The Biochemical Mechanism of Fork Regression in Prokaryotes and Eukaryotes—A Single Molecule Comparison
Abstract
1. Introduction
2. Fork Regression Enzymes
2.1. RecG
2.2. SMARCAL1
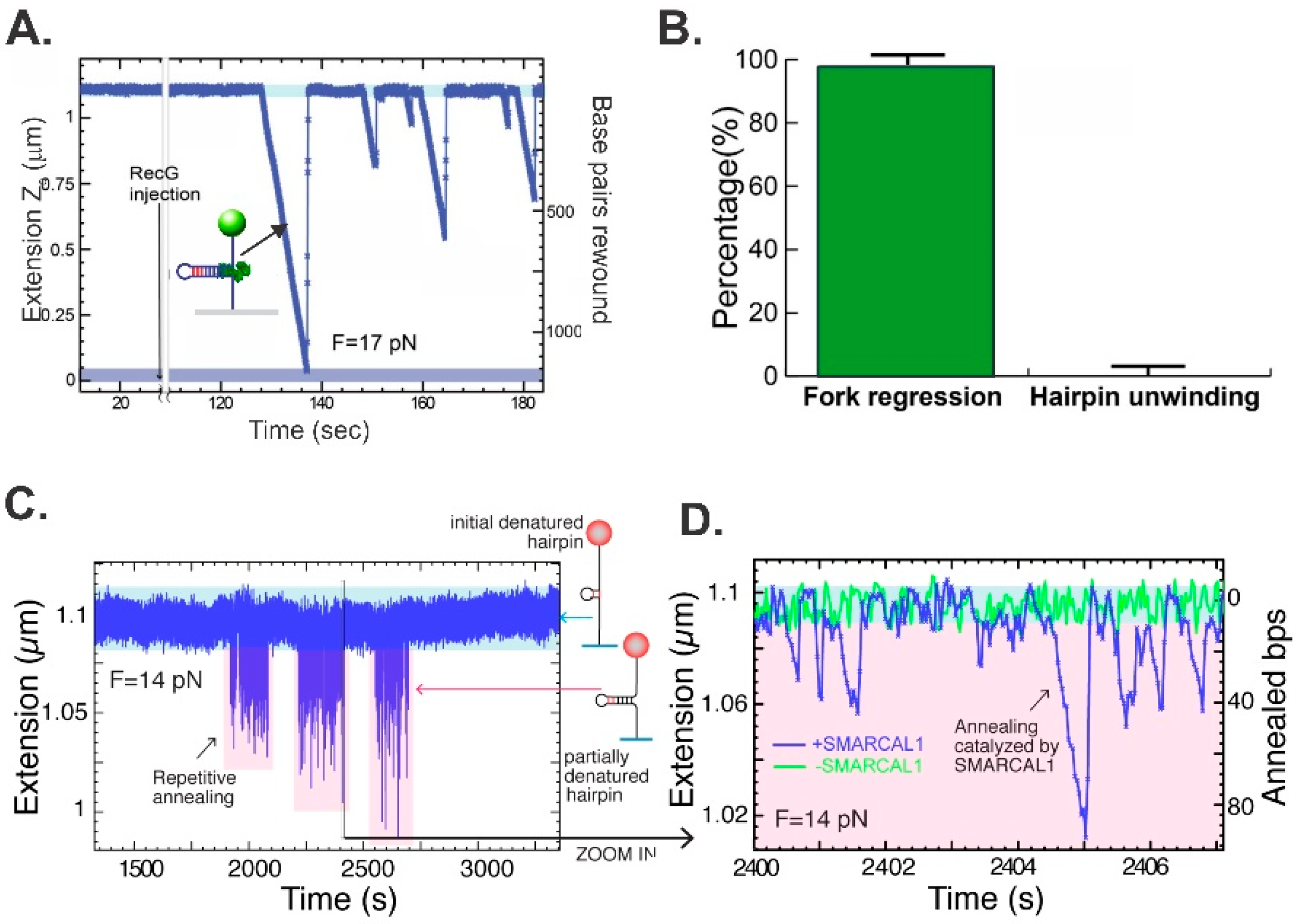
3. Method to Study Fork Regression at the Single-Molecule Level
4. Fork Arm Rewinding—An Essential Step in Fork Regression
5. Effects of Single-Stranded DNA Binding Proteins
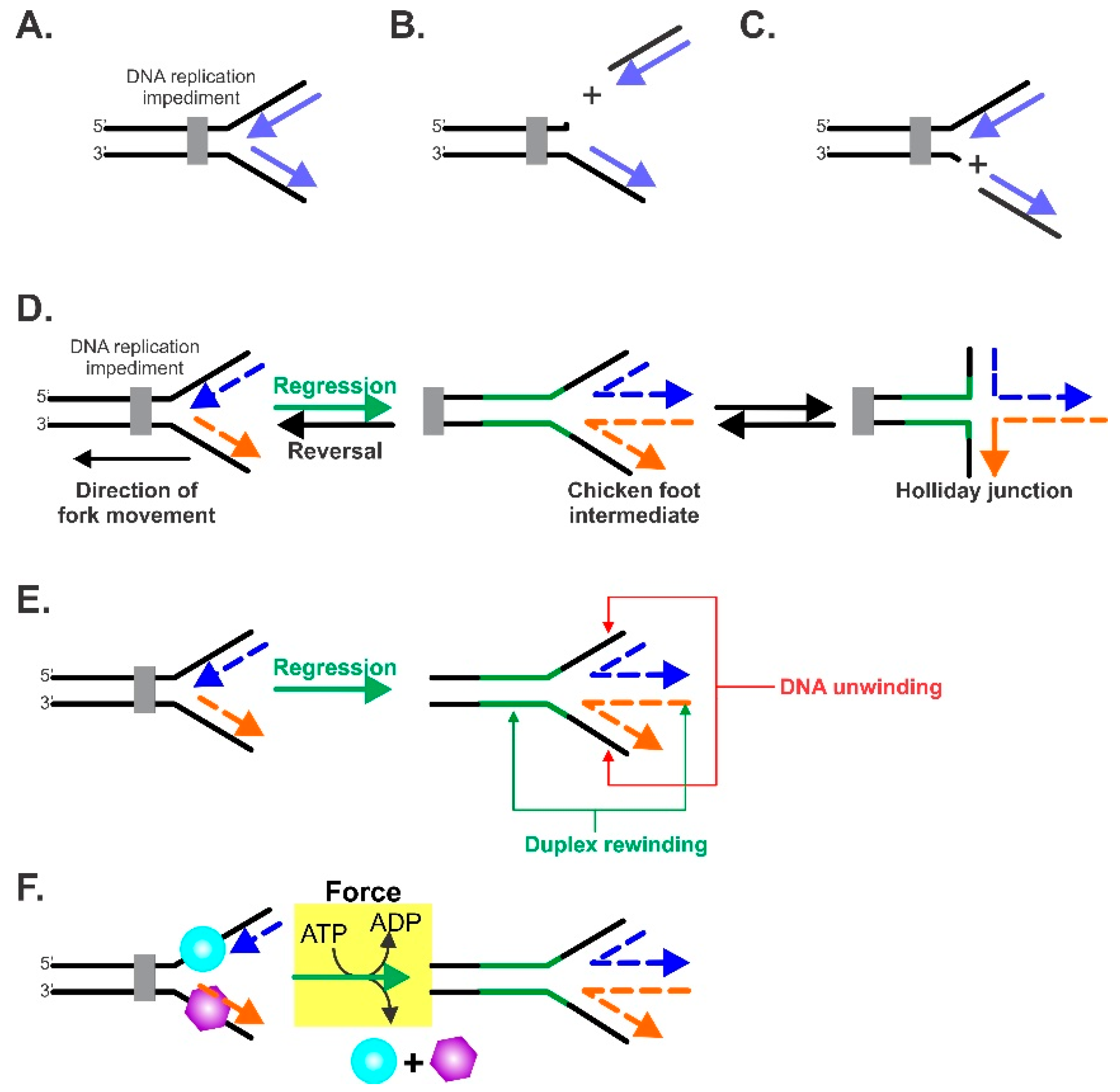
6. Formation of the Central Intermediate of Fork Regression
7. Conclusions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Fijalkowska, I.J.; Schaaper, R.M.; Jonczyk, P. DNA replication fidelity in Escherichia coli: A multi-DNA polymerase affair. FEMS Microbiol. Rev. 2012, 36, 1105–1121. [Google Scholar] [CrossRef]
- Merchut-Maya, J.M.; Bartek, J.; Maya-Mendoza, A. Regulation of replication fork speed: Mechanisms and impact on genomic stability. DNA Repair 2019, 81, 102654. [Google Scholar] [CrossRef] [PubMed]
- Skarstad, K.; Katayama, T. Regulating DNA replication in bacteria. Cold Spring Harb Perspect. Biol. 2013, 5, a012922. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S.; Jergic, S.; Dixon, N.E. The E. coli DNA Replication Fork. Enzymes 2016, 39, 31–88. [Google Scholar] [PubMed]
- Dewar, J.M.; Walter, J.C. Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 2017, 18, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Dimude, J.U.; Midgley-Smith, S.L.; Stein, M.; Rudolph, C.J. Replication Termination: Containing Fork Fusion-Mediated Pathologies in Escherichia coli. Genes 2016, 7, 40. [Google Scholar] [CrossRef]
- Higgins, N.P. The Bacterial Chromosome; ASM Press: Washington, DC, USA, 2005. [Google Scholar]
- Labib, K.; Hodgson, B. Replication fork barriers: Pausing for a break or stalling for time? EMBO Rep. 2007, 8, 346–353. [Google Scholar] [CrossRef]
- Calzada, A.; Hodgson, B.; Kanemaki, M.; Bueno, A.; Labib, K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005, 19, 1905–1919. [Google Scholar] [CrossRef]
- Diffley, J.F.; Labib, K. The chromosome replication cycle. J. Cell Sci. 2002, 115, 869–872. [Google Scholar] [CrossRef]
- Leman, A.R.; Noguchi, E. The replication fork: Understanding the eukaryotic replication machinery and the challenges to genome duplication. Genes 2013, 4, 1–32. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Pearl, L.H.; Carr, A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 2016, 16, 35–42. [Google Scholar] [CrossRef]
- Cox, M.M.; Goodman, M.F.; Kreuzer, K.N.; Sherratt, D.J.; Sandler, S.J.; Marians, K.J. The importance of repairing stalled replication forks. Nature 2000, 404, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.; Poli, J.; Marians, K.J.; Pasero, P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect. Biol. 2013, 5, a012815. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M. Recombinational DNA repair of damaged replication forks in Escherichia coli: Questions. Annu. Rev. Genet. 2001, 35, 53–82. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, D.; Vos, J. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: Fork uncoupling or gap formation. Proc. Natl. Acad. Sci. USA 1995, 92, 11975–11979. [Google Scholar] [CrossRef]
