903; cancer: p = 0.01, r = 0.488) as well as MCT4 and LAT1 (control: p = 0.027, r = 0.596; cancer: p = 0.044, r = 0.398) (Figure 3).
Additionally, in control samples we noticed a negative relationship between TFAM and FABP4 (p = 0.042, r = −0.556), COX4/1 and FATP4 (p = 0.011, r = −0.666), COX4/1 and MCT4 (p = 0.011, r = −0.789), as well as FASN and FABP4 (p = 0.008, r = −0.692). Positive relationships were observed for β-HAD with FATP1 (p = 0.038, r = 0.565), MCT1 (p = 0.002, r = 0.767) and ASCT2 (p = 0.01, r = 0.675), as well as for TFAM with GLUT1 (p = 0.005, r = 0.719), MCT1 (p = 0.044, r = 0.552) and SNAT1 (p = 0.042, r = 0.556). In OC, there were positive relationships between PGC-1α and CD36/SR-B2 (p = 0.035, r = 0.407), FABPpm (p = 0.005, r = 0.526), FATP4 (p = 0.004, r = 0.536), FABP4 (p = 0.006, r = 0.517) and GLUT4 (p = 0.025, r = 0.431). Correlations common to control and OC were between β-HAD and FATP4 (control: p = 0.011, r = 0.666; cancer: p = 0.003, r = 0.546) and FASN and GLUT1 (control: p = 0.001, r = 0.824; cancer: p = 0.002, r = 0.563; Figure 3).
Moreover, positive correlations were found between the expression of FABPpm and BMI, GLUT1 and plasma glucose concentration, and LAT1 and tumor volume (Figure S1).
2.6. Genetic Alterations in Metabolism-Related Genes Based on TCGA and GTEx Datasets
In the next step, we verified whether our results correspond with data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) projects, which provide comprehensive transcriptomic data in a large number of patients with OC compared to normal ovarian tissue. We noticed only a few discrepancies between our data and TCGA data; namely, in a large OC cohort there were no alterations in CD36/SR-B2, ASCT2 and β-HAD level (a decrease in our study) and FABPpm (an increase in our study). Additionally, while we did not notice alterations in FASN expression, in the TCGA-OC cohort this gene expression was elevated (Figure 4).
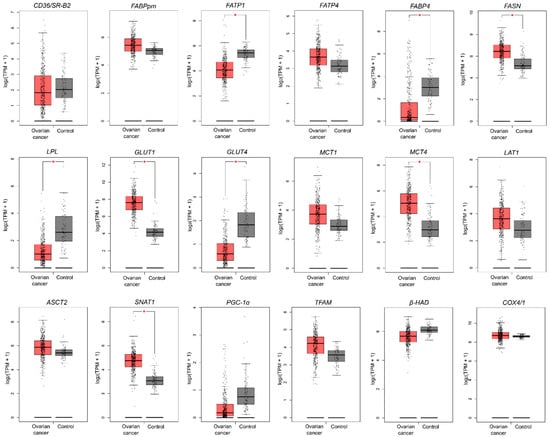
Figure 4.
The transcription level of metabolism-related genes that significantly differed between ovarian cancer (n = 426) and control tissue (n = 88). Data are based on the TCGA cancer cohort and GTEx control samples. Data derive from The Gene Expression Profiling Interactive Analysis (GEPIA) database based on the TCGA-OC dataset and normal ovarian tissues (GTEx, Genotype-Tissue Expression project). Differences statistically significant at * p < 0.05.
2.7. Prognostic Value of the Metabolic Pathway Genes Based on TCGA Cohort
To investigate the relationship between the expression of metabolism-related genes in OC and the clinical data, we analyzed overall survival, progression-free survival and clinical stage-related level for each gene. For overall survival analysis, higher FABP4 and LPL and lower TFAM indicated poorer prognosis (Figure 5a and Figure S3). In the case of progression-free survival, higher expression of FABP4, PGC-1α and COX4/1 indicated worse, while higher levels of GLUT4 and TFAM correlated with better patient progression-free survival (Figure 5b and Figure S4). Furthermore, the expression of FABPpm, FABP4, FASN, GLUT1 and TFAM correlated with the clinical stage of OC (Figure 6 and Figure S5).
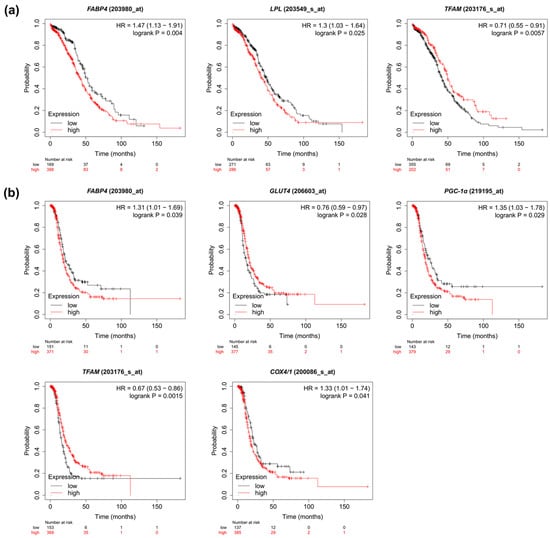
Figure 5.
Kaplan–Meier curve analysis of (a) overall and (b) progression-free survival comparing high and low levels of the metabolism-associated genes.
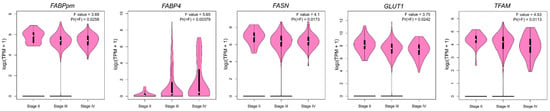
Figure 6.
Violin plots representing significant correlations (p < 0.05) between gene expression and clinical stage based on the TCGA-OC dataset.
3. Discussion
Metabolic reprogramming has been recognized as a hallmark of tumor development because it is required to sustain the energy supply for cancer progression and metastatic dissemination. The use of nutrients depends on their availability in the tumor microenvironment and reflects the capacity for cellular genetic alterations in response to stress conditions. To explore metabolite reliance in OC, we assessed the mRNA expression of numerous proteins required for transport of fatty acids (CD36/SR-B2, FABPpm, FATP1, FATP4), glucose (GLUT1, GLUT4), monocarboxylates (MCT1, MCT4) and amino acids (LAT1, ASCT2, SNAT1), and also evaluated the levels of several lipid metabolism-related (FASN, LPL) and mitochondrial genes (PGC-1α, TFAM, β-HAD and COX4/1). Moreover, association analysis of the above parameters with the clinical and histological characteristics of patients was performed. Finally, the transcriptome profiles from the TCGA-OC dataset were used to associate the above-mentioned genes’ expression with the clinical prognosis of OC patients.
