Endoglin and MMP14 Contribute to Ewing Sarcoma Spreading by Modulation of Cell–Matrix Interactions
Abstract
:1. Introduction
2. Results
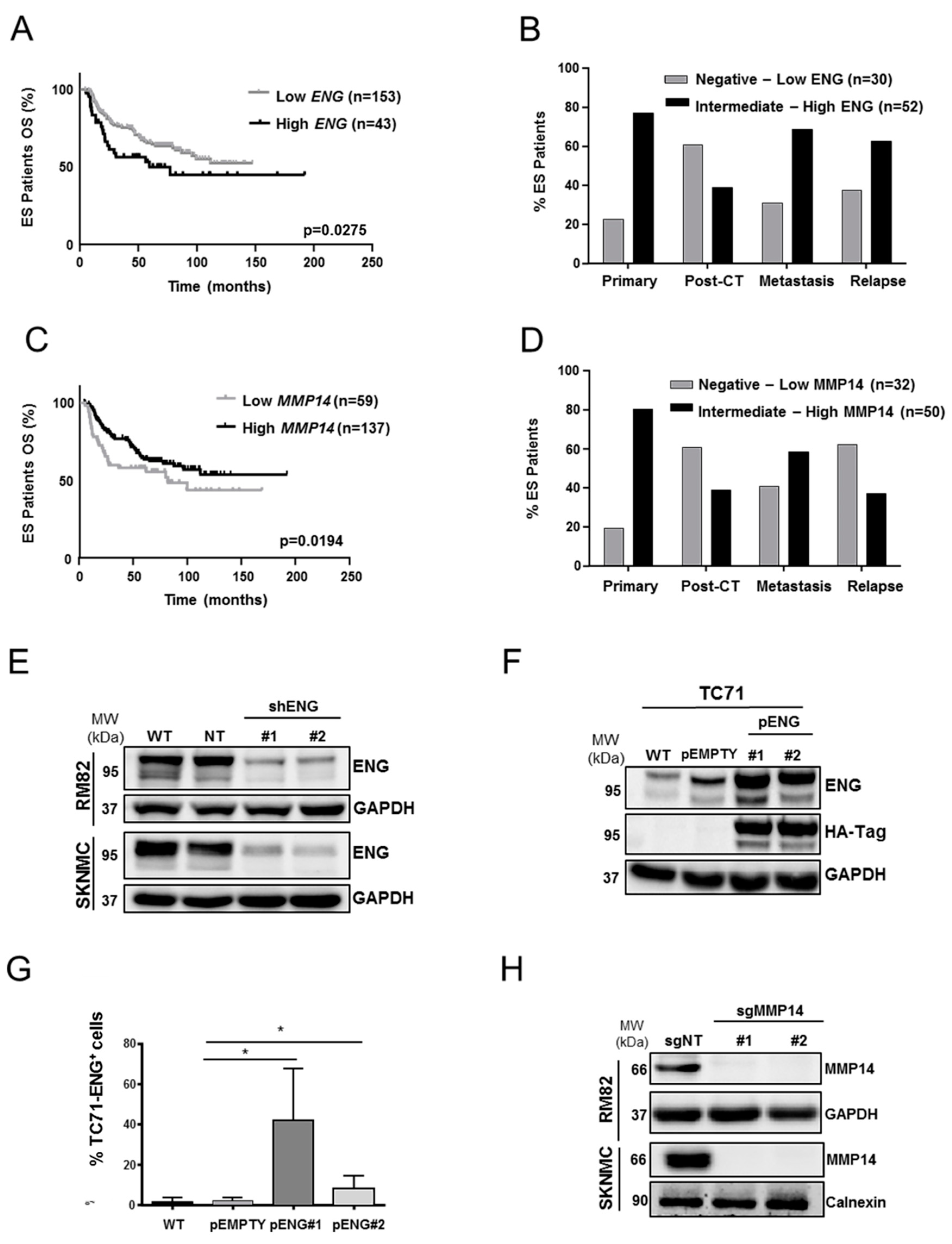
2.1. ENG and MMP14 Expression Predicts Prognosis of ES Patients
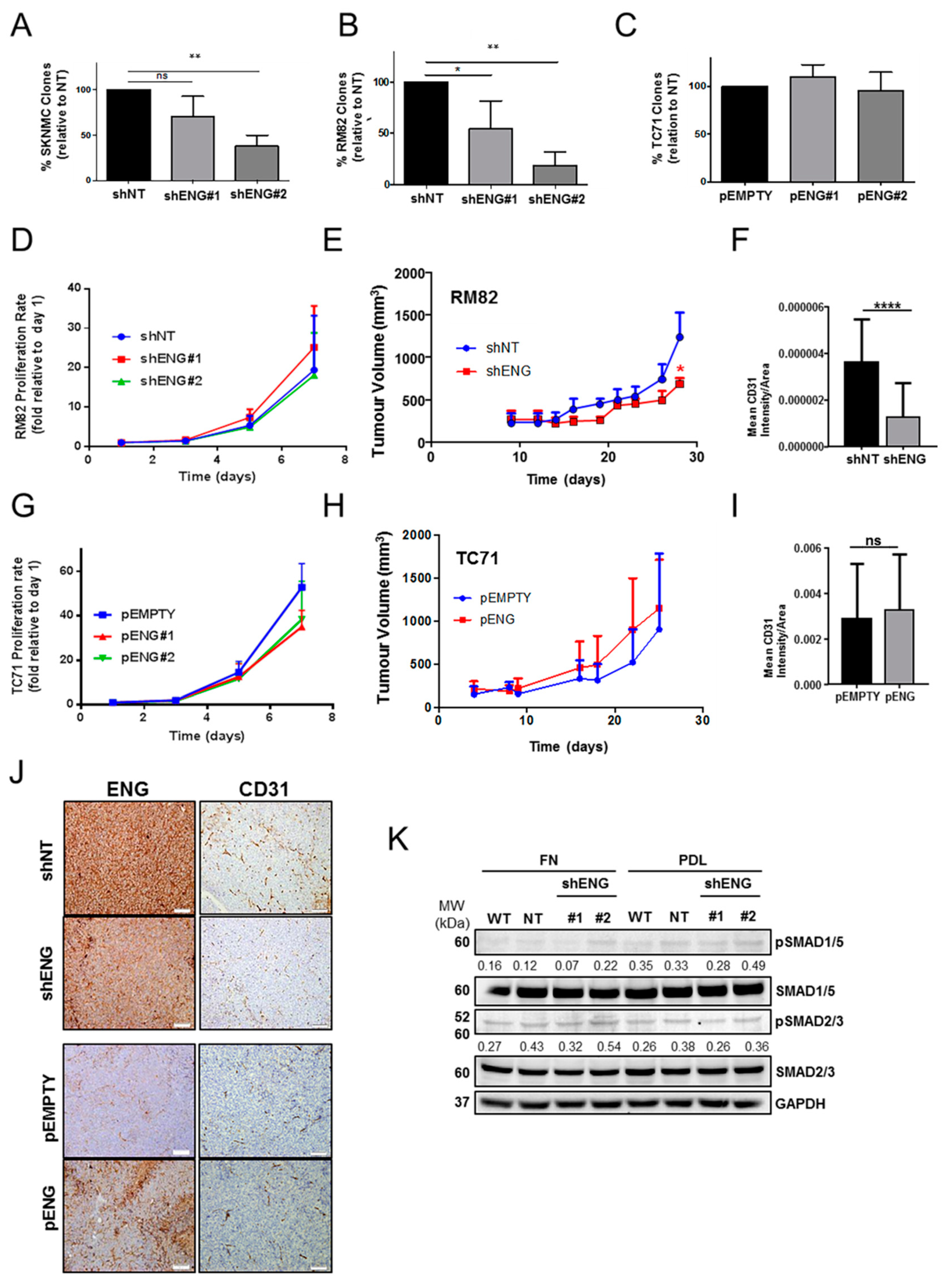
2.2. ENG Affects the Clonogenic Capacity and Neo-Angiogenesis of ES Cells
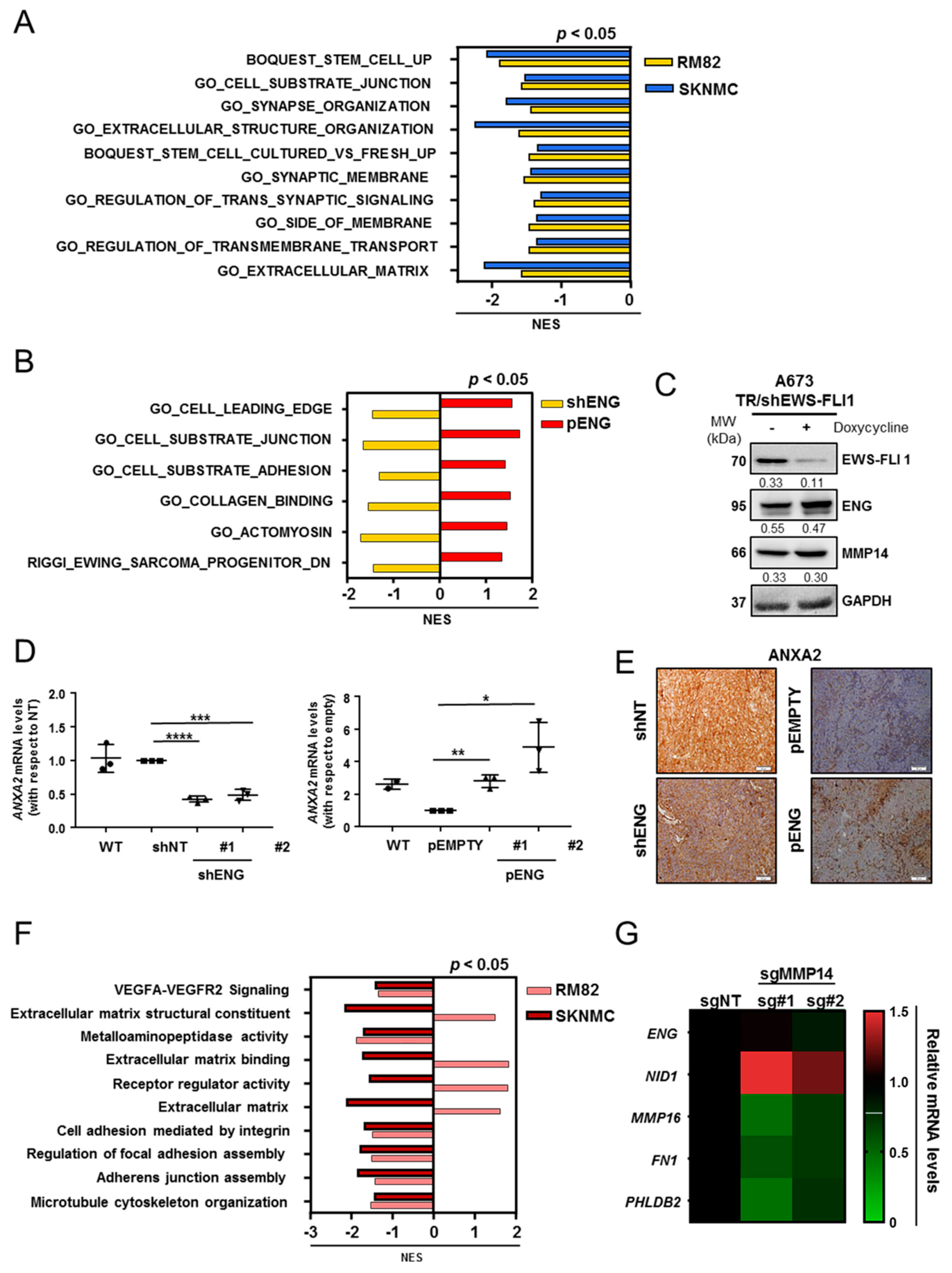
2.3. Comprehensive Transcriptomic Studies Suggest That ENG Is Associated with ES Cell Migration and Substrate Adhesion
2.4. MMP14 Is Involved in ECM Organization and Cell–Substrate Adhesion in ES
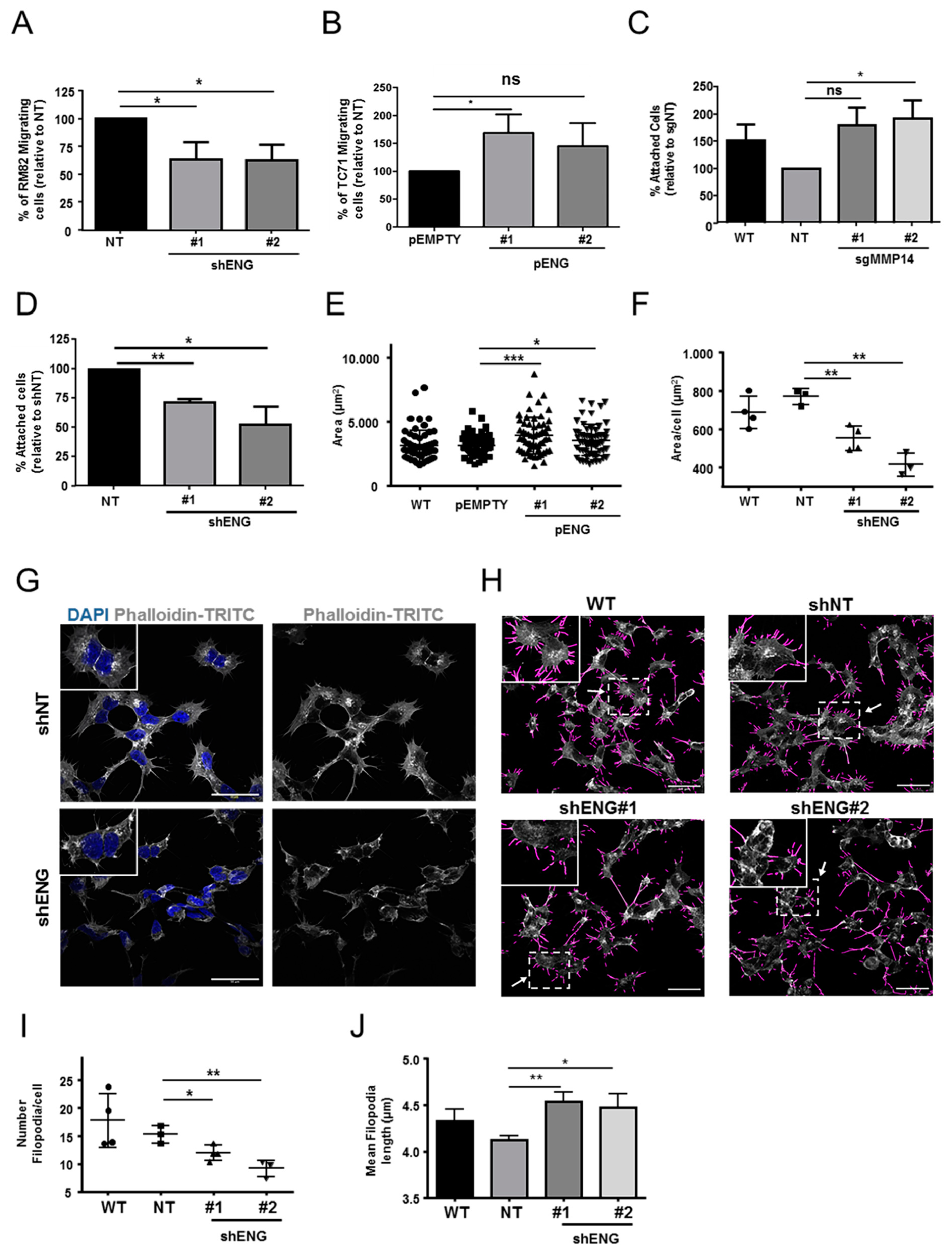
2.5. ENG Contributes to the Process of ES Cell Migration and Adhesion, whereas Loss of MMP14 Enhances Cell–Substrate Adhesion
2.6. ENG Participates in ES Actin Remodeling
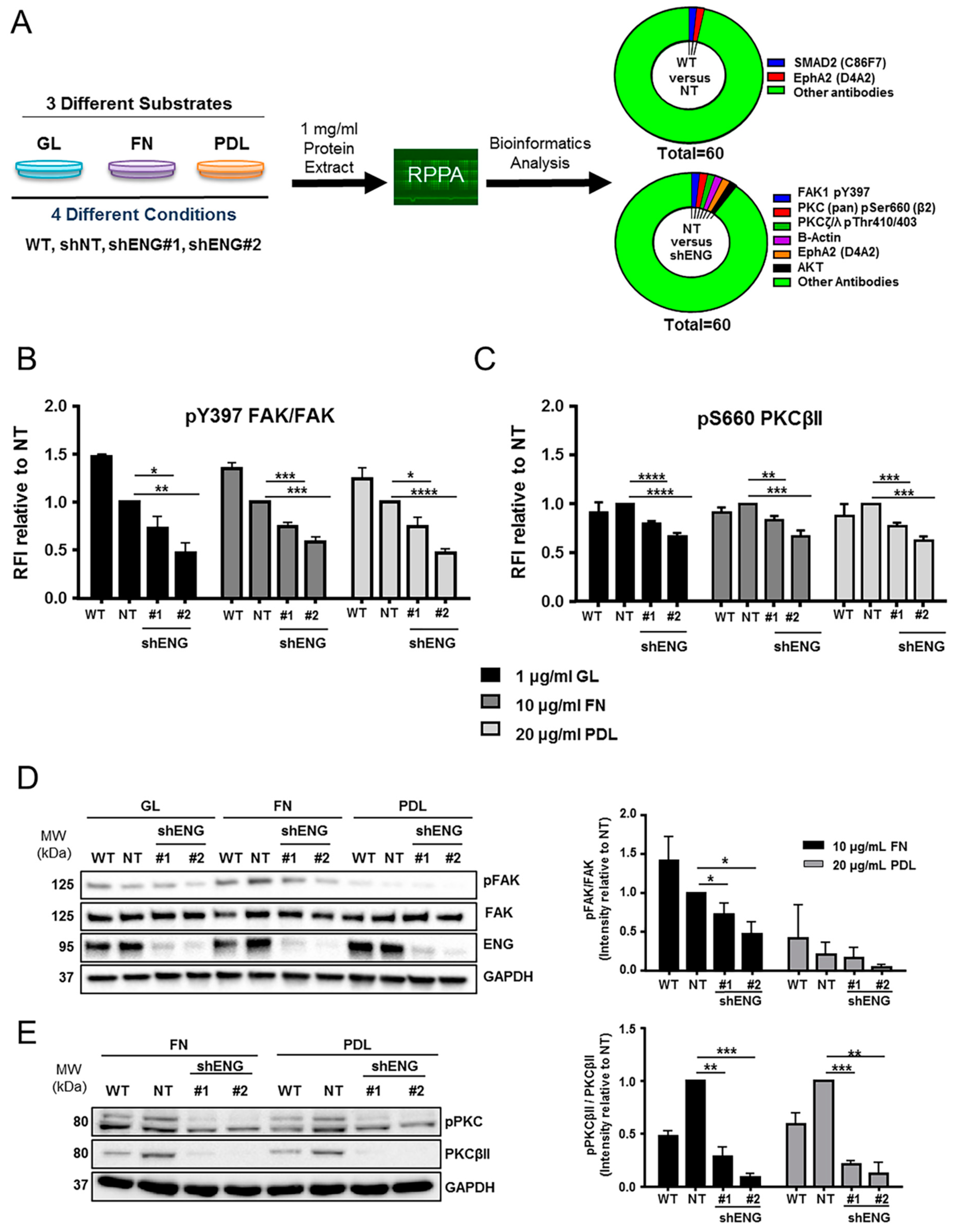
2.7. ENG Participates in the Activation of Signaling Pathways Related to Migration
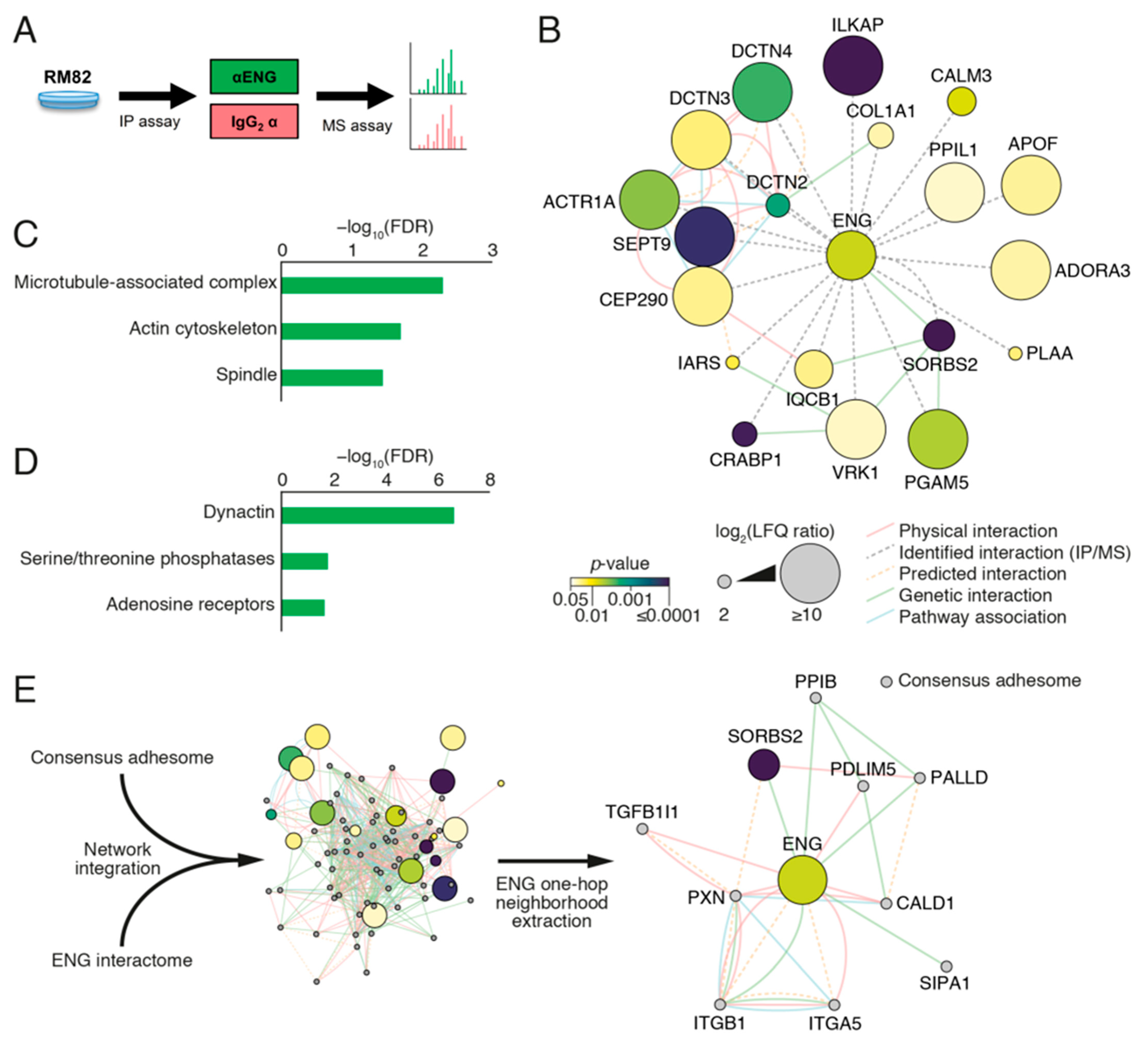
2.8. ENG Physically Interacts with Proteins Associated with Focal Adhesion Organization in ES Cells
