Molecular Dynamics Simulations of Curved Lipid Membranes
Abstract
1. Introduction
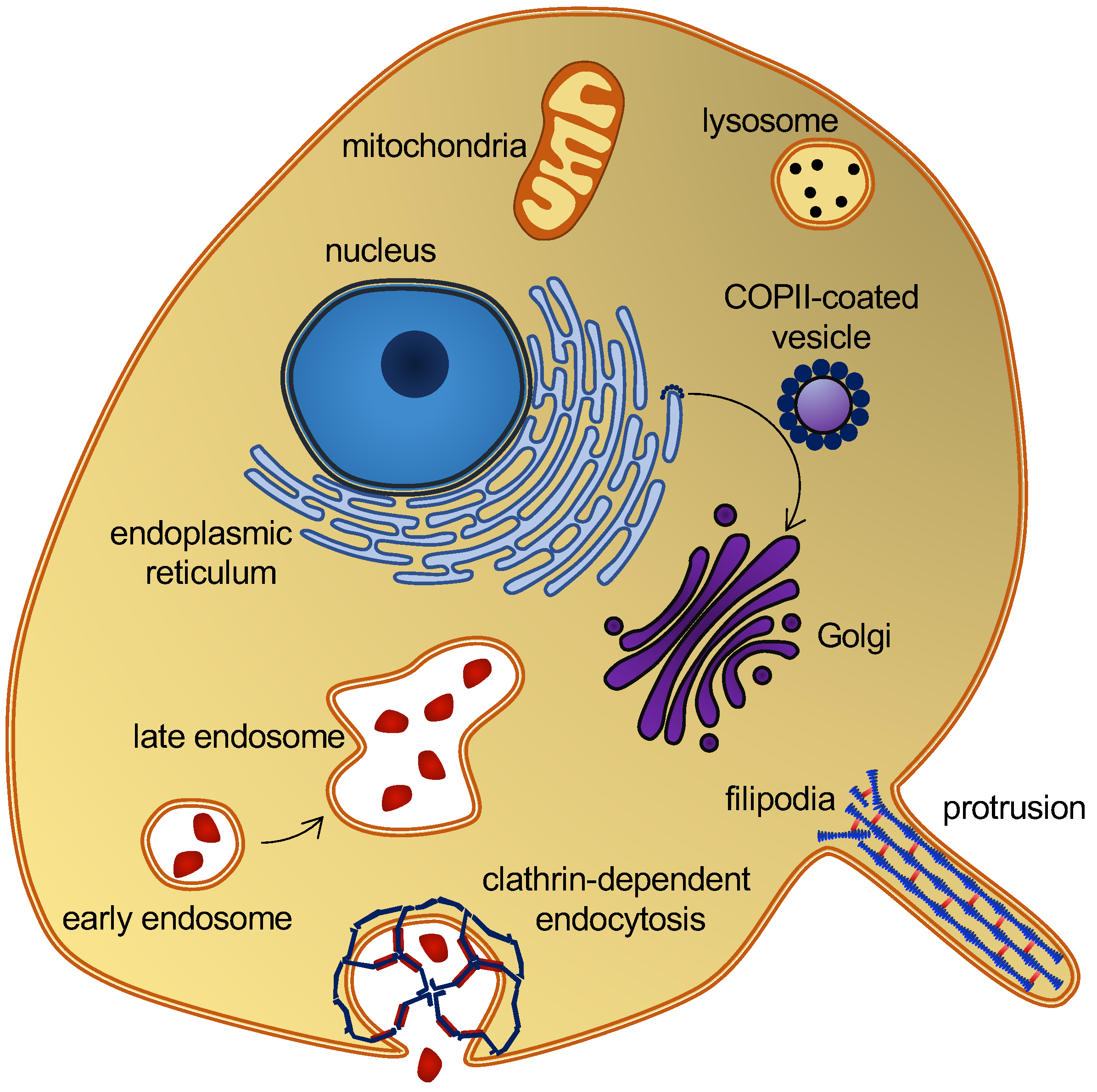
2. Biological Background
3. Spontaneously Induced Membrane Curvature
3.1. Self-Assembly
3.2. Bending of Plane Bilayers with Periodic Boundary Conditions
3.3. Bicelles and Bicelle-to-Vesicle Transitions
3.4. Budding of Plane Membranes
3.5. Lipid Bilayers with Free Edges in One Direction
4. Vesicle Formation
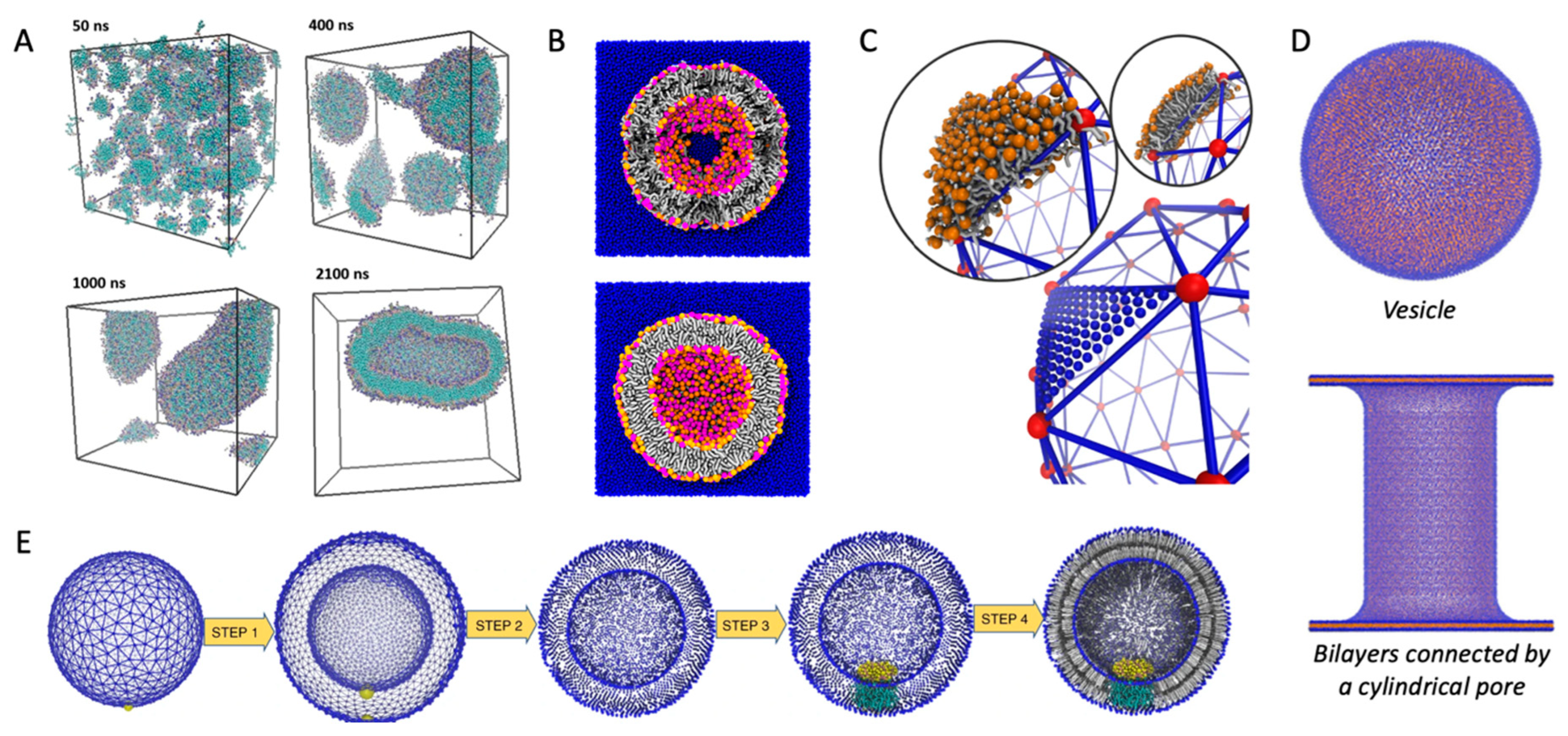
4.1. Self-Assembled Vesicles
4.2. CHARMM-GUI Vesicle Builders
4.3. From Triangular Surfaces to Lipid Vesicles
4.4. Meshless Grid
4.5. Transformation from Flat Membranes to Vesicles
5. Non-Spontaneous Membrane Curvature Sustained by Scaffolding Beads or Virtual Forces
5.1. Virtual Beads and Solid Supports
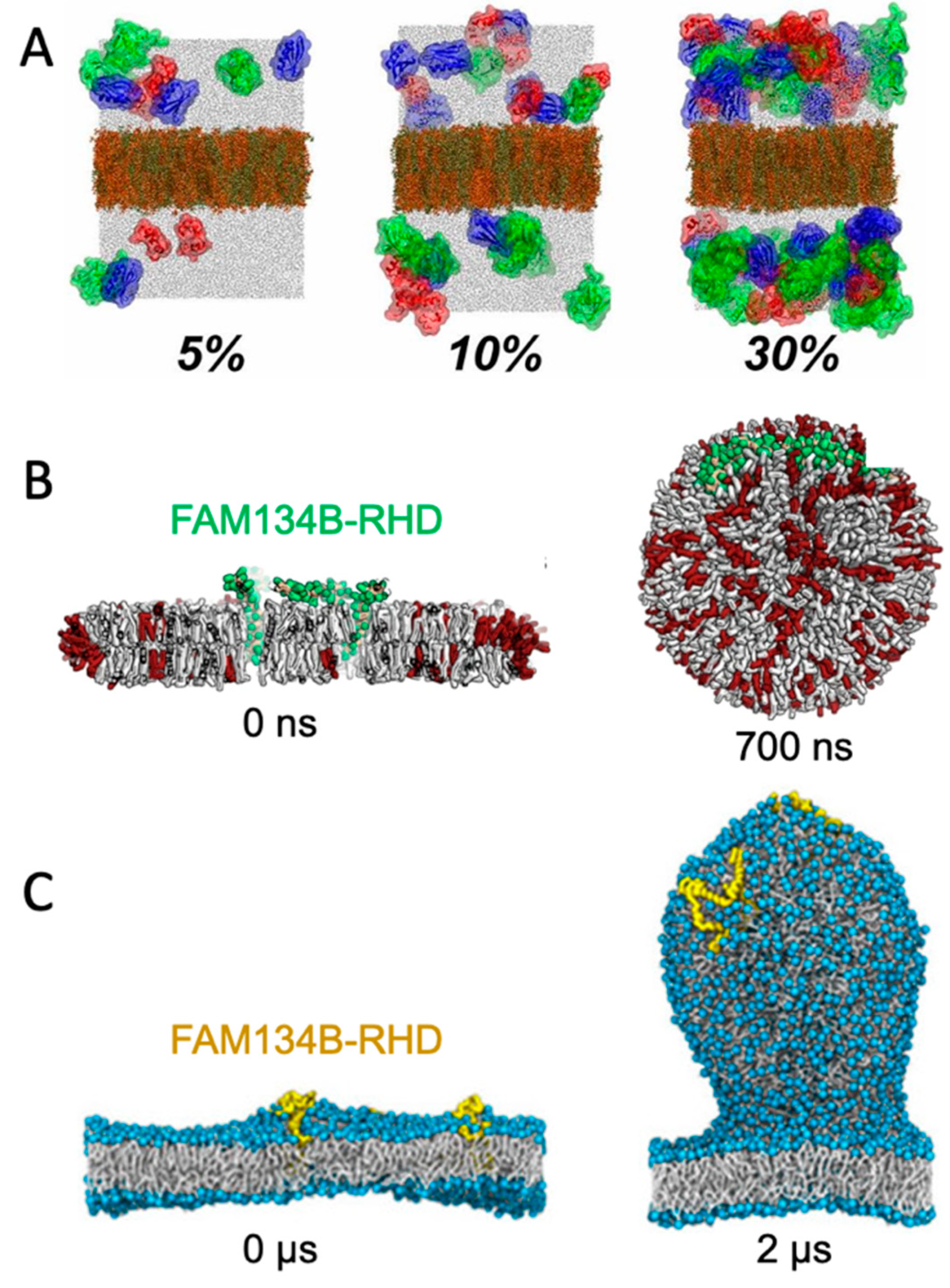
5.2. Curvature-Inducing Molecules
5.3. Virtual Forces
5.4. Free Energy Calculations
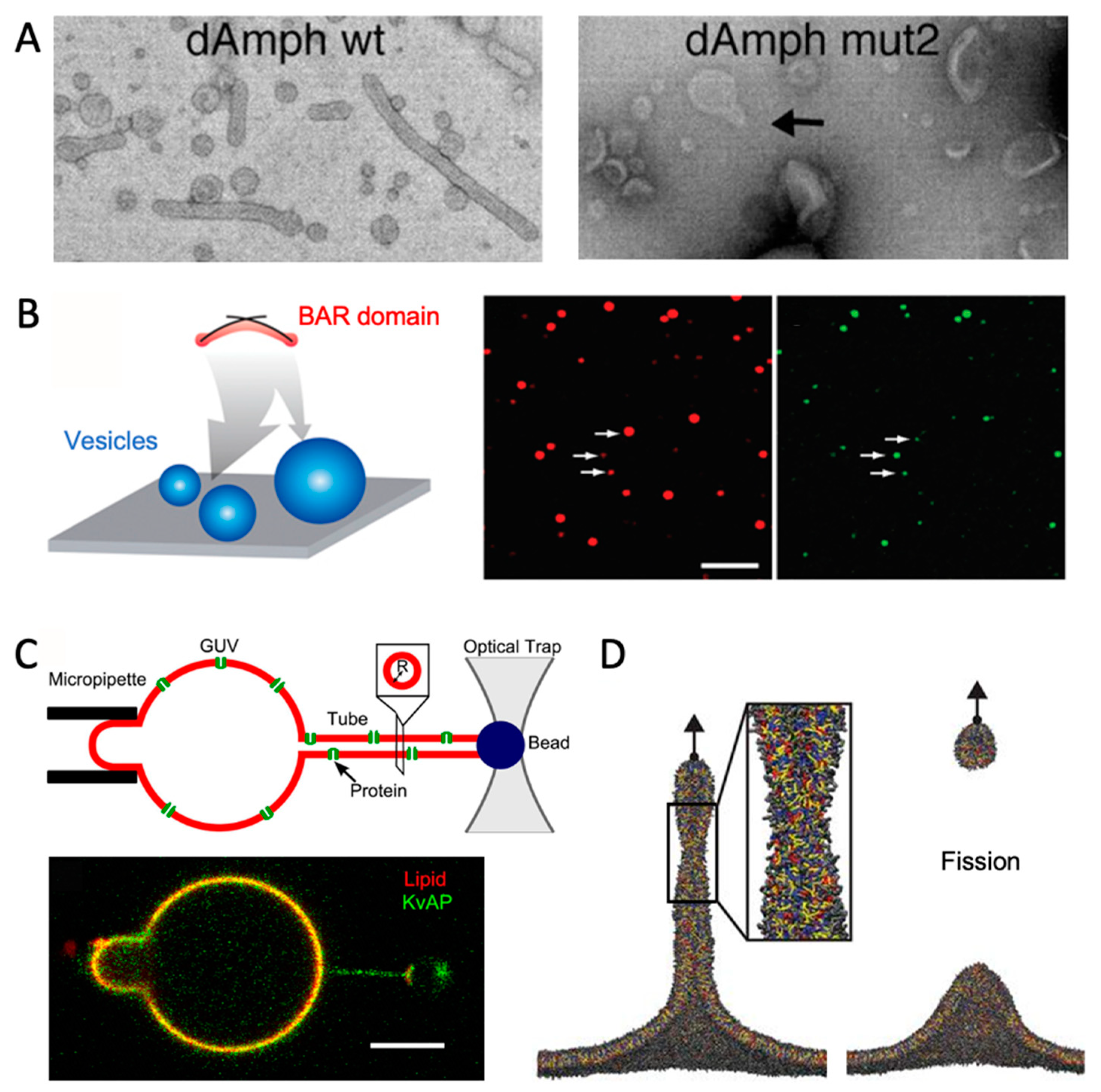
6. Comparison with Experiments
6.1. Vesicles
6.2. GUVs with Tubules
6.3. Solid Support
6.4. Example: Combining MD and Experiments for a Curved Membrane
7. Challenges and Future Directions
8. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Has, C.; Das, S.L. Recent Developments in Membrane Curvature Sensing and Induction by Proteins. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129971. [Google Scholar] [CrossRef] [PubMed]