3.1. Glucose Transporters
In our study, the GLUT1 transcription level exerted a significantly higher expression in HGSC tissues compared to controls. This result is consistent with previous studies, which revealed that GLUT1 mRNA and protein expression were enhanced in primary OC compared to other glucose transporters [,]. Moreover, concerning the stage of OC lesions, Cantuaria et al. revealed that there is a considerable correlation between the stage of OC and the expression of GLUT1, indicating that GLUT1 level is enhanced in malignant tumors compared to borderline tumors []. The hallmark feature of cancer cells is elevated glucose utilization by anaerobic processes such as glycolysis, even in the presence of oxygen, which is known as Warburg effect []. As a result of inefficient ATP production in anaerobic processes compared to oxidative phosphorylation, cancer cells are highly dependent on an enhanced influx of glucose []. In order to facilitate glucose uptake to maintain adequate energy supply, cancer cells enhance the expression of key factors of the glycolytic pathway, including glucose transporters (GLUTs) as well as other glycolytic enzymes []. The regulation of GLUT1 expression may be the reason why this glucose transporter is most prominently expressed in OC compared to other GLUTs. OC has the propensity to metastasize to the peritoneum; thus, OC cells and intraperitoneal metastases are susceptible to oxygen deprivation in the cancer microenvironment, and therefore hypoxia influences adaptive mechanisms in OC []. Hypoxia-inducible factor (HIF) is the main triggering factor responsible for the upregulation of GLUT1 []. Therefore, adaptive mechanisms to hypoxia may be a possible reason for the augmentation of GLUT1 expression in OC. In addition, genetic alterations such as P53 mutation, which is a triggering factor in developing OC, also contribute to GLUT1 upregulation []. On the other hand, the regulation mechanism of GLUT4 expression is not dependent on HIF-1α but rather is triggered by insulin. In our study, the expression of GLUT4 was decreased and there is scarce information regarding the role and regulation of GLUT4 and insulin in OC []. Finally, the overexpression of GLUT1 is related to high-grade and poorly differentiated tumors in OC and is associated with a detrimental malignant nature of the cancer and a dismal prognosis [,]. Interestingly, the application of GLUT1 inhibitor BAY-876 in OC diminished the tumor growth by 50–71%, and decreased glucose influx and consumption in both in vitro and in vivo murine models []. Interestingly, ciglitazone, an antidiabetic drug, exerted an anti-proliferating effect on OC cells in vitro by altering GLUT1 abundance in the plasma membrane, thus regulating glucose utilization and mitigating the rapid growth of OC []. Other research revealed that the GLUT1 inhibitor STF 31 and metformin, combined with chemotherapy, increased therapy efficiency in sensitive and resistant OC []. In addition, targeting GLUT4 appears to have therapeutic potential to hinder the development of OC. A recent study reported that apatinib, which inhibits VEGFR2/AKT1/GSK3β/SOX5/GLUT4 pathway, decreased glucose utilization in animal models in OC and exerted an anti-proliferating effect [].
3.2. Monocarboxylate Transporters
The enhanced expression of MCT4 mRNA in OC observed in our study is consistent with previous data that outline the augmented expression of MCT4 under hypoxic conditions and the prognostic value of MCT4 in several human cancers, e.g., cervical cancer, osteosarcoma, prostate cancer, gliomas, bladder cancer [,,]. MCT4 is an H+-coupled symporter that exports an excessive amount of lactate from cancer cells and is prominently expressed in metastatic tumors in a highly hypoxic environment []. Due to the Warburg effect, the neoplastic microenvironment is abundant in lactate and pyruvate []. Recently, it was revealed that MCT4 has a higher affinity for lactate compared to pyruvate and can export lactate even to a high lactate environment (MCT1 and MCT2 do not present this feature). These characteristics indicate that the expression of MCT4 takes precedence in metastatic cancers because their cells divide extensively and produce huge amounts of lactate, leading to increased acidity in the extracellular environment. Thus, the overexpression of MCT4 is an adaptive mechanism to prevent the deleterious effects of an acidic environment and hypoxia in a neoplastic environment []. Furthermore, recent studies demonstrated that MCT4 expression is triggered by hypoxia and mediated by HIF-1α, while MCT1 expression is not enhanced in this case []. Highly hypoxic areas are usually increased in large tumor lesions, along with inadequate blood flow and ascites in the tumor milieu []. Therefore, HIF-1α can be a plausible factor contributing to resistance to carboplatin via alterations in cancer cell metabolism by enhanced MCT4 expression []. The observation mentioned above highlights that the microenvironment of OC and its concomitant hypoxia contribute to the metabolic switch that results in cancer progression and treatment implications. Moreover, MCT4 expression and activity in OC are Influenced by chaperone CD147, and both are associated with dismal overall survival in several cancers []. The expression of CD147 was enhanced in most ovarian tumors induced by hypoxia, wherein CD147 served as an ancillary molecule for MCT-mediated lactate transport []. It has been shown that increased levels of MCT1, MCT4 and chaperone CD147 contribute to the development of EOC, and were correlated with high-grade tumors and ascites but not with histological type. However, only enhanced expression of MCT4, not of MCT1 or CD147, correlated with the likelihood of OC recurrence. Additionally, augmented expression of MCT4, MCT1 and chaperone CD147 was coupled with elevated levels of MDR1–multidrug resistance marker in OC. The expression of these molecules was detected in all metastatic lesions of OC after chemotherapy, so it can be stated that these molecule-mediated processes can lead to chemotherapy resistance []. Therefore, the MCT4-dependent mechanisms that contribute to EOC relapse need to be elucidated, because this transporter can potentially be used in targeted therapy in drug-resistant OC. The study conducted on mice with OC revealed that elevated MCT4 concentration yielded increased mouse mortality []. Interestingly, enhanced expression of MCT4 was observed in tumor stroma, especially in cancer-associated fibroblasts in breast cancer, whereas MCT1 was preferentially expressed in epithelial cancer cells and contributed to the influx of lactate to cancer cells. Thus, catabolic stromal cells transport lactate to epithelial cells, which have to fulfill high requirements for energy substances [].
Recent studies demonstrate that synergy of the Warburg effect and reverse Warburg (lactate efflux) is thought to be a basis of cancer metabolism; however, data on the alterations in expression of metabolism-related transporters in OC are limited []. There is compelling evidence that enhanced GLUT1 expression and MCT4 expression are strongly associated with cancer progression, while the augmented expression of these transporters may be an adaptive mechanism to increased anaerobic glycolysis even under normoxia in order to maintain an adequate energy supply to an intensely proliferating cancer cell []. To sum up, GLUT1 mediates glucose influx and MCT4 induces lactic acid efflux, which are interdependent processes in cancer metabolism []. The enhanced glycolysis and maintenance of pH equilibrium in the case of intensive lactate production are hallmark abilities that contribute to progression from in situ to invasive cancer []. While blocking an individual MCT has been ineffective to restrain cancer growth (i.e., cancer cells escape death using glucose and glutamine as energy sources, or alternatively overexpress the other MCT isoform), combined inhibition is cytotoxic when paralleled by loss of mitochondrial NAD+ regenerating capacity due to suppression of glycolytic ATP production [].