- Cordeiro-Stone, M.; Makhov, A.; Zaritskaya, L.; Griffith, J. Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 1999, 289, 1207–1218. [Google Scholar] [CrossRef]
- Rudolph, C.; Dhillon, P.; Moore, T.; Lloyd, R. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair 2007, 6, 981–993. [Google Scholar] [CrossRef]
- Usdin, K.; Woodford, K. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995, 23, 4202–4209. [Google Scholar] [CrossRef] [PubMed]
- Samadashwily, G.M.; Raca, G.; Mirkin, S.M. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 1997, 17, 298–304. [Google Scholar] [CrossRef]
- Kowalczykowski, S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000, 25, 156–165. [Google Scholar] [CrossRef]
- McGlynn, P.; Lloyd, R.G. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 2002, 3, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Marians, K.J. Mechanisms of replication fork restart in Escherichia coli. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 71–77. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mirkin, E.V.; Mirkin, S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007, 71, 13–35. [Google Scholar] [CrossRef] [PubMed]
- Voineagu, I.; Narayanan, V.; Lobachev, K.S.; Mirkin, S.M. Replication stalling at unstable inverted repeats: Interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 9936–9941. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, K.N. DNA damage responses in prokaryotes: Regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harb Perspect. Biol. 2013, 5, a012674. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.K.; Guy, C.P.; Yeeles, J.T.; Atkinson, J.; Bell, H.; Lloyd, R.G.; Marians, K.J.; McGlynn, P. Protein-DNA complexes are the primary sources of replication fork pausing in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 7252–7257. [Google Scholar] [CrossRef] [PubMed]
- Henderson, M.L.; Kreuzer, K.N. Functions that Protect Escherichia coli from Tightly Bound DNA-Protein Complexes Created by Mutant EcoRII Methyltransferase. PLoS ONE 2015, 10, e0128092. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef] [PubMed]
- Katou, Y.; Kanoh, Y.; Bando, M.; Noguchi, H.; Tanaka, H.; Ashikari, T.; Sugimoto, K.; Shirahige, K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 2003, 424, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Tercero, J.A.; Diffley, J.F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 2001, 412, 553–557. [Google Scholar] [CrossRef]
