Knockout of Katnal2 Leads to Autism-like Behaviors and Developmental Delay in Zebrafish
Abstract
:1. Introduction
2. Results
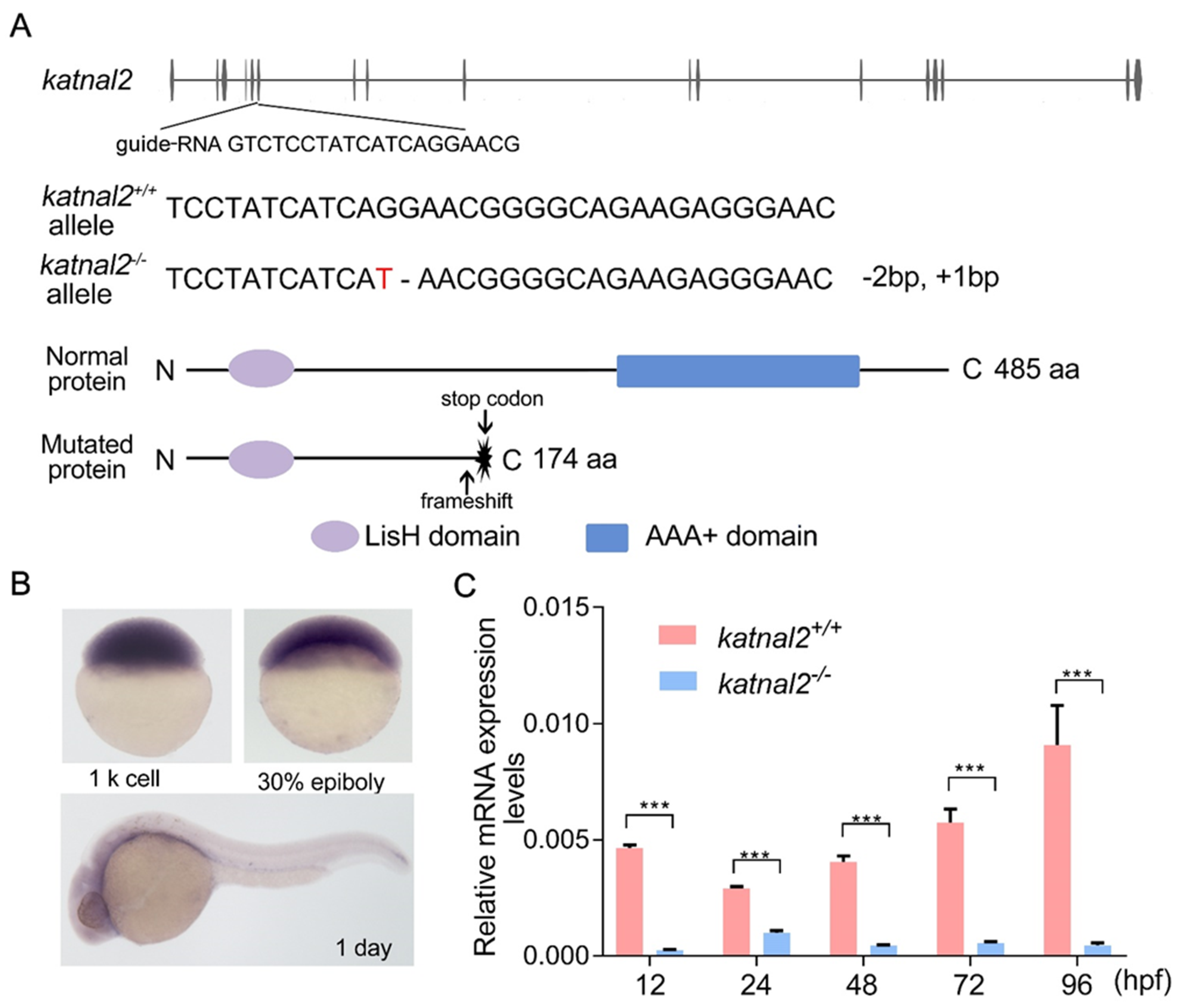
2.1. Generation of Katnal2-/- Zebrafish
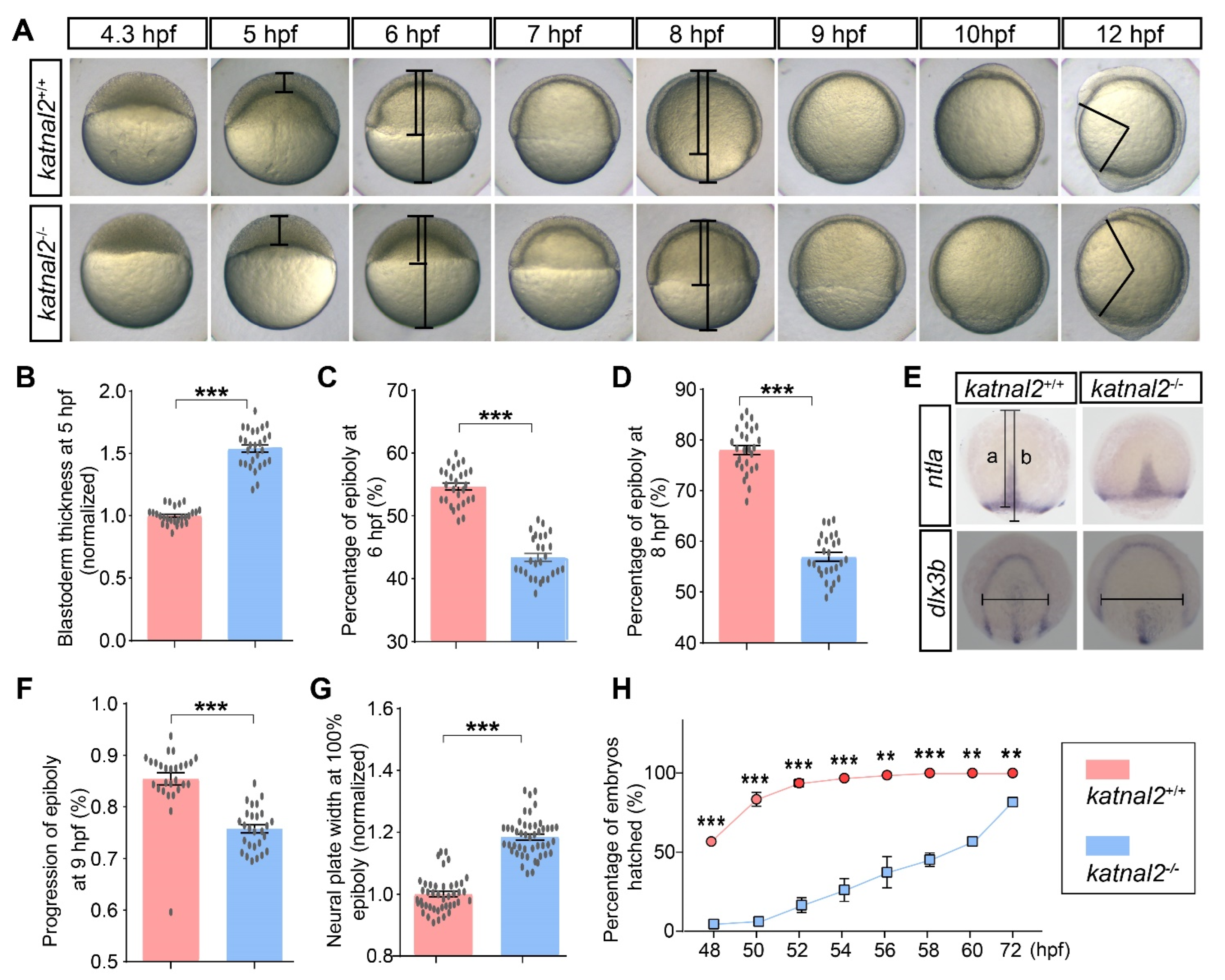
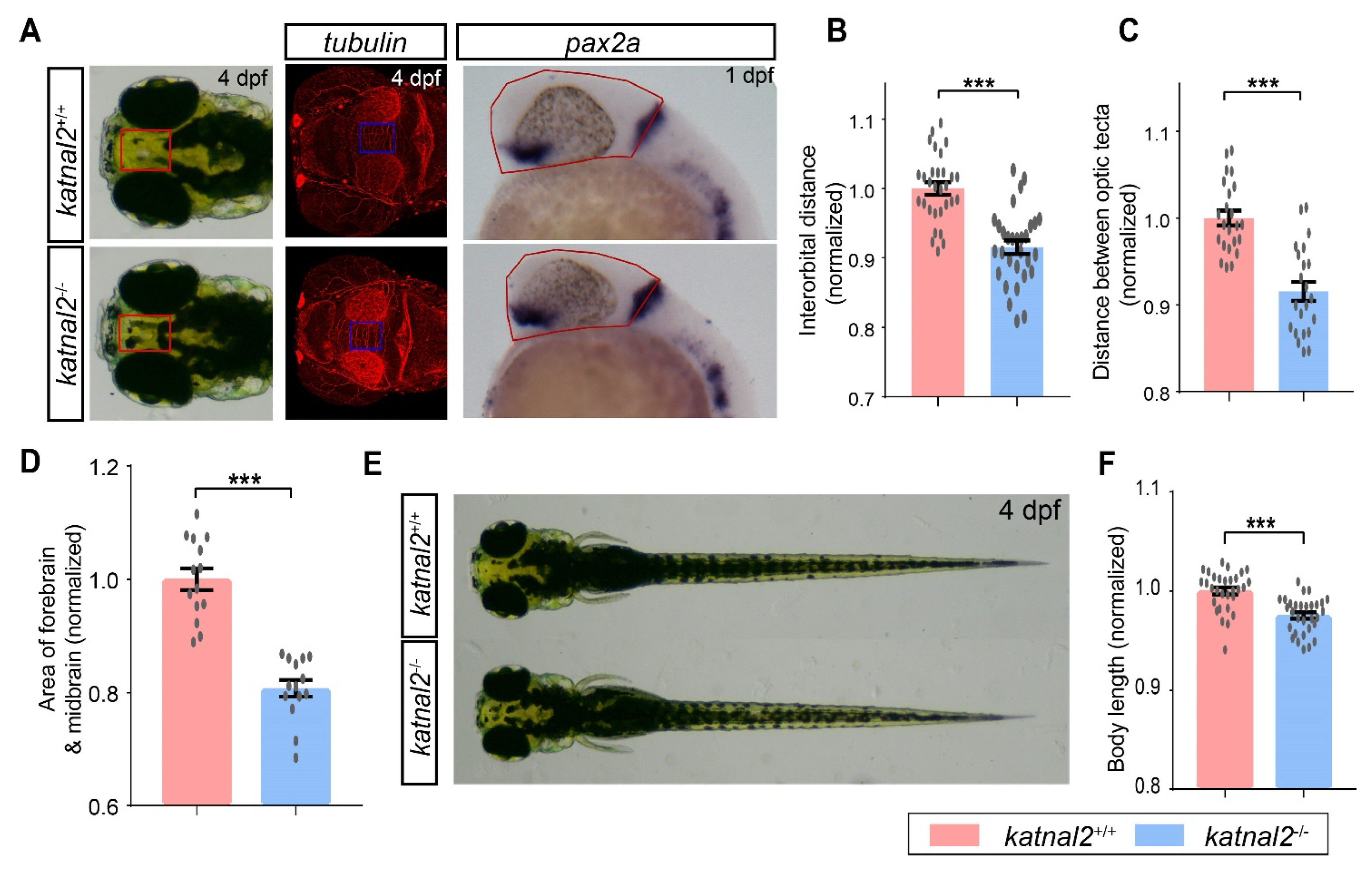
2.2. Katnal2-/- Zebrafish Displayed Developmental Delay during Embryogenesis
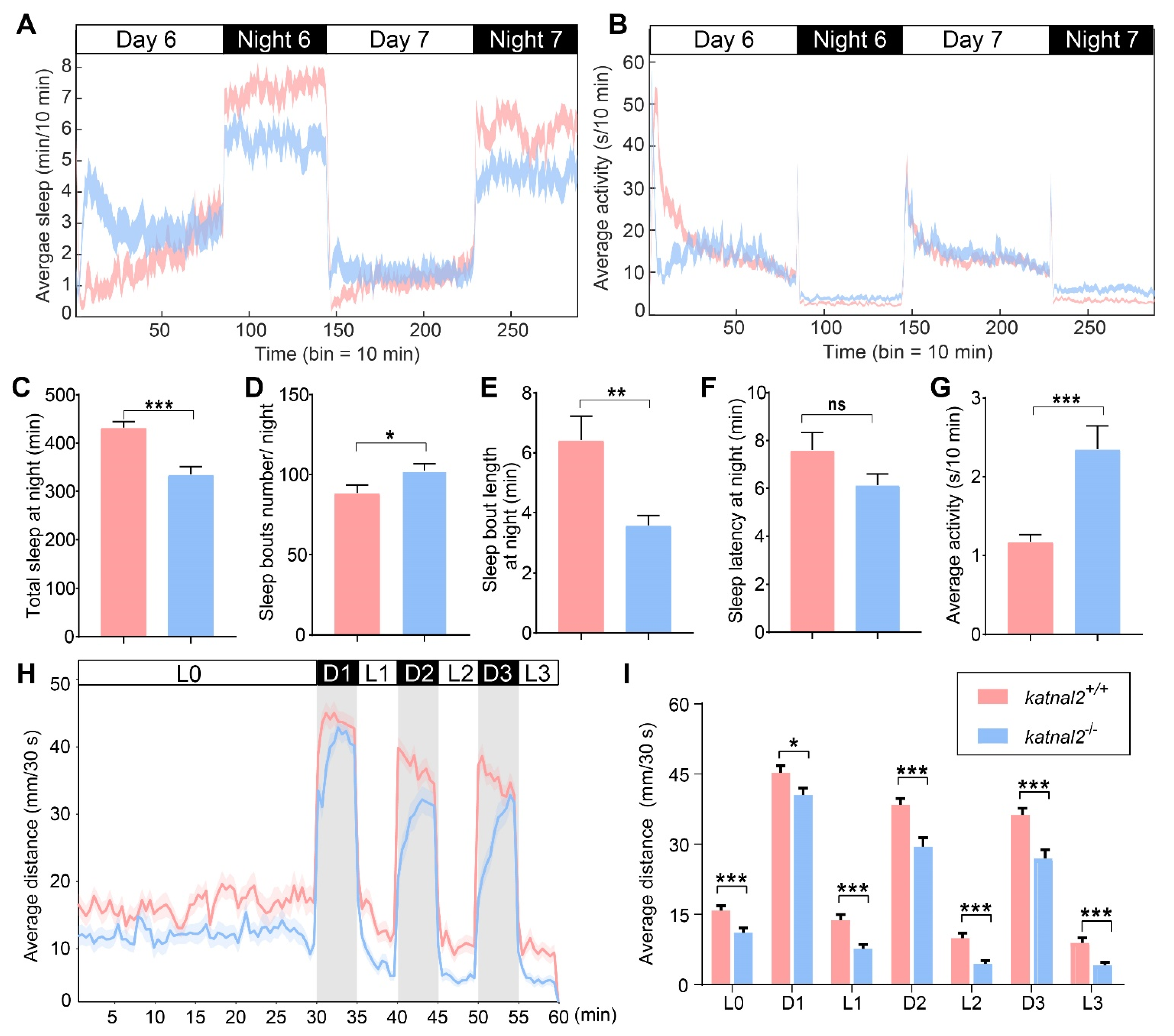
2.3. Katnal2-/- Larvae Displayed Abnormal Sleep States and Impaired Locomotion Activity
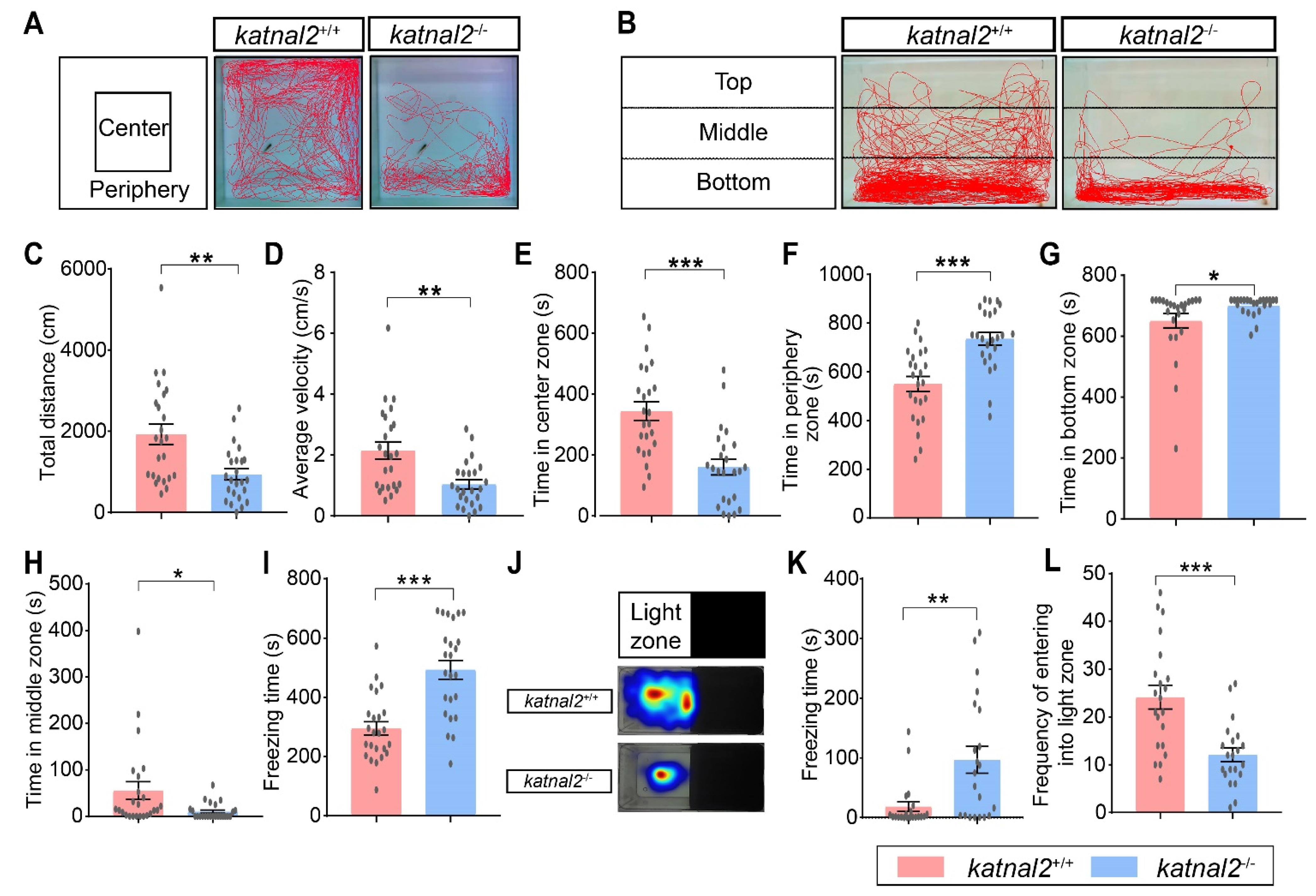
2.4. Adult Katnal2-/- Zebrafish Displayed Impaired Locomotion Activity and Elevated Anxiety Level
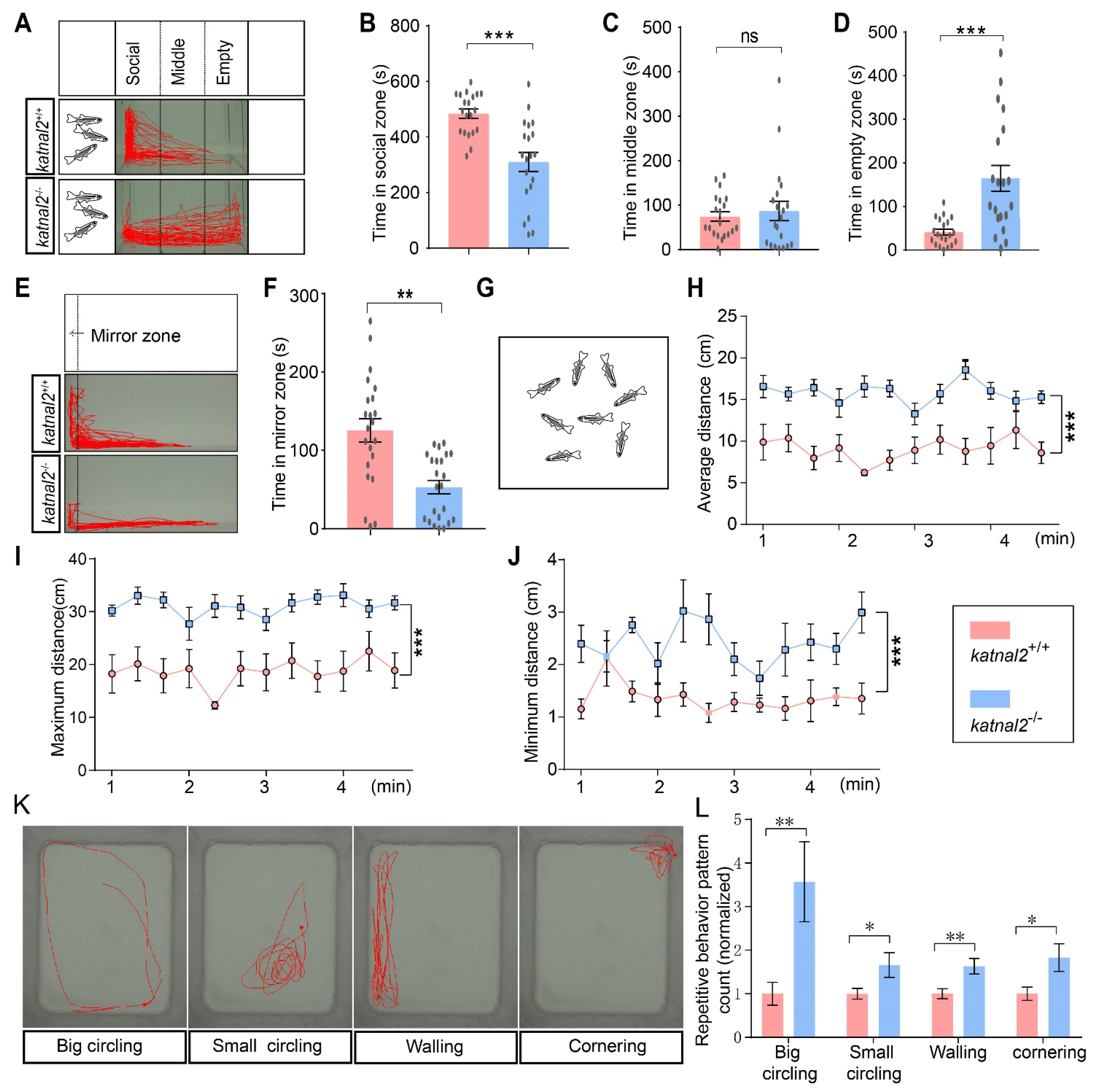
2.5. Katnal2-/- Zebrafish Exhibited ASD-like Behaviors
3. Discussion
4. Materials and Methods
4.1. Zebrafish Culture
4.2. Generation of Katnal2 Mutant Zebrafish
4.3. RNA Extraction and RT-PCR
4.4. Whole Mount mRNA In Situ Hybridization
4.5. Whole Mount Immunofluorescence
4.6. Larval Locomotion Activity Test
4.7. Sleep and Waking Activity of Zebrafish Larvae
4.8. Open Field Test
4.9. Novel Tank Assay
4.10. Light Dark Box Assay
4.11. Social Interaction Assay
4.12. Mirror Assay
4.13. Shoaling Assay
4.14. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iakoucheva, L.M.; Muotri, A.R.; Sebat, J. Getting to the Cores of Autism. Cell 2019, 178, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Brugha, T.S.; Charman, T.; Cusack, J.; Dumas, G.; Frazier, T.; Jones, E.J.H.; Jones, R.M.; Pickles, A.; State, M.W.; et al. Autism spectrum disorder. Nat. Rev. Dis. Primers 2020, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.J.; Brugha, T.S.; Erskine, H.E.; Scheurer, R.W.; Vos, T.; Scott, J.G. The