3.3. Fatty Acid Transporters
Despite the upregulation of glucose transporters and glucose-dependent metabolism in several human cancers, there is scarce data on fatty acid transporters in malignant cells. In a nutrient-deprived microenvironment (limited glucose availability) cancer cells’ metabolism shifts and fatty acid β-oxidation has a prominent role in maintaining an adequate energy supply for cancer cells []. Due to the evolving evidence for the cooperation between OC and adipocytes in the omentum, the alterations in lipid metabolism in HGSC are regarded as crucial factors in dissemination to the peritoneum and omentum []. In our study, however, we noticed reductions in the level of fatty acid transporters such as CD36/SR-B2, FATP1 and FABP4, which were associated with lower LPL and mitochondrial β-HAD levels. The upregulation of glucose-related transporters coupled with a decrease in the expression of fatty acid transporters could be the cause of malignant metabolic alteration in cancers. There is compelling evidence that FABP4 chaperone protein and CD36/SR-B2 constitute pivotal regulatory factors of fatty acid transport in human cancer and are upregulated in several human cancers, e.g., breast cancer, colon cancer, cervical cancer [,,]. FABP4 was observed in abundance in metastatic OC, which is discrepant with our study. Recent studies indicate that adipocytes and OC cells inevitably cooperate in the lipid supply and that the fatty acid transport between adipocytes and OC cells is mediated by FABP4 []. Congruently, the overexpression of FABP4 was associated with cancer-related adipocytes, and FABP4 mediated the transport of lipids from adipocytes to OC cells. Moreover, the overexpression of FABP4 increased the likelihood of formation of peritoneal metastases in OC []. In another study, the overexpression of FABP4 was associated with an adaptive mechanism and resulted in a decrease in lipid droplet formation coupled with ROS production. However, Nieman et al. did not demonstrate in their study enhanced expression of FABP4 in OC cells incubated without adipocytes, and the expression of FABP4 was enhanced in metastases compared to primary ovarian tumors []. The possible explanation for this discrepancy with our results is the fact that our study was conducted on OC at primary sites but not on metastases of OC from the peritoneum, which is surrounded by a lipid-abundant microenvironment. Indeed, some researchers reported that the suppression of FABP4 diminished the ability of an enhanced 5-hydroxymethylcytosine formation in DNA to invade the high-lipid cancer milieu, leading to decreased expression of genes involved in OC dissemination and enhanced susceptibility to chemotherapy both in vitro and in vivo. Thus, there is plausible evidence that targeting fatty acid transporters can be used as an alternative therapy in OC metastases []. Taken together, a prominent role of FABP4 is associated with metastases in the peritoneum, which develop in a microenvironment abundant in lipids that necessitates a linkage with the bioavailability of lipids in visceral fat.
CD36/SR-B2 is involved in binding and subsequently mediating the influx of long-chain fatty acids, oxidized lipids and phospholipids to cancer cells []. The findings of recent studies showed that cancers with enhanced expression of CD36/SR-B2, induced by a high-fat diet or by palmitic acid in mouse models of human oral cancer, have a proclivity for metastasizing []. Abundant presence of CD36/SR-B2 in OC was observed with concomitant adipocytes in the microenvironment. Previous research proved that OC cell lines co-cultured with human adipocytes have enhanced expression of CD36/SR-B2, which augments fatty acid influx to cancer cells. However, other FA transporters, such as FABPpm, FATP1 and FATP4, remained unaffected. Accordingly, CD36/SR-B2 is overexpressed in peritoneal metastases in OC compared to primary OC [], whereas our study was mostly conducted on OC at primary sites and diminished expression of CD36/SR-B2 was detected. Nevertheless, promising results were demonstrated in animal models with the use of anti-CD36/SR-B2 antibodies, which exerted growth-inhibitory effects and considerably limited the number of metastases [,].
Unlike other fatty acid transporters, FABPpm showed increased transcript content in OC tissue compared to the control group, with a positive correlation between the expression of FABPpm and BMI. It is hard to interpret these data since our study is one of the first to investigate the expression of FABPpm mRNA in OC. Therefore, future studies are necessary to determine FABPpm’s role in cancer progression.
3.4. Amino Acid Transporters
A vast number of studies have demonstrated that the expression of amino acid transporters LAT1 and ASCT2 is enhanced in tumor tissues, e.g., colorectal cancer, suggesting a crucial role of amino acid supply in fulfilling the high proliferation rate in cancer cells []. However, scarce information is available concerning the relationship of amino acid transporter expression with cell growth in human ovarian cancer. In our study, we showed that the level of ASCT2 expression was decreased, while LAT1 mRNA expression remained similar in both control and cancer groups. Previous studies demonstrated that LAT1 expression was increased in vitro in OC cell lines. However, the suppression of LAT1 did not cause a significant decrease in cell anchorage-dependent growth of both SKOV3 and IGROV1 cells in a liquid medium []. This finding suggests that the increase in LAT1 expression in the ovarian cell lines is not triggered by the OC cell proliferation mechanism, but rather that there are some other pathophysiological mechanisms that cause the increase in LAT1 expression in this cell line. These results also indicate that other energy substrate transporters play a more prominent role in OC cell proliferation, because the inhibition of LAT1 action does not alter the growth of OC cells. The lack of LAT1 alterations in our study may be due to the fact that study [] was conducted on OC cell lines, whereas ours was based on surgical specimens. Moreover, another study revealed that LAT1 shows enhanced expression in clear cell carcinoma and low presence in serous carcinoma compared to other histological types. This result is consistent with results of our study conducted on HGSC. These findings could prove that the overexpression of LAT1 is associated with certain OC types, especially clear cell carcinoma [,]. Furthermore, LAT1 expression was enhanced in G1 adenocarcinoma compared to G3 adenocarcinoma. These results are consistent with our studies, conducted mostly on advanced OC (FIGO III and IV in 23 out of 27 patients), which reported no differences in LAT1 mRNA expression. That study also proved the inverse correlation between LAT1 expression and p53 expression []. This phenomenon acknowledges that high LAT1 expression is not associated with the genetic basis of HGSC.
ASCT2 provides the compounds for tumor growth and progression but also maintains an adequate energy supply []. Literature results describing the relationships involving ASCT2 mRNA expression in OC are inconsistent. Some studies reported that ASCT2 expression has a linkage with FIGO stage and a correlation with unfavorable overall survival in OC patients []. TCGA and other studies demonstrated no alterations in ASCT2 mRNA expression in OC cell lines [], while herein we observed lowered ASCT2 levels in OC.
Unlike the above transporters, which are important for rapid amino acid exchange between the cell and its milieu to maintain amino acid homeostasis, SNAT1 is known to mediate net glutamine uptake to sustain glutaminolysis at a level that supports malignant hallmarks []. In line with that, we showed that primary ovarian tumors exhibited elevated SNAT1 expression to fulfill their proliferative drive. SNAT1 level was associated with survival time and metastasis status, while silencing of the gene in human melanoma [] and breast cancer cell lines [] promoted senescence and diminished cell migration rate, thus indicating it as an important target in anticancer therapy. Indeed, recent in vitro studies with functional inhibition of SNAT1 [] or glutamine deprivation [] reduced cancer cell proliferation and migration.
3.5. Mitochondrial Genes
Mitochondrial adaptive mechanisms are centrally important for cancer cell survival in the face of environmental stresses, including hypoxia and chemotherapeutic drugs. The role of PGC-1α, a major regulator of mitochondrial biosynthesis and antioxidant activator, in OC remains controversial, since some studies reported a significant increase in PGC-1α and its target (i.e., TFAM) protein levels in OC samples relative to controls [], which could mediate chemoresistance by reducing apoptosis [,]. The knockdown of PGC-1α or TFAM thereby increased sensitivity to cisplatin []. Contradicting these observations, other studies showed that HGSCs exhibited higher expression of PGC-1α/TFAM compared to clear cell carcinoma, which was related to better prognosis and chemosensitivity to initial platin and taxane therapy []. These data were also supported by in vitro experiments, wherein PGC-1α overexpression led to the apoptosis of OC cells mediated by a decreased Bcl-2/Bax ratio []; therefore, high expression of mitochondrial markers (TFAM and TIMM23) was considered beneficial in HGSC cell lines []. Although, in the TCGA cohort and in our study, there were no significant differences between PGC-1α expression in control and cancer tissue, this transcriptional co-activator level varied relative to BMI and cancer characteristics (Table A1 and Table S1). Therapeutic implications of these associations are still unclear, but the involvement of mitochondrial proteins in the pathogenesis of HGSC and its chemosensitivity are evident.