- Pacek, M.; Tutter, A.V.; Kubota, Y.; Takisawa, H.; Walter, J.C. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006, 21, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Kogoma, T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 1997, 61, 212–238. [Google Scholar] [PubMed]
- Kuzminov, A.; Stahl, F.W. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev. 1999, 13, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, K.N. Interplay between DNA replication and recombination in prokaryotes. Annu. Rev. Microbiol. 2005, 59, 43–67. [Google Scholar] [CrossRef]
- Neelsen, K.J.; Lopes, M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015, 16, 207–220. [Google Scholar] [CrossRef]
- Jeiranian, H.A.; Schalow, B.J.; Courcelle, C.T.; Courcelle, J. Fate of the replisome following arrest by UV-induced DNA damage in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 11421–11426. [Google Scholar] [CrossRef]
- Bianco, P.R.; Lu, Y. Single-molecule insight into stalled replication fork rescue in Escherichia coli. Nucleic Acids Res. 2021, 49, 4220–4238. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.; McGlynn, P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009, 37, 3475–3492. [Google Scholar] [CrossRef]
- Bianco, P.R. I came to a fork in the DNA and there was RecG. Prog. Biophys. Mol. Biol. 2015, 117, 166–173. [Google Scholar] [CrossRef]
- Manosas, M.; Perumal, S.K.; Bianco, P.R.; Ritort, F.; Benkovic, S.J.; Croquette, V. RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat. Commun. 2013, 4, 2368. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.G.; Rudolph, C.J. 25 years on and no end in sight: A perspective on the role of RecG protein. Curr. Genet. 2016, 62, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Wallet, C.; Le Ret, M.; Bergdoll, M.; Bichara, M.; Dietrich, A.; Gualberto, J.M. The RECG1 DNA Translocase Is a Key Factor in Recombination Surveillance, Repair, and Segregation of the Mitochondrial DNA in Arabidopsis. Plant. Cell. 2015, 27, 2907–2925. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Jiang, G.; Cao, L.; Huang, J. Replication Fork Reversal and Protection. Front. Cell Dev. Biol. 2021, 9, 670392. [Google Scholar] [CrossRef] [PubMed]
- Bansbach, C.E.; Betous, R.; Lovejoy, C.A.; Glick, G.G.; Cortez, D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009, 23, 2405–2414. [Google Scholar] [CrossRef] [PubMed]
- Betous, R.; Couch, F.B.; Mason, A.C.; Eichman, B.F.; Manosas, M.; Cortez, D. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep. 2013, 3, 1958–1969. [Google Scholar] [CrossRef] [PubMed]
- Betous, R.; Mason, A.C.; Rambo, R.P.; Bansbach, C.E.; Badu-Nkansah, A.; Sirbu, B.M.; Eichman, B.F.; Cortez, D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012, 26, 151–162. [Google Scholar] [CrossRef]
- Storm, P.K.; Hoekstra, W.P.; de Haan, P.G.; Verhoef, C. Genetic recombination in Escherichia coli. IV. Isolation and characterization of recombination-deficiency mutants of Escherichia coli K12. Mutat. Res. 1971, 13, 9–17. [Google Scholar] [CrossRef][Green Version]