3. Discussion
4. Materials and Methods
- IHC in ES samples: Intensity (scores 0-1-2-3) and area of staining (scores 0-1-2-3) derived from ENG/MMP14 expression were evaluated by an experienced pathologist as previously described by our group (19). CD31 intensity from tumoral samples was analyzed as described elsewhere (24). Analysis of variance (ANOVA test) between groups of patients according to means of intensity was also performed. Patient survival was evaluated by Kaplan–Meier curves. Overall survival (OS) statistical analysis on the local cohort was assessed by the log-rank (Mantel–Cox) test using GraphPad Prism (version 6.0).
- Flow cytometry-fluorescence-activated cell sorting-(FACS): Cells were seeded on 100-mm plates. After 24 h incubation, 80–90%-confluent cells were washed with PBS once and harvested either by trypsinization (in the case of intracellular protein analysis) or pipetting/scraping (in the case of transmembrane protein analysis). A total number of 5 × 105 cells were resuspended in 1 mL 2%-BSA PBS. Cells were centrifuged at 1500 rpm for 5 min. Cell pellets were mixed with Master Mix (anti-CD105-APC #105A-100T, Immunostep and PBS) for 30 min at room temperature, protected from light. Cells were centrifuged at 1500 rpm for 5 min and washed twice with 2%-BSA PBS. After the last wash, the suspension of cells in PBS was filtered with Falcon® 70 µm Cell Strainer (BD Falcon). For cell cycle analysis, cells were incubated with 100 μg/mL Propidium Iodide and 100 μg/mL RNAse A for 30 min before FACS. Cytometry was performed using the cell analyzer BD FACSCantoIITM Cell Analyzer (BD Biosciences) with FACS Diva software. FlowJo software was used for further analysis. At least 20,000 cells were analyzed per sample from three independent experiments.
- Immunofluorescence (IF): Glass coverslips were pre-treated with 10 µg/mL human FN (Corning) at 4 °C overnight in 24-well plates. Cells were seeded on coverslips at a cell confluence of 70%. After 24 h incubation, cells