The overt limitation of our study was the limited number of tested samples, which could impede adequate interpretation of the energy substrate transporter expression in OC. Another obstacle is the composition of tested tissue samples. In our research, ovarian tissue contained both epithelial and stromal cells. However, OC grows from the ovarian epithelium. Thus, analysis of the discrepancies in expressed genes in OC compared to control samples may pose a threat of misinterpretation because of an abundance of ovarian stromal cells, which may differ in the expression of diverse energy substrate transporters [].
4. Materials and Methods
4.1. Study and Control Group
The present study conforms with the guidelines delineated in the Declaration of Helsinki and was approved by the Ethics Committee at the Medical University of Bialystok (permission number APK.002.221.2021). The samples were obtained by Biobank team at Medical University of Bialystok immediately after primary tumor removal. The biobanking serves as a unique entity dedicated to the collection of patients’ biological material according to the Standard Operating Procedures, which ensure the highest quality systematic biobanking, novel imaging techniques, and advanced molecular analysis for precise tumor diagnosis and therapy: The Polish MOBIT project. Scraps were precisely selected, cut into pieces that consisted of endothelial and stromal cells, snap-frozen in liquid nitrogen and thereafter stored in tanks with liquid nitrogen. In total we obtained 158 OC tissues during surgical procedures. Samples were screened for eligibility, but 119 patients met the exclusion criteria (other histological type of OC, comorbidities such as diabetes, L-thyroxine intake, hyperlipidemia, BMI > 35, other metabolic disorders). In 12 cases technical problems occurred, or the quality of the material was unsatisfactory. Ultimately, the study cohort included 27 patients with HGSC (cases were diagnosed from 2017 to 2021). The control group included healthy ovarian tissues obtained from non-oncological patients (14 met the inclusion criteria). Additionally, we analyzed clinical parameters obtained before surgery and patient questionnaires (diet, smoking history, family history, ECOG Performance Status Scale (WHO-Zubrod score), signs and symptoms). None of the patients received any treatment (chemotherapy, radiotherapy, or hormone therapy) before surgery.
4.2. Real-Time PCR Analysis
Total RNA was extracted using the NucleoSpin RNA Plus Kit with RNase-free DNase I treatment (Ambion, Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s protocol. RNA quantity and quality measurements were performed using spectrophotometry (at an absorbance OD ratio of 260/280 and 260/230). Total RNA (1 µg) served as a template for first-strand cDNA synthesis using the EvoScript universal cDNA master kit (Roche Molecular Systems, Boston, MA, USA). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the LightCycler 96 System real-time thermal cycler with FastStart essential DNA green master (Roche Molecular Systems). The following reaction parameters were applied: 15 s denaturation at 94 °C, 30 s annealing at 60 °C for CD36/SR-B2, FATP1, FATP4, FABPpm, FABP4, GLUT1, GLUT4, FASN and β-actin or 61 °C for MCT1, MCT4, LAT1, ASCT2, SNAT1, PGC-1α, TFAM, β-HAD, COX4/1 and LPL, followed by 30 s extension at 72 °C for 45 cycles. PCR product specificity was verified by melting curve analysis. Reactions were run in duplicates and the expression was normalized against the housekeeper gene (β-actin). Results were calculated using the relative quantification method modified by Pfaffl []. The primers used in the study are listed in Table 4.

Table 4.
Primer sequences used for real-time PCR.
4.3. Public Data Mining
To screen for transcriptional metabolic dysregulation in ovarian cancer from a large patient cohort, we used TCGA RNA-sequencing data from The Gene Expression Profiling Interactive Analysis (GEPIA) database. GEPIA contains validated gene expression data from the TCGA-OC cohort and the Genotype-Tissue Expression project (GTEx) based on normal ovarian tissues []. The association between the mRNA expression of metabolism-related genes in TCGA and overall survival and progression-free survival in ovarian cancer patients was evaluated using Kaplan–Meier plots [].
4.4. Statistical Analysis
Statistical analyses were performed with GraphPad Prism software version 8.0 (GraphPad Software, Inc., San Diego, CA, USA). The Shapiro–Wilk test (test for normality) and Levene tests (test for homogeneity of variances) were used to determine the application of parametric or non-parametric methods. Afterwards, Student’s t-test or Mann–Whitney U test was used to compare the differences between the groups. For multiple comparisons, the Kruskal–Wallis test followed by Dunn’s post-hoc test was applied. The relationships between the analyzed parameters were assayed with Spearman’s correlation coefficient. A log-rank test was applied to compare high and low levels of metabolism-related gene expression in Kaplan–Meier curves. Statistical hypotheses were verified at the 0.05 significance level.
5. Conclusions
The successful outgrowth and aggressiveness of OC relies on cellular metabolic flexibility as a response to the microenvironmental context and nutrient availability (Figure 7). Our results suggest that the metabolism of ovarian cancer is highly reliant on glucose influx mediated by GLUT1. Moreover, given the fact that the OC environment is acidic [], the concomitant increase in MCT4 plays a pivotal role in maintaining Ph balance. Glycolysis also provides indirect metabolic precursors for the biosynthesis of nonglucidic acids crucial in amino acid and lipid transformation []; therefore, it may be a principal metabolic process in cancer cells. At the same time, the upregulation of amino acid transporter SNAT1 suggests high glutamine dependence in ovarian cancer cells. Overall, our results and TCGA data analysis reveal differences in the metabolic biology of healthy and cancer cells that indicate potential therapeutic intervention targets to impede ovarian cancer cells’ proliferation and metastasis.
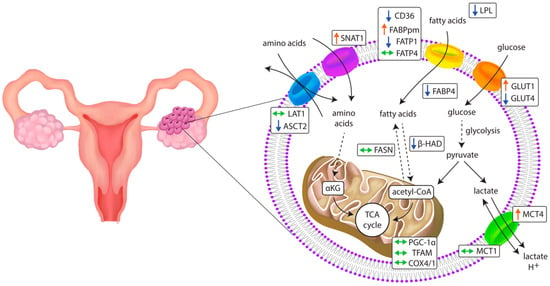
Figure 7.
Changes in the expression of genes related to metabolic pathways in primary ovarian cancer tissue. ↑: increase; ↓: decrease; ↔: unchanged.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23168968/s1.
Author Contributions
Conceptualization, M.B., E.S. and P.K.; methodology, M.B. and A.C.; formal analysis, E.S. and K.K.; investigation, M.B. and E.S.; resources, M.B., P.M., J.D. and E.S.; data curation, M.B., E.S. and K.K.; writing—original draft preparation, K.B., M.B. and E.S.; writing—review and editing, P.K. and P.G.; visualization, M.B. and E.S.; supervision, P.K. and A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Medical University of Bialystok grant number SUB/1/DN/22/007/1118.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee at the Medical University of Bialystok (permission number APK.002.221.2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga (accessed on 1 August 2022).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Appendix A

Table A1.
Relative gene expression in relation to clinical–pathological parameters. The values are presented as median (min–max).
Table A1.