epidemiology and global burden of autism spectrum disorders. Psychol. Med. 2015, 45, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.; Yip, B.H.K.; Windham, G.C.; Sourander, A.; Francis, R.; Yoffe, R.; Glasson, E.; Mahjani, B.; Suominen, A.; Leonard, H.; et al. Association of Genetic and Environmental Factors With Autism in a 5-Country Cohort. JAMA Psychiatry 2019, 76, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Vorstman, J.A.S.; Parr, J.R.; Moreno-De-Luca, D.; Anney, R.J.L.; Nurnberger Jr, J.I.; Hallmayer, J.F. Autism genetics: Opportunities and challenges for clinical translation. Nature Rev. Genet. 2017, 18, 362–376. [Google Scholar] [CrossRef] [PubMed]
- O’Roak, B.J.; Vives, L.; Girirajan, S.; Karakoc, E.; Krumm, N.; Coe, B.P.; Levy, R.; Ko, A.; Lee, C.; Smith, J.D.; et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012, 485, 246–250. [Google Scholar] [CrossRef] [Green Version]
- Sanders, S.J.; Murtha, M.T.; Gupta, A.R.; Murdoch, J.D.; Raubeson, M.J.; Willsey, A.J.; Ercan-Sencicek, A.G.; DiLullo, N.M.; Parikshak, N.N.; Stein, J.L.; et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012, 485, 237–241. [Google Scholar] [CrossRef]
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Ercument Cicek, A.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef]
- Stessman, H.A.F.; Xiong, B.; Coe, B.P.; Wang, T.; Hoekzema, K.; Fenckova, M.; Kvarnung, M.; Gerdts, J.; Trinh, S.; Cosemans, N.; et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017, 49, 515–526. [Google Scholar] [CrossRef]