- Warner, H.; Wilson, B.J.; Caswell, P.T. Control of Adhesion and Protrusion in Cell Migration by Rho GTPases. Curr. Opin. Cell Biol. 2019, 56, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Pezeshkian, W.; Marrink, S.J. Simulating Realistic Membrane Shapes. Curr. Opin. Cell Biol. 2021, 71, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; Corradi, V.; Souza, P.C.T.; Ingólfsson, H.I.; Tieleman, D.P.; Sansom, M.S.P. Computational Modeling of Realistic Cell Membranes. Chem. Rev. 2019, 119, 6184–6226. [Google Scholar] [CrossRef]
- Pezeshkian, W.; König, M.; Wassenaar, T.A.; Marrink, S.J. Backmapping Triangulated Surfaces to Coarse-Grained Membrane Models. Nat. Commun. 2020, 11, 2296. [Google Scholar] [CrossRef]
- Moqadam, M.; Tubiana, T.; Moutoussamy, E.E.; Reuter, N. Membrane Models for Molecular Simulations of Peripheral Membrane Proteins. Adv. Phys. X 2021, 6, 1932589. [Google Scholar] [CrossRef]
- Bar, L.; Perissinotto, F.; Redondo-Morata, L.; Giannotti, M.I.; Goole, J.; Losada-Pérez, P. Interactions of Hydrophilic Quantum Dots with Defect-Free and Defect Containing Supported Lipid Membranes. Colloids Surf. B Biointerfaces 2022, 210, 112239. [Google Scholar] [CrossRef]
- Lee, H. Membrane Penetration and Curvature Induced by Single-Walled Carbon Nanotubes: The Effect of Diameter, Length, and Concentration. Phys. Chem. Chem. Phys. 2013, 15, 16334–16340. [Google Scholar] [CrossRef]