were washed twice with PBS. Cell fixation was performed with 15 min incubation of 3.7% paraformaldehyde (PFA). PFA was removed, and possible remaining free aldehyde was quenched by additional 15 min incubation with 100 mM glycine. Cells were washed once with PBS and permeabilized by 0.1%-Triton X-100 PBS for 30 min at room temperature. Cells were washed twice with 0.2%-BSA PBS, and samples were prepared for incubating with primary antibodies. Free primary antibody was removed, and cells were washed twice with 0.2%-BSA PBS. Secondary antibody was added for 1 h. In the case of phalloidin staining (Sigma), no secondary antibody was required, and samples were treated as follows. Samples were washed twice with 0.2%-BSA PBS, and after wash with PBS, they were maintained in milliQ water until mounting. A drop of mounting medium (Dako) was used for coverslip mounting on slides. Images from IF were taken in a confocal microscope Nikon A1R+.
- sgMMP14 cell development: Human MMP14 sequence (Gene ID 4323), located at chromosome 14, was used to design single-guide RNAs (sgRNA). The sgRNAs were designed by using Benchling software. Design and cloning were performed as previously described by our group. Briefly, the sgRNA oligo duplexes were cloned into the plasmid CBh-hfCas9-2A-eGFP, kindly provided by Dr. Trond Aasen. The sgRNA oligo duplexes were ligated into the CBh-hfCas9-2A-eGFP vector. The plasmid was digested with Bbs1 to generate compatible ends. Gene editing was confirmed by using EnGen™ Mutation Detection Kit (NEB). Clone validation was assessed by WB. Genomic DNA was extracted, and the edited locus was amplified by PCR. Sanger sequencing was used to determine gene editing. To detect indels, the deconvolution tool CRISPR-ID was used (http://crispid.gbiomed.kuleuven.be, accessed on October 2019).
- Sanger sequencing: Sanger sequencing was performed at the Genomics and Sequencing Service from the IBiS using the automatic sequencer Applied Biosystems 3500 Genetic Analyzer (Applied Biosystems, Waltham, MA, USA).