- Aspromonte, M.C.; Bellini, M.; Gasparini, A.; Carraro, M.; Bettella, E.; Polli, R.; Cesca, F.; Bigoni, S.; Boni, S.; Carlet, O.; et al. Characterization of intellectual disability and autism comorbidity through gene panel sequencing. Hum. Mutat. 2019, 40, 1346–1363. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Wang, T.; Wu, H.; Long, M.; Coe, B.P.; Li, H.; Xun, G.; Ou, J.; Chen, B.; Duan, G.; et al. Inherited and multiple de novo mutations in autism/developmental delay risk genes suggest a multifactorial model. Mol. Autism 2018, 9, 64. [Google Scholar] [CrossRef]
- Husson, T.; Lecoquierre, F.; Cassinari, K.; Charbonnier, C.; Quenez, O.; Goldenberg, A.; Guerrot, A.-M.; Richard, A.-C.; Drouin-Garraud, V.; Brehin, A.-C.; et al. Rare genetic susceptibility variants assessment in autism spectrum disorder: Detection rate and practical use. Transl. Psychiatry 2020, 10, 77. [Google Scholar] [CrossRef]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Hoekzema, K.; Vecchio, D.; Wu, H.; Sulovari, A.; Coe, B.P.; Gillentine, M.A.; Wilfert, A.B.; Perez-Jurado, L.A.; Kvarnung, M.; et al. Large-scale targeted sequencing identifies risk genes for neurodevelopmental disorders. Nat. Commun. 2020, 11, 4932. [Google Scholar] [CrossRef]
- Yuen, R.K.C.; Thiruvahindrapuram, B.; Merico, D.; Walker, S.; Tammimies, K.; Hoang, N.; Chrysler, C.; Nalpathamkalam, T.; Pellecchia, G.; Liu, Y.; et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015, 21, 185–191. [Google Scholar] [CrossRef]
- Ververis, A.; Christodoulou, A.; Christoforou, M.; Kamilari, C.; Lederer, C.W.; Santama, N. A novel family of katanin-like 2 protein isoforms (KATNAL2), interacting with nucleotide-binding proteins Nubp1 and Nubp2, are key regulators of different MT-based processes in mammalian cells. Cell. Mol. Life Sci. 2016, 73, 163–184. [Google Scholar] [CrossRef] [Green Version]