- Duden, R. ER-to-Golgi Transport: COP I and COP II Function. Mol. Membr. Biol. 2003, 20, 197–207. [Google Scholar] [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of Clathrin-Mediated Endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
- Frost, A.; Unger, V.M.; De Camilli, P. The BAR Domain Superfamily: Membrane-Molding Macromolecules. Cell 2009, 137, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Simunovic, M.; Evergren, E.; Callan-Jones, A.; Bassereau, P. Curving Cells inside and out: Roles of BAR Domain Proteins in Membrane Shaping and Its Cellular Implications. Annu. Rev. Cell Dev. Biol. 2019, 35, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Simunovic, M.; Voth, G.A.; Callan-Jones, A.; Bassereau, P. When Physics Takes Over: BAR Proteins and Membrane Curvature. Trends Cell Biol. 2015, 25, 780–792. [Google Scholar] [CrossRef] [PubMed]
- Jao, C.C.; Hegde, B.G.; Gallop, J.L.; Hegde, P.B.; McMahon, H.T.; Haworth, I.S.; Langen, R. Roles of Amphipathic Helices and the Bin/Amphiphysin/Rvs (BAR) Domain of Endophilin in Membrane Curvature Generation. J. Biol. Chem. 2010, 285, 20164–20170. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing Antimicrobial Peptides: Form Follows Function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Xu, K.; Zhou, T.; Acharya, P.; Lemmin, T.; Liu, K.; Ozorowski, G.; Soto, C.; Taft, J.D.; Bailer, R.T.; et al. Fusion Peptide of HIV-1 as a Site of Vulnerability to Neutralizing Antibody. Nat. Microbiol. 2016, 352, 828–832. [Google Scholar] [CrossRef]