- Bioinformatics analysis: performed by the Bioinformatics Facility at IBiS. Briefly, array data were quantile normalized by the Robust Multichip Average (RMA) method using the oligo package from R/Bioconductor. Custom ClariomSHuman_Hs_ENSG CDF files from Brainarray (version 24) were used to avoid unspecific and bad quality probe sets.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar]
- Gougos, A.; Letarte, M. Primary structure of endoglin, an RGD-containing glycoprotein of human endothelial cells. J. Biol. Chem. 1990, 265, 8361–8364. [Google Scholar]
- ten Dijke, P.; Goumans, M.J.; Pardali, E. Endoglin in angiogenesis and vascular diseases. Angiogenesis 2008, 11, 79–89. [Google Scholar]
- Lopez-Novoa, J.M.; Bernabeu, C. The physiological role of endoglin in the cardiovascular system. American journal of physiology. Heart Circ. Physiol. 2010, 299, H959–H974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catchpoole, D.; Lail, A.; Guo, D.; Chen, Q.R.; Khan, J. Gene expression profiles that segregate patients with childhood acute lymphoblastic leukaemia: An independent validation study identifies that endoglin associates with patient outcome. Leuk. Res. 2007, 31, 1741–1747. [Google Scholar] [CrossRef]
- Davidson, B.; Stavnes, H.T.; Forsund, M.; Berner, A.; Staff, A.C. CD105 (Endoglin) expression in breast carcinoma effusions is a marker of poor survival. Breast 2010, 19, 493–498. [Google Scholar]
- Liu, P.; Sun, Y.L.; Du, J.; Hou, X.S.; Meng, H. CD105/Ki67 coexpression correlates with tumor progression and poor prognosis in epithelial ovarian cancer. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 2012, 22, 586–592. [Google Scholar] [CrossRef]
- Pardali, E.; van der Schaft, D.W.; Wiercinska, E.; Gorter, A.; Hogendoorn, P.C.; Griffioen, A.W.; ten Dijke, P. Critical role of endoglin in tumor cell plasticity of Ewing sarcoma and melanoma. Oncogene 2011, 30, 334–345. [Google Scholar]
- Zakrzewski, P.K.; Cygankiewicz, A.I.; Mokrosinski, J.; Nowacka-Zawisza, M.; Semczuk, A.; Rechberger, T.; Krajewska, W.M. Expression of endoglin in primary endometrial cancer. Oncology 2011, 81, 243–250. [Google Scholar]
- Koleva, R.I.; Conley, B.A.; Romero, D.; Riley, K.S.; Marto, J.A.; Lux, A.; Vary, C.P. Endoglin structure and function: Determinants of endoglin phosphorylation by transforming growth factor-beta receptors. J. Biol. Chem. 2006, 281, 25110–25123. [Google Scholar] [CrossRef] [Green Version]
- Lebrin, F.; Goumans, M.J.; Jonker, L.; Carvalho, R.L.; Valdimarsdottir, G.; Thorikay, M.; Mummery, C.; Arthur, H.M.; ten Dijke, P. Endoglin promotes endothelial cell proliferation and TGF-beta/ALK1 signal transduction. EMBO J. 2004, 23, 4018–4028. [Google Scholar] [CrossRef] [Green Version]
- Conley, B.A.; Koleva, R.; Smith, J.D.; Kacer, D.; Zhang, D.; Bernabeu, C.; Vary, C.P. Endoglin controls cell migration and composition of focal adhesions: Function of the cytosolic domain. J. Biol. Chem. 2004, 279, 27440–27449. [Google Scholar]
- Sanz-Rodriguez, F.; Guerrero-Esteo, M.; Botella, L.M.; Banville, D.; Vary, C.P.; Bernabeu, C. Endoglin regulates cytoskeletal organization through binding to ZRP-1, a member of the Lim family of proteins. J. Biol. Chem. 2004, 279, 32858–32868. [Google Scholar] [PubMed] [Green Version]
- Naumanen, P.; Lappalainen, P.; Hotulainen, P. Mechanisms of actin stress fibre assembly. J. Microsc. 2008, 231, 446–454. [Google Scholar] [CrossRef]
- Carragher, N.O.; Frame, M.C. Focal adhesion and actin dynamics: A place where kinases and proteases meet to promote invasion. Trends Cell Biol. 2004, 14, 241–249. [Google Scholar]
- Tian, H.; Mythreye, K.; Golzio, C.; Katsanis, N.; Blobe, G.C. Endoglin mediates fibronectin/alpha5beta1 integrin and TGF-beta pathway crosstalk in endothelial cells. EMBO J. 2012, 31, 3885–3900. [Google Scholar] [CrossRef] [Green Version]
- Puerto-Camacho, P.; Amaral, A.T.; Lamhamedi-Cherradi, S.E.; Menegaz, B.A.; Castillo-Ecija, H.; Ordonez, J.L.; Dominguez, S.; Jordan-Perez, C.; Diaz-Martin, J.; Romero-Perez, L.; et al. Preclinical Efficacy of Endoglin-Targeting Antibody-Drug Conjugates for the Treatment of Ewing Sarcoma. Clin. Cancer Res. 2019, 25, 2228–2240. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Alava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Primers 2018, 4, 5. [Google Scholar]
- Hawinkels, L.J.; Kuiper, P.; Wiercinska, E.; Verspaget, H.W.; Liu, Z.; Pardali, E.; Sier, C.F.; ten Dijke, P. Matrix metalloproteinase-14 (MT1-MMP)-mediated endoglin shedding inhibits tumor angiogenesis. Cancer Res. 2010, 70, 4141–4150. [Google Scholar] [PubMed] [Green Version]
- Hofmann, U.B.; Westphal, J.R.; Zendman, A.J.; Becker, J.C.; Ruiter, D.J.; van Muijen, G.N. Expression and activation of matrix metalloproteinase-2 (MMP-2) and its co-localization with membrane-type 1 matrix metalloproteinase (MT1-MMP) correlate with melanoma progression. J. Pathol. 2000, 191, 245–256. [Google Scholar] [CrossRef]