- Willsey, H.R.; Walentek, P.; Exner, C.R.T.; Xu, Y.; Lane, A.B.; Harland, R.M.; Heald, R.; Santama, N. Katanin-like protein Katnal2 is required for ciliogenesis and brain development in Xenopus embryos. Dev. Biol. 2018, 442, 276–287. [Google Scholar] [CrossRef]
- Joachimiak, E.; Waclawek, E.; Niziolek, M.; Osinka, A.; Fabczak, H.; Gaertig, J.; Wloga, D. The LisH Domain-Containing N-Terminal Fragment is Important for the Localization, Dimerization, and Stability of Katnal2 in Tetrahymena. Cells 2020, 9, 292. [Google Scholar] [CrossRef] [Green Version]
- Mariella A., M. Franker, Casper C. Hoogenraad. Microtubule-based transport—Basic mechanisms, traffic rules and role in neurological pathogenesis. J. Cell Sci. 2013, 126, 2319–2329. [Google Scholar] [CrossRef] [Green Version]
- Williams, M.R.; Fricano-Kugler, C.J.; Getz, S.A.; Skelton, P.D.; Lee, J.; Rizzuto, C.P.; Geller, J.S.; Li, M.; Luikart, B.W. A Retroviral CRISPR-Cas9 System for Cellular Autism-Associated Phenotype Discovery in Developing Neurons. Sci. Rep. 2016, 6, 25611. [Google Scholar] [CrossRef] [Green Version]
- Banks, G.; Lassi, G.; Hoerder-Suabedissen, A.; Tinarelli, F.; Simon, M.M.; Wilcox, A.; Lau, P.; Lawson, T.N.; Johnson, S.; Rutman, A.; et al. A missense mutation in Katnal1 underlies behavioural, neurological and ciliary anomalies. Mol. Psychiatry 2018, 23, 713–722. [Google Scholar] [CrossRef]