Relative gene expression in relation to clinical–pathological parameters. The values are presented as median (min–max).
| Gene | FIGO I and II vs. FIGO III and IV 1 | N0 vs. N1+N2 (Lymph Node Metastasis) 2 | Lack of vs. ‘Omental-Cake’ | Nodal Invasion > Omental Invasion vs. Nodal Invasion < Omental Invasion 3 |
|---|---|---|---|---|
| CD36/SR-B2 | p = 0.5756 0.61 (0.125–5.529) vs. 0.296 (0.077–2.746) | p = 0.2773 0.717 (0.077–5.529) vs. 0.283 (0.135–1.427) | p = 0.5806 0.482 (0.125–5.529) vs. 0.29 (0.077–2.746) | p = 0.2371 0.268 (0.135–0.486) vs. 0.472 (0.17–2.746) |
| FABPpm | p = 0.5756 1.488 (1.021–4.368) vs. 1.266 (0.273–4.859) | p = 0.2562 1.49 (0.961–4.802) vs. 1.226 (0.273–4.859) | p = 0.3473 1.125 (0.273–4.859) vs. 1.422 (0.594–4.802) | p = 0.6636 1.294 (1.198–1.389) vs. 1.472 (0.594–4.029) |
| FATP1 | p = 0.9215 0.69 (0.345–1.828) vs. 0.642 (0.272–3.788) | p = 0.4559 0.849 (0.307–3.788) vs. 0.637 (0.272–3.403) | p = 0.4856 0.6 (0.307–3.788) vs. 0.868 (0.272–3.403) | p = 0.5608 0.436 (0.436–0.436) vs. 0.9442 (0.2719–3.403) |
| FATP4 | p = 0.9737 1.193 (0.314–3.066) vs. 0.867 (0.234–2.811) | p = 0.5480 0.98 (0.314–3.066) vs. 0.867 (0.234–2.445) | p = 0.2773 0.831 (0.234–3.066) vs. 1.053 (0.429–2.811) | p = 0.1607 0.665 (0.234–0.899) vs. 1.259 (0.513–2.811) |
| FABP4 | p ≥ 0.9999 0.881 (0.046–2.094) vs. 0.2 (0.013–26.74) | p = 0.2773 0.79 (0.046–26.74) vs. 0.161 (0.013–22.11) | p = 0.2773 0.199 (0.013–10.11) vs. 0.474 (0.015–26.74) | p = 0.3083 0.0472 (0.0429–1.354) vs. 1.416 (0.114–26.74) |
| GLUT1 | p = 0.5314 1.112 (0.157–2.911) vs. 0.76 (0.076–4.324) | p = 0.1138 1.109 (0.157–2.911) vs. 0.519 (0.076–4.324) | p = 0.4856 1.044 (0.076–4.324) vs. 0.626 (0.125–2.495) | p = 0.698 0.155 (0.076–1.17) vs. 0.3119 (0.125–2.397) |
| GLUT4 | p = 0.2719 0.209 (0.155–2.327) vs. 0.141 (0.013–1.957) | p = 0.1385 0.334 (0.028–2.327) vs. 0.06 (0.013–1.957) | p = 0.8291 0.155 (0.013–2.327) vs. 0.155 (0.013–1.957) | p = 0.344 0.034 (0.017–0.126) vs. 0.169 (0.037–1.957) |
| MCT1 | p = 0.5314 1.895 (0.295–2.387) vs. 1.025 (0.315–6.052) | p = 0.2562 1.569 (0.295–6.052) vs. 0.952 (0.315–3.801) | p = 0.4271 1.256 (0.295–6.052) vs. 0.93 (0.375–3.801) | p = 0.7989 1.115 (0.921–1.256) vs. 0.835 (0.375–3.801) |
| MCT4 | p = 0.9183 0.812 (0.317–8.742) vs. 0.777 (0.259–2.225) | p = 0.5267 0.869 (0.259–8.742) vs. 0.605 (0.316–2.225) | p = 0.0869 0.995 (0.316–8.742) vs. 0.516 (0.259–1.538) | p = 0.0419 1.497 (0.357–1.609) vs. 0.481 (0.259–0.688) |
| LAT1 | p = 0.8693 0.549 (0.05–3.221) vs. 0.466 (0.139–8.853) | p = 0.7551 0.416 (0.051–3.221) vs. 0.47 (0.179–8.853) | p = 0.5164 0.47 (0.051–8.853) vs. 0.416 (0.139–1.300) | p = 0.1864 0.47 (0.179–8.853) vs. 0.474 (0.191–1.289) |
| ASCT2 | p = 0.3715 0.86 (0.456–1.663) vs. 0.566 (0.066–2.349) | p = 0.4559 0.777 (0.249–2.349) vs. 0.546 (0.066–1.278) | p = 0.6833 0.566 (0.066–2.349) vs. 0.737 (0.22–1.278) | p = 0.5431 0.522 (0.254–1.135) vs. 0.841 (0.296–1.153) |
| SNAT1 | p = 0.1910 1.951 (1.582–3.27) vs. 1.363 (0.516–3.996) | p = 0.1385 1.94 (0.516–3.27) vs. 1.099 (0.645–3.996) | p = 0.3473 1.589 (0.822–3.398) vs. 1.300 (0.516–3.996) | p = 0.4262 0.994 (0.847–1.021) vs. 1.034 (0.516–3.996) |
| PGC-1α | p = 0.6217 0.49 (0.153–1.252) vs. 0.196 (0.024–6.298) | p = 0.7919 0.373 (0.024–6.298) vs. 0.181 (0.05–6.156) | p = 0.4559 0.177 (0.024–6.156) vs. 0.363 (0.098–6.298) | p = 0.0359 0.024 (0.05–0.077) vs. 1.002 (0.098–6.298) |
| TFAM | p = 0.2158 2.513 (1.643–2.950) vs. 1.516 (0.464–5.620) | p = 0.6833 1.838 (0.647–3.301) vs. 1.514 (0.464–5.62) | p = 0.8667 1.643 (0.513–4.308) vs. 1.838 (0.464–5.62) | p = 0.9088 1.514 (1.007–4.308) vs. 1.831 (0.464–5.62) |
| β-HAD | p = 0.8693 0.662 (0.374–2.543) vs. 0.685 (0.259–2.649) | p = 0.3420 0.94 (0.298–2.543) vs. 0.62 (0.259–2.649) | p = 0.9046 0.725 (0.259–2.649) vs. 0.62 (0.365–1.685) | p = 0.1967 0.349 (0.259–0.865) vs. 0.873 (0.372–1.685) |
| COX4/1 | p = 0.1243 2.088 (1.021–4.29) vs. 0.894 (0.287–6.43) | p = 0.2827 1.415 (0.401–4.47) vs. 0.894 (0.287–6.43) | p = 0.7826 1.021 (0.287–4.29) vs. 0.873 (0.305–6.43) | p = 0.7839 0.952 (0.8–2.14) vs. 0.736 (0.305–6.43) |
| FASN | p = 0.4085 0.576 (0.375–1.636) vs. 0.895 (0.341–7.296) | p = 0.6483 0.904 (0.374–2.334) vs. 0.654 (0.341–7.296) | p = 0.7919 0.822 (0.341–3.03) vs. 0.858 (0.374–7.296) | p = 0.2619 0.506 (0.341–0.602) vs. 0.668 (0.414–7.296) |
| LPl | p = 0.6688 0.541 (0.39–0.763) vs. 0.46 (0.072–4.689) | p = 0.0044 0.668 (0.348–4.689) vs. 0.276 (0.072–1.399) | p = 0.3229 0.39 (0.072–4.689) vs. 0.578 (0.12–3.437) | p = 0.1833 0.182 (0.072–1.06) vs. 0.748 (0.275–3.437) |
1 FIGO, International Federation of Gynecology and Obstetrics system. 2 American Joint Committee on Cancer (AJCC) TNM staging system. Spread to nearby lymph nodes, also called para-aortic lymph nodes (N). 3 Refer to Table 2.