- Benson, F.; Collier, S.; Lloyd, R.G. Evidence of abortive recombination in ruv mutants of Escherichia coli K12. Mol. Gen. Genet. 1991, 225, 266–272. [Google Scholar] [CrossRef]
- Lloyd, R.G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J. Bacteriol. 1991, 173, 5414–5418. [Google Scholar] [CrossRef] [PubMed]
- Kalman, M.; Murphy, H.; Cashel, M. The nucleotide sequence of recG, the distal spo operon gene in Escherichia coli K-12. Gene 1992, 110, 95–99. [Google Scholar] [CrossRef]
- Singleton, M.; Dillingham, M.; Wigley, D. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Vincent, S.D.; Mahdi, A.A.; Lloyd, R.G. The RecG branch migration protein of Escherichia coli dissociates R-loops. J. Mol. Biol. 1996, 264, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Sharples, G.J.; Ingleston, S.M.; Lloyd, R.G. Holliday junction processing in bacteria: Insights from the evolutionary conservation of RuvABC, RecG, and RusA. J. Bacteriol. 1999, 181, 5543–5550. [Google Scholar] [CrossRef] [PubMed]
- Abd Wahab, S.; Choi, M.; Bianco, P.R. Characterization of the ATPase activity of RecG and RuvAB proteins on model fork structures reveals insight into stalled DNA replication fork repair. J. Biol. Chem. 2013, 288, 26397–26409. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Yeeles, J.T.; Marians, K.J. Regression of replication forks stalled by leading-strand template damage: I. Both RecG and RuvAB catalyze regression, but RuvC cleaves the holliday junctions formed by RecG preferentially. J. Biol. Chem. 2014, 289, 28376–28387. [Google Scholar] [CrossRef] [PubMed]
- Whitby, M.C.; Vincent, S.D.; Lloyd, R.G. Branch migration of Holliday junctions: Identification of RecG protein as a junction specific DNA helicase. EMBO J. 1994, 13, 5220–5228. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Lloyd, R. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 1999, 27, 3049–3056. [Google Scholar] [PubMed]
- McGlynn, P.; Mahdi, A.; Lloyd, R. Characterisation of the catalytically active form of RecG helicase. Nucleic Acids Res. 2000, 28, 2324–2332. [Google Scholar] [CrossRef] [PubMed]
- Singleton, M.R.; Scaife, S.; Wigley, D.B. Structural analysis of DNA replication fork reversal by RecG. Cell 2001, 107, 79–89. [Google Scholar] [CrossRef]
- Baharoglu, Z.; Petranovic, M.; Flores, M.; Michel, B. RuvAB is essential for replication forks reversal in certain replication mutants. EMBO J. 2006, 25, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Seigneur, M.; Bidnenko, V.; Ehrlich, S.; Michel, B. RuvAB acts at arrested replication forks. Cell 1998, 95, 419–430. [Google Scholar] [CrossRef]
- Buss, J.; Kimura, Y.; Bianco, P. RecG interacts directly with SSB: Implications for stalled replication fork regression. Nucleic Acids Res. 2008, 36, 7029–7042. [Google Scholar] [CrossRef] [PubMed]