- Bartholdi, D.; Stray-Pedersen, A.; Azzarello-Burri, S.; Kibaek, M.; Kirchhoff, M.; Oneda, B.; Rødningen, O.; Schmitt-Mechelke, T.; Rauch, A.; Kjaergaard, S. A newly recognized 13q12.3 microdeletion syndrome characterized by intellectual disability, microcephaly, and eczema/atopic dermatitis encompassing the HMGB1 and KATNAL1 genes. Am. J. Med. Genet. Part A 2014, 164, 1277–1283. [Google Scholar] [CrossRef]
- Mishra-Gorur, K.; Çağlayan, A.O.; Schaffer, A.E.; Chabu, C.; Henegariu, O.; Vonhoff, F.; Akgümüş, G.T.; Nishimura, S.; Han, W.; Tu, S.; et al. Mutations in KATNB1 Cause Complex Cerebral Malformations by Disrupting Asymmetrically Dividing Neural Progenitors. Neuron 2014, 84, 1226–1239. [Google Scholar] [CrossRef]
- Cheung, K.; Senese, S.; Kuang, J.; Bui, N.; Ongpipattanakul, C.; Gholkar, A.; Cohn, W.; Capri, J.; Whitelegge, J.P.; Torres, J.Z. Proteomic Analysis of the Mammalian Katanin Family of Microtubule-severing Enzymes Defines Katanin p80 subunit B-like 1 (KATNBL1) as a Regulator of Mammalian Katanin Microtubule-severing. Mol. Cell. Proteom. MCP 2016, 15, 1658–1669. [Google Scholar] [CrossRef] [Green Version]
- Solnica-Krezel, L.; Driever, W. Microtubule arrays of the zebrafish yolk cell: Organization and function during epiboly. Development 1994, 120, 2443–2455. [Google Scholar] [CrossRef]
- Koh, A.; Tao, S.; Jing Goh, Y.; Chaganty, V.; See, K.; Purushothaman, K.; Orbán, L.; Mathuru, A.S.; Wohland, T.; Winkler, C. A Neurexin2aa deficiency results in axon pathfinding defects and increased anxiety in zebrafish. Hum. Mol. Genet. 2020, 29, 3765–3780. [Google Scholar] [CrossRef] [PubMed]