- Hanzal-Bayer, M.F.; Hancock, J.F. Lipid Rafts and Membrane Traffic. FEBS Lett. 2007, 581, 2098–2104. [Google Scholar] [CrossRef]
- Doktorova, M.; Symons, J.L.; Levental, I. Structural and Functional Consequences of Reversible Lipid Asymmetry in Living Membranes. Nat. Chem. Biol. 2020, 16, 1321–1330. [Google Scholar] [CrossRef]
- Cooke, I.R.; Deserno, M. Coupling between Lipid Shape and Membrane Curvature. Biophys. J. 2006, 91, 487–495. [Google Scholar] [CrossRef]
- Yu, H.; Schulten, K. Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations. PLoS Comput. Biol. 2013, 9, e1002892. [Google Scholar] [CrossRef]
- Blood, P.D.; Swenson, R.D.; Voth, G.A. Factors Influencing Local Membrane Curvature Induction by N-BAR Domains as Revealed by Molecular Dynamics Simulations. Biophys. J. 2008, 95, 1866–1876. [Google Scholar] [CrossRef] [PubMed]
- Kluge, C.; Pöhnl, M.; Böckmann, R.A. Spontaneous Local Membrane Curvature Induced by Transmembrane Proteins. Biophys. J. 2022, 121, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.H.; Hsu, H.J.; Bartenschlager, R.; Fischer, W.B. Membrane Undulation Induced by NS4A of Dengue Virus: A Molecular Dynamics Simulation Study. J. Biomol. Struct. Dyn. 2014, 32, 1552–1562. [Google Scholar] [CrossRef]