- Slocum, S.L.; Buss, J.A.; Kimura, Y.; Bianco, P.R. Characterization of the ATPase activity of the Escherichia coli RecG protein reveals that the preferred cofactor is negatively supercoiled DNA. J. Mol. Biol. 2007, 367, 647–664. [Google Scholar] [CrossRef][Green Version]
- Briggs, G.S.; Mahdi, A.A.; Wen, Q.; Lloyd, R.G. DNA binding by the substrate specificity (wedge) domain of RecG helicase suggests a role in processivity. J. Biol. Chem. 2005, 280, 13921–13927. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.R.; Lyubchenko, Y.L. SSB and the RecG DNA helicase: An intimate association to rescue a stalled replication fork. Protein Sci. 2017, 26, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Tan, H.Y.; Bianco, P.R.; Lyubchenko, Y.L. Remodeling of RecG Helicase at the DNA Replication Fork by SSB Protein. Sci. Rep. 2015, 5, 9625. [Google Scholar] [CrossRef]
- Yu, C.; Tan, H.Y.; Choi, M.; Stanenas, A.J.; Byrd, A.K.; Raney, K.; Cohan, C.S.; Bianco, P.R. SSB binds to the RecG and PriA helicases in vivo in the absence of DNA. Genes Cells 2016, 21, 163–184. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Tan, H.Y.; Zhang, J.X.; Wilczek, L.A.; Hsieh, K.R.; Mulkin, J.A.; Bianco, P.R. The mechanism of Single strand binding protein-RecG binding: Implications for SSB interactome function. Protein Sci. 2020, 29, 1211–1227. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, A.A.; McGlynn, P.; Levett, S.D.; Lloyd, R.G. DNA binding and helicase domains of the Escherichia coli recombination protein RecG. Nucleic Acids Res. 1997, 25, 3875–3880. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, Z.; Hashemi, M.; Warren, G.; Bianco, P.R.; Lyubchenko, Y.L. Dynamics of the Interaction of RecG Protein with Stalled Replication Forks. Biochemistry 2018, 57, 1967–1976. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Tan, H.Y.; Wilczek, L.A.; Pottinger, S.; Manosas, M.; Yu, C.; Nguyenduc, T.; Bianco, P.R. The intrinsically disordered linker of E. coli SSB is critical for the release from single-stranded DNA. Protein Sci. 2017, 26, 700–717. [Google Scholar] [CrossRef]
- Flaus, A.; Martin, D.M.; Barton, G.J.; Owen-Hughes, T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Koonin, E.V. One more conserved sequence motif in helicases. Nucleic Acids Res. 1988, 16, 7734. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peterson, C.L. Chromatin remodeling enzymes: Taming the machines. Third in review series on chromatin dynamics. EMBO Rep. 2002, 3, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Hockensmith, J.W.; Wahl, A.F.; Kowalski, S.; Bambara, R.A. Purification of a calf thymus DNA-dependent adenosinetriphosphatase that prefers a primer-template junction effector. Biochemistry 1986, 25, 7812–7821. [Google Scholar] [CrossRef] [PubMed]