- Kalueff, A.V.; Stewart, A.M.; Gerlai, R. Zebrafish as an emerging model for studying complex brain disorders. Trends Pharmacol. Sci. 2014, 35, 63–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, S.; Simonich, M.T.; Tanguay, R.L. Zebrafish Behavioral Assays in Toxicology. In High-Throughput Screening Assays in Toxicology; Zhu, H., Xia, M., Eds.; Springer: New York, NY, USA, 2022; pp. 109–122. [Google Scholar]
- Orger, M.B.; de Polavieja, G.G. Zebrafish Behavior: Opportunities and Challenges. Annu. Rev. Neurosci. 2017, 40, 125–147. [Google Scholar] [CrossRef] [Green Version]
- Peng, W.; Wu, Z.; Song, K.; Zhang, S.; Li, Y.; Xu, M. Regulation of sleep homeostasis mediator adenosine by basal forebrain glutamatergic neurons. Science 2020, 369, eabb0556. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.J.; Turner, K.J.; Fernandez, J.M.; Cifuentes, D.; Ghosh, M.; Ijaz, S.; Jain, R.A.; Kubo, F.; Bill, B.R.; Baier, H.; et al. Estrogens Suppress a Behavioral Phenotype in Zebrafish Mutants of the Autism Risk Gene, CNTNAP2. Neuron 2016, 89, 725–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kokel, D.; Rennekamp, A.J.; Shah, A.H.; Liebel, U.; Peterson, R.T. Behavioral barcoding in the cloud: Embracing data-intensive digital phenotyping in neuropharmacology. Trends Biotechnol. 2012, 30, 421–425. [Google Scholar] [CrossRef] [Green Version]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt Kevin, M.; Aach, J.; Guell, M.; DiCarlo James, E.; Norville Julie, E.; Church George, M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef]
- Wilson, S.W.; Ross, L.S.; Parrett, T.; Easter, S.S. The development of a simple scaffold of axon tracts in the brain of the embryonic zebrafish, Brachydanio rerio. Development 1990, 108, 121–145. [Google Scholar] [CrossRef]