- Li, Z.; Takino, T.; Endo, Y.; Sato, H. Activation of MMP-9 by membrane type-1 MMP/MMP-2 axis stimulates tumor metastasis. Cancer Sci. 2017, 108, 347–353. [Google Scholar] [CrossRef] [Green Version]
- Hahm, K.B. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat. Genet. 1999, 23, 481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franzetti, G.A.; Laud-Duval, K.; van der Ent, W.; Brisac, A.; Irondelle, M.; Aubert, S.; Dirksen, U.; Bouvier, C.; de Pinieux, G.; Snaar-Jagalska, E.; et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 2017, 36, 3505–3514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Nunez, P.; Romero-Perez, L.; Amaral, A.T.; Puerto-Camacho, P.; Jordan, C.; Marcilla, D.; Grunewald, T.G.; Alonso, J.; de Alava, E.; Diaz-Martin, J. Hippo pathway effectors YAP1/TAZ induce an EWS-FLI1-opposing gene signature and associate with disease progression in Ewing sarcoma. J. Pathol. 2020, 250, 374–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomazou, E.M.; Sheffield, N.C.; Schmidl, C.; Schuster, M.; Schonegger, A.; Datlinger, P.; Kubicek, S.; Bock, C.; Kovar, H. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015, 10, 1082–1095. [Google Scholar] [CrossRef] [Green Version]
- Hidalgo, M.R.; Cubuk, C.; Amadoz, A.; Salavert, F.; Carbonell-Caballero, J.; Dopazo, J. High throughput estimation of functional cell activities reveals disease mechanisms and predicts relevant clinical outcomes. Oncotarget 2017, 8, 5160–5178. [Google Scholar] [CrossRef] [Green Version]
- Horton, E.R.; Byron, A.; Askari, J.A.; Ng, D.H.J.; Millon-Fremillon, A.; Robertson, J.; Koper, E.J.; Paul, N.R.; Warwood, S.; Knight, D.; et al. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 2015, 17, 1577–1587. [Google Scholar] [CrossRef] [Green Version]
- Li Mow Chee, F.; Byron, A. Network Analysis of Integrin Adhesion Complexes. Methods Mol. Biol. 2021, 2217, 149–179. [Google Scholar]
- Di Paolo, V.; Russo, I.; Boldrini, R.; Rava, L.; Pezzullo, M.; Benedetti, M.C.; Galardi, A.; Colletti, M.; Rota, R.; Orlando, D.; et al. Evaluation of Endoglin (CD105) expression in pediatric rhabdomyosarcoma. BMC Cancer 2018, 18, 31. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez Munoz, T.; Amaral, A.T.; Puerto-Camacho, P.; Peinado, H.; de Alava, E. Endoglin in the Spotlight to Treat Cancer. Int. J. Mol. Sci. 2021, 22, 3186. [Google Scholar] [CrossRef]
- Xiong, J.; Yan, L.; Zou, C.; Wang, K.; Chen, M.; Xu, B.; Zhou, Z.; Zhang, D. Integrins regulate stemness in solid tumor: An emerging therapeutic target. J. Hematol. Oncol. 2021, 14, 177. [Google Scholar] [CrossRef]
- Grange, C.; Tapparo, M.; Collino, F.; Vitillo, L.; Damasco, C.; Deregibus, M.C.; Tetta, C.; Bussolati, B.; Camussi, G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011, 71, 5346–5356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero-Esteo, M.; Sanchez-Elsner, T.; Letamendia, A.; Bernabeu, C. Extracellular and cytoplasmic domains of endoglin interact with the transforming growth factor-beta receptors I and II. J. Biol. Chem. 2002, 277, 29197–29209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaral, A.T.; Garofalo, C.; Frapolli, R.; Manara, M.C.; Mancarella, C.; Uboldi, S.; Di Giandomenico, S.; Ordonez, J.L.; Sevillano, V.; Malaguarnera, R.; et al. Trabectedin efficacy in Ewing sarcoma is greatly increased by combination with anti-IGF signaling agents. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 1373–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castonguay, R.; Werner, E.D.; Matthews, R.G.; Presman, E.; Mulivor, A.W.; Solban, N.; Sako, D.; Pearsall, R.S.; Underwood, K.W.; Seehra, J.; et al. Soluble endoglin specifically binds bone morphogenetic proteins 9 and 10 via its orphan domain, inhibits blood vessel formation, and suppresses tumor growth. J. Biol. Chem. 2011, 286, 30034–30046. [Google Scholar] [CrossRef] [Green Version]
- Knauper, V.; Bailey, L.; Worley, J.R.; Soloway, P.; Patterson, M.L.; Murphy, G. Cellular activation of proMMP-13 by MT1-MMP depends on the C-terminal domain of MMP-13. FEBS Lett. 2002, 532, 127–130. [Google Scholar] [CrossRef] [Green Version]
- Filipenko, N.R.; Waisman, D.M. The C terminus of annexin II mediates binding to F-actin. J. Biol. Chem. 2001, 276, 5310–5315. [Google Scholar]
- Jones, P.G.; Moore, G.J.; Waisman, D.M. A nonapeptide to the putative F-actin binding site of annexin-II tetramer inhibits its calcium-dependent activation of actin filament bundling. J. Biol. Chem. 1992, 267, 13993–13997. [Google Scholar] [CrossRef]
- Dubois, T.; Oudinet, J.P.; Mira, J.P.; Russo-Marie, F. Annexins and protein kinases C. Biochim. Biophys. Acta 1996, 1313, 290–294. [Google Scholar] [CrossRef] [Green Version]