- Nawrocki, G.; Im, W.; Sugita, Y.; Feig, M. Clustering and Dynamics of Crowded Proteins near Membranes and Their Influence on Membrane Bending. Proc. Natl. Acad. Sci. USA 2019, 116, 24562–24567. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, R.M.; Grumati, P.; Garcia-Pardo, J.; Kalayil, S.; Covarrubias-Pinto, A.; Chen, W.; Kudryashev, M.; Dikic, I.; Hummer, G. Curvature Induction and Membrane Remodeling by FAM134B Reticulon Homology Domain Assist Selective ER-Phagy. Nat. Commun. 2019, 10, 2370. [Google Scholar] [CrossRef]
- Siggel, M.; Bhaskara, R.M.; Moesser, M.K.; Dikić, I.; Hummer, G. FAM134B-RHD Protein Clustering Drives Spontaneous Budding of Asymmetric Membranes. J. Phys. Chem. Lett. 2021, 12, 1926–1931. [Google Scholar] [CrossRef]
- Raschle, T.; Hiller, S.; Etzkorn, M.; Wagner, G. Nonmicellar Systems for Solution NMR Spectroscopy of Membrane Proteins. Curr. Opin. Struct. Biol. 2010, 20, 471–479. [Google Scholar] [CrossRef]
- Pannuzzo, M.; Raudino, A.; Böckmann, R.A. Peptide-Induced Membrane Curvature in Edge-Stabilized Open Bilayers: A Theoretical and Molecular Dynamics Study. J. Chem. Phys. 2014, 141, 024901. [Google Scholar] [CrossRef]
- dos Santos Soares, R.D.O.; Bortot, L.O.; Van Der Spoel, D.; Caliri, A. Membrane Vesiculation Induced by Proteins of the Dengue Virus Envelope Studied by Molecular Dynamics Simulations. J. Phys. Condens. Matter 2017, 29, 504002. [Google Scholar] [CrossRef]
- Mahmood, M.I.; Noguchi, H.; Okazaki, K. Curvature Induction and Sensing of the F-BAR Protein Pacsin1 on Lipid Membranes via Molecular Dynamics Simulations. Sci. Rep. 2019, 9, 14557. [Google Scholar] [CrossRef]