References
- Huang, J.; Chan, W.C.; Ngai, C.H.; Lok, V.; Zhang, L.; Lucero-Prisno, D.E.; Xu, W.; Zheng, Z.J.; Elcarte, E.; Withers, M.; et al. Worldwide Burden, Risk Factors, and Temporal Trends of Ovarian Cancer: A Global Study. Cancers 2022, 14, 2230. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Prim. 2016, 2, 16061. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.M. The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am. J. Surg. Pathol. 2010, 34, 433–443. [Google Scholar] [CrossRef]
- Pereira, M.; Matuszewska, K.; Jamieson, C.; Petrik, J. Characterizing Endocrine Status, Tumor Hypoxia and Immunogenicity for Therapy Success in Epithelial Ovarian Cancer. Front. Endocrinol. 2021, 12, 772349. [Google Scholar] [CrossRef]
- Freimund, A.E.; Beach, J.A.; Christie, E.L.; Bowtell, D.D.L. Mechanisms of Drug Resistance in High-Grade Serous Ovarian Cancer. Hematol. Oncol. Clin. N. Am. 2018, 32, 983–996. [Google Scholar] [CrossRef]
- Cho, H.; Lee, Y.S.; Kim, J.; Chung, J.Y.; Kim, J.H. Overexpression of glucose transporter-1 (GLUT-1) predicts poor prognosis in epithelial ovarian cancer. Cancer Investig. 2013, 31, 607–615. [Google Scholar] [CrossRef]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef]
- Bose, S.; Le, A. Glucose Metabolism in Cancer. Adv. Exp. Med. Biol. 2018, 1063, 3–12. [Google Scholar]
- Yoon, H.; Lee, S. Fatty Acid Metabolism in Ovarian Cancer: Therapeutic Implications. Int. J. Mol. Sci. 2022, 23, 2170. [Google Scholar] [CrossRef] [PubMed]
- Corn, K.C.; Windham, M.A.; Rafat, M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog. Lipid Res. 2020, 80, 101055. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.; Duke, G.; Heuer, T.S.; O’Farrell, M.; Wagman, A.S.; McCulloch, W.; Kemble, G. Fatty acid synthase—Modern tumor cell biology insights into a classical oncology target. Pharmacol. Ther. 2017, 177, 23–31. [Google Scholar] [CrossRef]
- Deberardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Supruniuk, E.; Żebrowska, E.; Chabowski, A. Branched chain amino acids—Friend or foe in the control of energy substrate turnover and insulin sensitivity? Crit. Rev. Food Sci. Nutr. 2021, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Moss, T.; Mangala, L.S.; Marini, J.; Zhao, H.; Wahlig, S.; Armaiz-Pena, G.; Jiang, D.; Achreja, A.; Win, J.; et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol. Syst. Biol. 2014, 10, 728. [Google Scholar] [CrossRef]
- Li, N.; Li, H.; Wang, Y.; Cao, L.; Zhan, X. Quantitative proteomics revealed energy metabolism pathway alterations in human epithelial ovarian carcinoma and their regulation by the antiparasite drug ivermectin: Data interpretation in the context of 3P medicine. EPMA J. 2020, 11, 661–694. [Google Scholar] [CrossRef]
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; Disaia, P.; Gabra, H.; Glenn, P.; et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Miethe, C.; Torres, L.; Beristain, J.; Zamora, M.; Price, R.S. The role of visfatin and resistin in an in vitro model of obesityinduced invasive liver cancer. Can. J. Physiol. Pharmacol. 2021, 99, 839–846. [Google Scholar] [CrossRef]
- Li, Z.; Liu, H.; He, J.; Wang, Z.; Yin, Z.; You, G.; Wang, Z.; Davis, R.E.; Lin, P.; Bergsagel, P.L.; et al. Acetyl-CoA Synthetase 2: A Critical Linkage in Obesity-Induced Tumorigenesis in Myeloma. Cell Metab. 2021, 33, 78–93.e7. [Google Scholar] [CrossRef]
- Beyaz, S.; Mana, M.D.; Roper, J.; Kedrin, D.; Saadatpour, A.; Hong, S.J.; Bauer-Rowe, K.E.; Xifaras, M.E.; Akkad, A.; Arias, E.; et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 2016, 531, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Morley, T.S.; Kim, M.; Clegg, D.J.; Scherer, P.E. Obesity and cancer—Mechanisms underlying tumour progression and recurrence. Nat. Rev. Endocrinol. 2014, 10, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Tworoger, S.S.; Huang, T. Obesity and ovarian cancer. In Recent Results in Cancer Research; Springer: Berlin/Heidelberg, Germany, 2016; Volume 208, pp. 155–176. [Google Scholar]
- Liu, Z.; Zhang, T.T.; Zhao, J.J.; Qi, S.F.; Du, P.; Liu, D.W.; Tian, Q.B. The association between overweight, obesity and ovarian cancer: A meta-analysis. Jpn. J. Clin. Oncol. 2015, 45, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Foong, K.W.; Bolton, H. Obesity and ovarian cancer risk: A systematic review. Post Reprod. Health 2017, 23, 183–198. [Google Scholar] [CrossRef]
- Mukherjee, A.; Chiang, C.Y.; Daifotis, H.A.; Nieman, K.M.; Fahrmann, J.F.; Lastra, R.R.; Romero, I.L.; Fiehn, O.; Lengyel, E. Adipocyte-induced FABP4 expression in ovarian cancer cells promotes metastasis and mediates carboplatin resistance. Cancer Res. 2020, 80, 1748–1761. [Google Scholar] [CrossRef]
- Rudlowski, C.; Moser, M.; Becker, A.J.; Rath, W.; Buttner, R.; Schroder, W.; Schurmann, A. GLUT1 mRNA and protein expression in ovarian borderline tumors and cancer. Oncology 2004, 66, 404–410. [Google Scholar] [CrossRef]
- Pizzuti, L.; Sergi, D.; Mandoj, C.; Antoniani, B.; Sperati, F.; Chirico, A.; Di Lauro, L.; Valle, M.; Garofalo, A.; Vizza, E.; et al. GLUT 1 receptor expression and circulating levels of fasting glucose in high grade serous ovarian cancer. J. Cell. Physiol. 2018, 233, 1396–1401. [Google Scholar] [CrossRef]
- Cantuaria, G.; Magalhaes, A.; Penalver, M.; Angioli, R.; Braunschweiger, P.; Gomez-Marin, O.; Kanhoush, R.; Gomez-Fernandez, C.; Nadji, M. Expression of GLUT-1 glucose transporter in borderline and malignant epithelial tumors of the ovary. Gynecol. Oncol. 2000, 79, 33–37. [Google Scholar] [CrossRef]
- Vaupel, P.; Multhoff, G. Revisiting the Warburg effect: Historical dogma versus current understanding. J. Physiol. 2021, 599, 1745–1757. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef]
- Barron, C.C.; Bilan, P.J.; Tsakiridis, T.; Tsiani, E. Facilitative glucose transporters: Implications for cancer detection, prognosis and treatment. Metabolism 2016, 65, 124–139. [Google Scholar] [CrossRef] [PubMed]