- Koese, M.; Rentero, C.; Kota, B.P.; Hoque, M.; Cairns, R.; Wood, P.; Vila de Muga, S.; Reverter, M.; Alvarez-Guaita, A.; Monastyrskaya, K.; et al. Annexin A6 is a scaffold for PKCalpha to promote EGFR inactivation. Oncogene 2013, 32, 2858–2872. [Google Scholar] [CrossRef] [Green Version]
- Kheifets, V.; Bright, R.; Inagaki, K.; Schechtman, D.; Mochly-Rosen, D. Protein kinase C delta (deltaPKC)-annexin V interaction: A required step in deltaPKC translocation and function. J. Biol. Chem. 2006, 281, 23218–23226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lastres, P.; Martin-Perez, J.; Langa, C.; Bernabeu, C. Phosphorylation of the human-transforming-growth-factor-beta-binding protein endoglin. Biochem. J. 1994, 301 Pt 3, 765–768. [Google Scholar] [CrossRef]
- Ng, T.; Shima, D.; Squire, A.; Bastiaens, P.I.; Gschmeissner, S.; Humphries, M.J.; Parker, P.J. PKCalpha regulates beta1 integrin-dependent cell motility through association and control of integrin traffic. EMBO J. 1999, 18, 3909–3923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buensuceso, C.S.; Obergfell, A.; Soriani, A.; Eto, K.; Kiosses, W.B.; Arias-Salgado, E.G.; Kawakami, T.; Shattil, S.J. Regulation of outside-in signaling in platelets by integrin-associated protein kinase C beta. J. Biol. Chem. 2005, 280, 644–653. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.S.; Naruszewicz, I.; Wang, P.; Leung-Hagesteijn, C.; Hannigan, G.E. ILKAP regulates ILK signaling and inhibits anchorage-independent growth. Oncogene 2004, 23, 3454–3461. [Google Scholar] [CrossRef] [Green Version]
- Leung-Hagesteijn, C.; Mahendra, A.; Naruszewicz, I.; Hannigan, G.E. Modulation of integrin signal transduction by ILKAP, a protein phosphatase 2C associating with the integrin-linked kinase, ILK1. EMBO J. 2001, 20, 2160–2170. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Cui, S.; Han, S.; Cheng, B.; Zheng, Y.; Zhang, Y. Palladin is a novel binding partner of ILKAP in eukaryotic cells. Biochem. Biophys. Res. Commun. 2011, 411, 768–773. [Google Scholar] [CrossRef]
- Sannino, G.; Marchetto, A.; Ranft, A.; Jabar, S.; Zacherl, C.; Alba-Rubio, R.; Stein, S.; Wehweck, F.S.; Kiran, M.M.; Holting, T.L.B.; et al. Gene expression and immunohistochemical analyses identify SOX2 as major risk factor for overall survival and relapse in Ewing sarcoma patients. EBioMedicine 2019, 47, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Turriziani, B.; Garcia-Munoz, A.; Pilkington, R.; Raso, C.; Kolch, W.; von Kriegsheim, A. On-beads digestion in conjunction with data-dependent mass spectrometry: A shortcut to quantitative and dynamic interaction proteomics. Biology 2014, 3, 320–332. [Google Scholar] [CrossRef] [Green Version]
- Humphries, M.J. Cell adhesion assays. Methods Mol. Biol. 2009, 522, 203–210. [Google Scholar] [PubMed]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [Green Version]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puerto-Camacho, P.; Díaz-Martín, J.; Olmedo-Pelayo, J.; Bolado-Carrancio, A.; Salguero-Aranda, C.; Jordán-Pérez, C.; Esteban-Medina, M.; Álamo-Álvarez, I.; Delgado-Bellido, D.; Lobo-Selma, L.; et al. Endoglin and MMP14 Contribute to Ewing Sarcoma Spreading by Modulation of Cell–Matrix Interactions. Int. J. Mol. Sci. 2022, 23, 8657. https://doi.org/10.3390/ijms23158657
Puerto-Camacho P, Díaz-Martín J, Olmedo-Pelayo J, Bolado-Carrancio A, Salguero-Aranda C, Jordán-Pérez C, Esteban-Medina M, Álamo-Álvarez I, Delgado-Bellido D, Lobo-Selma L, et al. Endoglin and MMP14 Contribute to Ewing Sarcoma Spreading by Modulation of Cell–Matrix Interactions. International Journal of Molecular Sciences. 2022; 23(15):8657. https://doi.org/10.3390/ijms23158657
Chicago/Turabian StylePuerto-Camacho, Pilar, Juan Díaz-Martín, Joaquín Olmedo-Pelayo, Alfonso Bolado-Carrancio, Carmen Salguero-Aranda, Carmen Jordán-Pérez, Marina Esteban-Medina, Inmaculada Álamo-Álvarez, Daniel Delgado-Bellido, Laura Lobo-Selma, and et al. 2022. "Endoglin and MMP14 Contribute to Ewing Sarcoma Spreading by Modulation of Cell–Matrix Interactions" International Journal of Molecular Sciences 23, no. 15: 8657. https://doi.org/10.3390/ijms23158657
APA StylePuerto-Camacho, P., Díaz-Martín, J., Olmedo-Pelayo, J., Bolado-Carrancio, A., Salguero-Aranda, C., Jordán-Pérez, C., Esteban-Medina, M., Álamo-Álvarez, I., Delgado-Bellido, D., Lobo-Selma, L., Dopazo, J., Sastre, A., Alonso, J., Grünewald, T. G. P., Bernabeu, C., Byron, A., Brunton, V. G., Amaral, A. T., & Álava, E. D. (2022). Endoglin and MMP14 Contribute to Ewing Sarcoma Spreading by Modulation of Cell–Matrix Interactions. International Journal of Molecular Sciences, 23(15), 8657. https://doi.org/10.3390/ijms23158657