- Mesner, L.D.; Truman, P.A.; Hockensmith, J.W. DNA-dependent adenosinetriphosphatase A: Immunoaffinity purification and characterization of immunological reagents. Biochemistry 1993, 32, 7772–7778. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.A.; Eisen, J.A.; Mohrenweiser, H.W. Cloning and characterization of HARP/SMARCAL1: A prokaryotic HepA-related SNF2 helicase protein from human and mouse. Genomics 2000, 65, 274–282. [Google Scholar] [CrossRef]
- Bansal, R.; Hussain, S.; Chanana, U.B.; Bisht, D.; Goel, I.; Muthuswami, R. SMARCAL1, the annealing helicase and the transcriptional co-regulator. IUBMB Life 2020, 72, 2080–2096. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Mazumder, M.; Dhatchinamoorthy, K.; Nongkhlaw, M.; Haokip, D.T.; Gourinath, S.; Komath, S.S.; Muthuswami, R. Ligand-induced conformation changes drive ATP hydrolysis and function in SMARCAL1. FEBS J. 2015, 282, 3841–3859. [Google Scholar] [CrossRef]
- Muthuswami, R.; Truman, P.A.; Mesner, L.D.; Hockensmith, J.W. A eukaryotic SWI2/SNF2 domain, an exquisite detector of double-stranded to single-stranded DNA transition elements. J. Biol. Chem. 2000, 275, 7648–7655. [Google Scholar] [CrossRef] [PubMed]
- Yusufzai, T.; Kadonaga, J.T. HARP is an ATP-driven annealing helicase. Science 2008, 322, 748–750. [Google Scholar] [CrossRef]
- Yuan, J.; Ghosal, G.; Chen, J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009, 23, 2394–2399. [Google Scholar] [CrossRef] [PubMed]
- Kolinjivadi, A.M.; Sannino, V.; De Antoni, A.; Zadorozhny, K.; Kilkenny, M.; Techer, H.; Baldi, G.; Shen, R.; Ciccia, A.; Pellegrini, L.; et al. Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments. Mol. Cell. 2017, 67, 867–881.e867. [Google Scholar] [CrossRef] [PubMed]
- Poole, L.A.; Cortez, D. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 696–714. [Google Scholar] [CrossRef]
- Ciccia, A.; Bredemeyer, A.L.; Sowa, M.E.; Terret, M.E.; Jallepalli, P.V.; Harper, J.W.; Elledge, S.J. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009, 23, 2415–2425. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Lu, Y.; Jakoncic, J.; Sun, H.; Xia, J.; Qian, C. Structure of RPA32 bound to the N-terminus of SMARCAL1 redefines the binding interface between RPA32 and its interacting proteins. FEBS J. 2014, 281, 3382–3396. [Google Scholar] [CrossRef] [PubMed]
- Delagoutte, E.; Heneman-Masurel, A.; Baldacci, G. Single-stranded DNA binding proteins unwind the newly synthesized double-stranded DNA of model miniforks. Biochemistry 2011, 50, 932–944. [Google Scholar] [CrossRef] [PubMed]
- De Laat, W.L.; Appeldoorn, E.; Sugasawa, K.; Weterings, E.; Jaspers, N.G.; Hoeijmakers, J.H. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998, 12, 2598–2609. [Google Scholar] [CrossRef]
- Manosas, M.; Perumal, S.K.; Croquette, V.; Benkovic, S.J. Direct observation of stalled fork restart via fork regression in the T4 replication system. Science 2012, 338, 1217–1220. [Google Scholar] [CrossRef] [PubMed]