- Kalir, T.; Wang, B.Y.; Goldfischer, M.; Haber, R.S.; Reder, I.; Demopoulos, R.; Cohen, C.J.; Burstein, D.E. Immunohistochemical staining of GLUT1 in benign, borderline, and malignant ovarian epithelia. Cancer 2002, 94, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Ebert, B.L.; Firth, J.D.; Ratcliffe, P.J. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct cis-acting sequences. J. Biol. Chem. 1995, 270, 29083–29089. [Google Scholar] [CrossRef] [PubMed]
- Watson, R.T.; Pessin, J.E. Intracellular organization of insulin signaling and GLUT4 translocation. Recent Prog. Horm. Res. 2001, 56, 175–193. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, W.; Idowu, M.O.; Oh, U.; Wang, X.Y.; Temkin, S.M.; Fang, X. Ovarian cancer relies on glucose transporter 1 to fuel glycolysis and growth: Anti-tumor activity of BAY-876. Cancers 2019, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.J.; Kim, J.Y.; Kwon, S.Y.; Mun, K.-C.; Cho, C.H.; Ha, E. Ciglitazone Enhances Ovarian Cancer Cell Death via Inhibition of Glucose Transporter-1. Eur. J. Pharmacol. 2014, 743, 17–23. [Google Scholar] [CrossRef]
- Xintaropoulou, C.; Ward, C.; Wise, A.; Queckborner, S.; Turnbull, A.; Michie, C.O.; Williams, A.R.W.; Rye, T.; Gourley, C.; Langdon, S.P. Expression of Glycolytic Enzymes in Ovarian Cancers and Evaluation of the Glycolytic Pathway as a Strategy for Ovarian Cancer Treatment. BMC Cancer 2018, 18, 636. [Google Scholar] [CrossRef]
- Chen, L.; Cheng, X.; Tu, W.; Qi, Z.; Li, H.; Liu, F.; Yang, Y.; Zhang, Z.; Wang, Z. Apatinib Inhibits Glycolysis by Suppressing the VEGFR2/AKT1/SOX5/GLUT4 Signaling Pathway in Ovarian Cancer Cells. Cell. Oncol. 2019, 42, 679–690. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, X.; Huo, C.; Sun, C.; Zhu, J. Monocarboxylate Transporter 4 (MCT4) Overexpression Is Correlated with Poor Prognosis of Osteosarcoma. Med. Sci. Monit. 2019, 25, 4278–4284. [Google Scholar] [CrossRef]
- Reuss, A.M.; Groos, D.; Ghoochani, A.; Buchfelder, M.; Savaskan, N. MCT4 Promotes Tumor Malignancy in F98 Glioma Cells. J. Oncol. 2021, 2021, 6655529. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, B.; Yan, W.H.; Xia, Y.; Wang, Z.H.; Zheng, G.Y.; Wang, W.D.; Zhang, Y.S. Integrative Analysis Identified MCT4 as an Independent Prognostic Factor for Bladder Cancer. Front. Oncol. 2021, 11, 704857. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Baeza, Y.; Sandoval, P.Y.; Alarcón, R.; Galaz, A.; Cortés-Molina, F.; Alegriá, K.; Baeza-Lehnert, F.; Arce-Molina, R.; Guequén, A.; Flores, C.A.; et al. Monocarboxylate transporter 4 (MCT4) is a high affinity transporter capable of exporting lactate in high-lactate microenvironments. J. Biol. Chem. 2019, 294, 20135–20147. [Google Scholar] [CrossRef]
- San-Millán, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2017, 38, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.S.; Davies, A.J.; Halestrap, A.P. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1α-dependent mechanism. J. Biol. Chem. 2006, 281, 9030–9037. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. HIF-1: Upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 2010, 20, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Bovenzi, C.D.; Hamilton, J.; Tassone, P.; Johnson, J.; Cognetti, D.M.; Luginbuhl, A.; Keane, W.M.; Zhan, T.; Tuluc, M.; Bar-Ad, V.; et al. Prognostic Indications of Elevated MCT4 and CD147 across Cancer Types: A Meta-Analysis. BioMed Res. Int. 2015, 2015, 242437. [Google Scholar] [CrossRef]
- Yang, H.; Zou, W.; Chen, B. Overexpression of CD147 in ovarian cancer is initiated by the hypoxic microenvironment. Cell Biol. Int. 2013, 37, 1139–1142. [Google Scholar] [CrossRef]
- Chen, H.; Wang, L.; Beretov, J.; Hao, J.; Xiao, W.; Li, Y. Co-expression of CD147/EMMPRIN with monocarboxylate transporters and multiple drug resistance proteins is associated with epithelial ovarian cancer progression. Clin. Exp. Metastasis 2010, 27, 557–569. [Google Scholar] [CrossRef]
- Piga, I.; Verza, M.; Montenegro, F.; Nardo, G.; Zulato, E.; Zanin, T.; Del Bianco, P.; Esposito, G.; Indraccolo, S. In situ Metabolic Profiling of Ovarian Cancer Tumor Xenografts: A Digital Pathology Approach. Front. Oncol. 2020, 10, 1277. [Google Scholar] [CrossRef]
- Whitaker-Menezes, D.; Martinez-Outschoorn, U.E.; Lin, Z.; Ertel, A.; Flomenberg, N.; Witkiewicz, A.K.; Birbe, R.C.; Howell, A.; Pavlides, S.; Gandara, R.; et al. Evidence for a stromal-epithelial “lactate shuttle” in human tumors: MCT4 is a marker of oxidative stress in cancer-associated fibroblasts. Cell Cycle 2011, 10, 1772–1783. [Google Scholar] [CrossRef]
- Kim, S.; Jung, W.H.; Koo, J.S. The expression of Glut-1, CAIX, and MCT4 in Mucinous carcinoma. J. Breast Cancer 2013, 16, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Mogi, A.; Koga, K.; Aoki, M.; Hamasaki, M.; Uesugi, N.; Iwasaki, A.; Shirakusa, T.; Tamura, K.; Nabeshima, K. Expression and role of GLUT-1, MCT-1, and MCT-4 in malignant pleural mesothelioma. Virchows Arch. 2013, 462, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.; Robay, D.; Hindupur, S.K.; Pohlmann, J.; Colombi, M.; El-Shemerly, M.Y.; Maira, S.M.; Moroni, C.; Lane, H.A.; Hall, M.N. Dual Inhibition of the Lactate Transporters MCT1 and MCT4 Is Synthetic Lethal with Metformin due to NAD+ Depletion in Cancer Cells. Cell Rep. 2018, 25, 3047–3058.e4. [Google Scholar] [CrossRef]
- Massari, F.; Ciccarese, C.; Santoni, M.; Iacovelli, R.; Mazzucchelli, R.; Piva, F.; Scarpelli, M.; Berardi, R.; Tortora, G.; Lopez-Beltran, A.; et al. Metabolic phenotype of bladder cancer. Cancer Treat. Rev. 2016, 45, 46–57. [Google Scholar] [CrossRef]
- Zhao, G.; Cardenas, H.; Matei, D. Ovarian cancer—why lipids matter. Cancers 2019, 11, 1870. [Google Scholar] [CrossRef]
- Zeng, J.; Sauter, E.R.; Li, B. FABP4: A New Player in Obesity-Associated Breast Cancer. Trends Mol. Med. 2020, 26, 437–440. [Google Scholar] [CrossRef]
- Tian, W.; Zhang, W.; Zhang, Y.; Zhu, T.; Hua, Y.; Li, H.; Zhang, Q.; Xia, M. FABP4 promotes invasion and metastasis of colon cancer by regulating fatty acid transport. Cancer Cell Int. 2020, 20, 512. [Google Scholar] [CrossRef]