- Bernier, R.; Golzio, C.; Xiong, B.; Stessman, H.A.; Coe, B.P.; Penn, O.; Witherspoon, K.; Gerdts, J.; Baker, C.; Vulto-van Silfhout, A.T.; et al. Disruptive CHD8 Mutations Define a Subtype of Autism Early in Development. Cell 2014, 158, 263–276. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-x.; Li, C.-y.; Hu, C.-c.; Wang, Y.; Lin, J.; Jiang, Y.-h.; Li, Q.; Xu, X. CRISPR/Cas9-induced shank3b mutant zebrafish display autism-like behaviors. Mol. Autism 2018, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Rihel, J.; Prober, D.A.; Schier, A.F. Chapter 11—Monitoring Sleep and Arousal in Zebrafish. In Methods in Cell Biology; Detrich, H.W., Westerfield, M., Zon, L.I., Eds.; Academic Press: Cambridge, MA, USA, 2010; Volume 100, pp. 281–294. [Google Scholar]
- Lee, D.A.; Oikonomou, G.; Prober, D.A. Large-scale Analysis of Sleep in Zebrafish. Bio-Protocol 2022, 12, e4313. [Google Scholar] [CrossRef]
- Rihel, J.; Prober, D.A.; Arvanites, A.; Lam, K.; Zimmerman, S.; Jang, S.; Haggarty, S.J.; Kokel, D.; Rubin, L.L.; Peterson, R.T.; et al. Zebrafish Behavioral Profiling Links Drugs to Biological Targets and Rest/Wake Regulation. Science 2010, 327, 348–351. [Google Scholar] [CrossRef] [Green Version]
- Cachat, J.; Stewart, A.; Grossman, L.; Gaikwad, S.; Kadri, F.; Chung, K.M.; Wu, N.; Wong, K.; Roy, S.; Suciu, C.; et al. Measuring behavioral and endocrine responses to novelty stress in adult zebrafish. Nat. Protoc. 2010, 5, 1786–1799. [Google Scholar] [CrossRef]
- Maximino, C.; de Brito, T.M.; Colmanetti, R.; Pontes, A.A.A.; de Castro, H.M.; de Lacerda, R.I.T.; Morato, S.; Gouveia, A. Parametric analyses of anxiety in zebrafish scototaxis. Behavioural Brain Res. 2010, 210, 1–7. [Google Scholar] [CrossRef]