- Gosse, C.; Croquette, V. Magnetic tweezers: Micromanipulation and force measurement at the molecular level. Biophys. J. 2002, 82, 3314–3329. [Google Scholar] [CrossRef]
- Neuert, G.; Albrecht, C.; Pamir, E.; Gaub, H.E. Dynamic force spectroscopy of the digoxigenin-antibody complex. FEBS Lett. 2006, 580, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Chen, A.; Kolb, P.; Moy, V.T. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry 2000, 39, 10219–10223. [Google Scholar] [CrossRef]
- Martinez-Senac, M.M.; Webb, M.R. Mechanism of translocation and kinetics of DNA unwinding by the helicase RecG. Biochemistry 2005, 44, 16967–16976. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Lu, C.H.; Li, H.W. RecA-SSB Interaction Modulates RecA Nucleoprotein Filament Formation on SSB-Wrapped DNA. Sci. Rep. 2017, 7, 11876. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.C.; Liu, B.; Kowalczykowski, S.C. Imaging and energetics of single SSB-ssDNA molecules reveal intramolecular condensation and insight into RecOR function. eLife 2015, 4, e08646. [Google Scholar] [CrossRef]
- Liu, J.; Choi, M.; Stanenas, A.G.; Byrd, A.K.; Raney, K.D.; Cohan, C.; Bianco, P.R. Novel, fluorescent, SSB protein chimeras with broad utility. Protein Sci. 2011, 20, 1005–1020. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Kozlov, A.G.; Roy, R.; Zhang, J.; Korolev, S.; Lohman, T.M.; Ha, T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell 2011, 146, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Liu, Y.; Wu, X.; Shell, S.M. Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J. Cell Physiol. 2006, 208, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Paulus, B.F.; Wold, M.S. Interactions of human replication protein A with oligonucleotides. Biochemistry 1994, 33, 14197–14206. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Le, S.; Basu, A.; Chazin, W.J.; Yan, J. Mechanochemical regulations of RPA’s binding to ssDNA. Sci. Rep. 2015, 5, 9296. [Google Scholar] [CrossRef] [PubMed]
- Kemmerich, F.E.; Daldrop, P.; Pinto, C.; Levikova, M.; Cejka, P.; Seidel, R. Force regulated dynamics of RPA on a DNA fork. Nucleic Acids Res. 2016, 44, 5837–5848. [Google Scholar] [CrossRef] [PubMed]
- Chrysogelos, S.; Griffith, J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc. Natl. Acad. Sci. USA 1982, 79, 5803–5807. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, S.; Kozlov, A.; Lohman, T.; Ansari, A. Microsecond dynamics of protein-DNA interactions: Direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J. Mol. Biol. 2006, 359, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Naufer, M.N.; Morse, M.; Moller, G.B.; McIsaac, J.; Rouzina, I.; Beuning, P.J.; Williams, M.C. Multiprotein E. coli SSB-ssDNA complex shows both stable binding and rapid dissociation due to interprotein interactions. Nucleic Acids Res. 2021, 49, 1532–1549. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.R.; Pottinger, S.; Tan, H.Y.; Nguyenduc, T.; Rex, K.; Varshney, U. The IDL of E. coli SSB links ssDNA and protein binding by mediating protein-protein interactions. Protein Sci. 2017, 26, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.; Betous, R.; Cortez, D. High-affinity DNA-binding domains of replication protein A (RPA) direct SMARCAL1-dependent replication fork remodeling. J. Biol. Chem. 2015, 290, 4110–4117. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Lloyd, R.G. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 2000, 101, 35–45. [Google Scholar] [CrossRef]
- McGlynn, P.; Lloyd, R. Rescue of stalled replication forks by RecG: Simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA 2001, 98, 8227–8234. [Google Scholar] [CrossRef]
- Gregg, A.; McGlynn, P.; Jaktaji, R.; Lloyd, R. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell. 2002, 9, 241–251. [Google Scholar] [CrossRef]
- Postow, L.; Ullsperger, C.; Keller, R.W.; Bustamante, C.; Vologodskii, A.V.; Cozzarelli, N.R. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 2001, 276, 2790–2796. [Google Scholar] [CrossRef] [PubMed]
- Malacaria, E.; Pugliese, G.M.; Honda, M.; Marabitti, V.; Aiello, F.A.; Spies, M.; Franchitto, A.; Pichierri, P. Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation. Nat. Commun. 2019, 10, 1412. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Lloyd, R.G.; Marians, K.J. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. USA 2001, 98, 8235–8240. [Google Scholar] [CrossRef]
- Bianco, P.R.; Bradfield, J.J.; Castanza, L.R.; Donnelly, A.N. Rad54 oligomers translocate and cross-bridge double-stranded DNA to stimulate synapsis. J. Mol. Biol. 2007, 374, 618–640. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bianco, P.R.; Tracy, R.B.; Kowalczykowski, S.C. DNA strand exchange proteins: A biochemical and physical comparison. Front. Biosci. 1998, 3, D570–D603. [Google Scholar]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianco, P.R. The Biochemical Mechanism of Fork Regression in Prokaryotes and Eukaryotes—A Single Molecule Comparison. Int. J. Mol. Sci. 2022, 23, 8613. https://doi.org/10.3390/ijms23158613
Bianco PR. The Biochemical Mechanism of Fork Regression in Prokaryotes and Eukaryotes—A Single Molecule Comparison. International Journal of Molecular Sciences. 2022; 23(15):8613. https://doi.org/10.3390/ijms23158613
Chicago/Turabian StyleBianco, Piero R. 2022. "The Biochemical Mechanism of Fork Regression in Prokaryotes and Eukaryotes—A Single Molecule Comparison" International Journal of Molecular Sciences 23, no. 15: 8613. https://doi.org/10.3390/ijms23158613
APA StyleBianco, P. R. (2022). The Biochemical Mechanism of Fork Regression in Prokaryotes and Eukaryotes—A Single Molecule Comparison. International Journal of Molecular Sciences, 23(15), 8613. https://doi.org/10.3390/ijms23158613