- Li, G.; Wu, Q.; Gong, L.; Xu, X.; Cai, J.; Xu, L.; Zeng, Y.; He, X.; Wang, Z. FABP4 is an independent risk factor for lymph node metastasis and poor prognosis in patients with cervical cancer. Cancer Cell Int. 2021, 21, 568. [Google Scholar] [CrossRef]
- Gharpure, K.M.; Pradeep, S.; Sans, M.; Rupaimoole, R.; Ivan, C.; Wu, S.Y.; Bayraktar, E.; Nagaraja, A.S.; Mangala, L.S.; Zhang, X.; et al. FABP4 as a key determinant of metastatic potential of ovarian cancer. Nat. Commun. 2018, 9, 2923. [Google Scholar] [CrossRef]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y. CD36 tango in cancer: Signaling pathways and functions. Theranostics 2019, 9, 4893–4908. [Google Scholar] [CrossRef] [PubMed]
- Pascual, G.; Avgustinova, A.; Mejetta, S.; Martín, M.; Castellanos, A.; Attolini, C.S.O.; Berenguer, A.; Prats, N.; Toll, A.; Hueto, J.A.; et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017, 541, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Ladanyi, A.; Mukherjee, A.; Kenny, H.A.; Johnson, A.; Mitra, A.K.; Sundaresan, S.; Nieman, K.M.; Pascual, G.; Benitah, S.A.; Montag, A.; et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 2018, 37, 2285–2301. [Google Scholar] [CrossRef] [PubMed]
- Dias, F.; Almeida, C.; Teixeira, A.L.; Morais, M.; Medeiros, R. Lat1 and asct2 related micrornas as potential new therapeutic agents against colorectal cancer progression. Biomedicines 2021, 9, 195. [Google Scholar] [CrossRef]
- Fan, X.; Ross, D.D.; Arakawa, H.; Ganapathy, V.; Tamai, I.; Nakanishi, T. Impact of system L amino acid transporter 1 (LAT1) on proliferation of human ovarian cancer cells: A possible target for combination therapy with anti-proliferative aminopeptidase inhibitors. Biochem. Pharmacol. 2010, 80, 811–818. [Google Scholar] [CrossRef]
- Sato, K.; Miyamoto, M.; Takano, M.; Furuya, K.; Tsuda, H. Significant relationship between the LAT1 expression pattern and chemoresistance in ovarian clear cell carcinoma. Virchows Arch. 2019, 474, 701–710. [Google Scholar] [CrossRef]
- Kaira, K.; Nakamura, K.; Hirakawa, T.; Imai, H.; Tominaga, H.; Oriuchi, N.; Nagamori, S.; Kanai, Y.; Tsukamoto, N.; Oyama, T.; et al. Prognostic significance of l-type amino acid transporter 1 (Lat1) expression in patients with ovarian tumors. Am. J. Transl. Res. 2015, 7, 1161–1171. [Google Scholar]
- Watanabe, J.; Yokoyama, Y.; Futagami, M.; Mizunuma, H.; Yoshioka, H.; Washiya, K.; Hana, K.; Endou, H.; Okayasu, I. L-type amino acid transporter 1 expression increases in well-differentiated but decreases in poorly differentiated endometrial endometrioid adenocarcinoma and shows an inverse correlation with p53 expression. Int. J. Gynecol. Cancer 2014, 24, 659–663. [Google Scholar] [CrossRef]
- Choi, Y.K.; Park, K.G. Targeting glutamine metabolism for cancer treatment. Biomol. Ther. 2018, 26, 19–28. [Google Scholar] [CrossRef]
- Guo, H.; Xu, Y.; Wang, F.; Shen, Z.; Tuo, X.; Qian, H.; Wang, H.; Wang, K. Clinical associations between ASCT2 and p-mTOR in the pathogenesis and prognosis of epithelial ovarian cancer. Oncol. Rep. 2018, 40, 3725–3733. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, Y.; Fang, W.; Liu, K.; Jiao, X.; Wang, Z.; Wang, J.; Zang, Y.S. Increased SNAT1 is a marker of human osteosarcoma and potential therapeutic target. Oncotarget 2017, 8, 78930–78939. [Google Scholar] [CrossRef] [PubMed]
- Bröer, A.; Rahimi, F.; Bröer, S. Deletion of amino acid transporter ASCT2 (SLC1A5) Reveals an essential role for transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to sustain glutaminolysis in cancer cells. J. Biol. Chem. 2016, 291, 13194–13205. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Cao, F.; Fang, W.; Hu, Y.; Chen, Y.; Ding, H.; Yu, G. Activation of SNAT1/SLC38A1 in human breast cancer: Correlation with p-Akt overexpression. BMC Cancer 2013, 13, 343. [Google Scholar] [CrossRef] [PubMed]
- Böhme-Schäfer, I.; Lörentz, S.; Bosserhoff, A.K. Role of Amino Acid Transporter SNAT1/SLC38A1 in Human Melanoma. Cancers 2022, 14, 2151. [Google Scholar] [CrossRef]
- Gwangwa, M.V.; Joubert, A.M.; Visagie, M.H. Effects of glutamine deprivation on oxidative stress and cell survival in breast cell lines. Biol. Res. 2019, 52, 15. [Google Scholar] [CrossRef]
- Signorile, A.; De Rasmo, D.; Cormio, A.; Musicco, C.; Rossi, R.; Fortarezza, F.; Palese, L.L.; Loizzi, V.; Resta, L.; Scillitani, G.; et al. Human ovarian cancer tissue exhibits increase of mitochondrial biogenesis and cristae remodeling. Cancers 2019, 11, 1350. [Google Scholar] [CrossRef]
- Li, Y.; Kang, J.; Fu, J.; Luo, H.; Liu, Y.; Li, Y.; Sun, L. Pgc1α promotes cisplatin resistance in ovarian cancer by regulating the hsp70/hk2/vdac1 signaling pathway. Int. J. Mol. Sci. 2021, 22, 2537. [Google Scholar] [CrossRef]
- Kim, B.; Jung, J.W.; Jung, J.; Han, Y.; Suh, D.H.; Kim, H.S.; Dhanasekaran, D.N.; Song, Y.S. PGC1α induced by reactive oxygen species contributes to chemoresistance of ovarian cancer cells. Oncotarget 2017, 8, 60299–60311. [Google Scholar] [CrossRef]
- Gabrielson, M.; Björklund, M.Y.; Carlson, J.; Shoshan, M. Expression of mitochondrial regulators PGC1a and TFAM as putative markers of subtype and chemoresistance in epithelial ovarian carcinoma. PLoS ONE 2014, 9, e107109. [Google Scholar] [CrossRef]
- Zhang, Y.; Ba, Y.; Liu, C.; Sun, G.; Ding, L.; Gao, S.; Hao, J.; Yu, Z.; Zhang, J.; Zen, K.; et al. PGC-1α induces apoptosis in human epithelial ovarian cancer cells through a PPARγ-dependent pathway. Cell Res. 2007, 17, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Kleih, M.; Böpple, K.; Dong, M.; Gaißler, A.; Heine, S.; Olayioye, M.A.; Aulitzky, W.E.; Essmann, F. Direct impact of cisplatin on mitochondria induces ROS production that dictates cell fate of ovarian cancer cells. Cell Death Dis. 2019, 10, 851. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, E45. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Izumi, H.; Torigoe, T.; Ishiguchi, H.; Uramoto, H.; Yoshida, Y.; Tanabe, M.; Ise, T.; Murakami, T.; Yoshida, T.; Nomoto, M.; et al. Cellular pH regulators: Potentially promising molecular targets for cancer chemotherapy. Cancer Treat. Rev. 2003, 29, 541–549. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).