- Engeszer, R.E.; Ryan, M.J.; Parichy, D.M. Learned Social Preference in Zebrafish. Curr. Biol. 2004, 14, 881–884. [Google Scholar] [CrossRef] [Green Version]
- Gerlai, R.; Lahav, M.; Guo, S.; Rosenthal, A. Drinks like a fish: Zebra fish (Danio rerio) as a behavior genetic model to study alcohol effects. Pharmacol. Biochem. Behav. 2000, 67, 773–782. [Google Scholar] [CrossRef]
- Green, J.; Collins, C.; Kyzar, E.J.; Pham, M.; Roth, A.; Gaikwad, S.; Cachat, J.; Stewart, A.M.; Landsman, S.; Grieco, F.; et al. Automated high-throughput neurophenotyping of zebrafish social behavior. J. Neurosci. Methods 2012, 210, 266–271. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Long, F.; Cao, X.; Xiong, B.; Li, Y. Knockout of Katnal2 Leads to Autism-like Behaviors and Developmental Delay in Zebrafish. Int. J. Mol. Sci. 2022, 23, 8389. https://doi.org/10.3390/ijms23158389
Zheng J, Long F, Cao X, Xiong B, Li Y. Knockout of Katnal2 Leads to Autism-like Behaviors and Developmental Delay in Zebrafish. International Journal of Molecular Sciences. 2022; 23(15):8389. https://doi.org/10.3390/ijms23158389
Chicago/Turabian StyleZheng, Jing, Fei Long, Xu Cao, Bo Xiong, and Yu Li. 2022. "Knockout of Katnal2 Leads to Autism-like Behaviors and Developmental Delay in Zebrafish" International Journal of Molecular Sciences 23, no. 15: 8389. https://doi.org/10.3390/ijms23158389
APA StyleZheng, J., Long, F., Cao, X., Xiong, B., & Li, Y. (2022). Knockout of Katnal2 Leads to Autism-like Behaviors and Developmental Delay in Zebrafish. International Journal of Molecular Sciences, 23(15), 8389. https://doi.org/10.3390/ijms23158389