- Mandal, T.; Lough, W.; Spagnolie, S.E.; Audhya, A.; Cui, Q. Molecular Simulation of Mechanical Properties and Membrane Activities of the ESCRT-III Complexes. Biophys. J. 2020, 118, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Pabis, A.; Rawle, R.J.; Kasson, P.M. Influenza Hemagglutinin Drives Viral Entry via Two Sequential Intramembrane Mechanisms. Proc. Natl. Acad. Sci. USA 2020, 117, 7200–7207. [Google Scholar] [CrossRef] [PubMed]
- Kasson, P.M.; Pande, V.S. Control of Membrane Fusion Mechanism by Lipid Composition: Predictions from Ensemble Molecular Dynamics. PLoS Comput. Biol. 2007, 3, e220. [Google Scholar] [CrossRef] [PubMed]
- Chng, C.P.; Sadovsky, Y.; Hsia, K.J.; Huang, C. Curvature-Regulated Lipid Membrane Softening of Nano-Vesicles. Extrem. Mech. Lett. 2021, 43, 101174. [Google Scholar] [CrossRef]
- Parchekani, J.; Allahverdi, A.; Taghdir, M.; Naderi-Manesh, H. Design and Simulation of the Liposomal Model by Using a Coarse-Grained Molecular Dynamics Approach towards Drug Delivery Goals. Sci. Rep. 2022, 12, 2371. [Google Scholar] [CrossRef]
- Qi, Y.; Ingólfsson, H.I.; Cheng, X.; Lee, J.; Marrink, S.J.; Im, W. CHARMM-GUI Martini Maker for Coarse-Grained Simulations with the Martini Force Field. J. Chem. Theory Comput. 2015, 11, 4486–4494. [Google Scholar] [CrossRef]
- Boyd, K.J.; May, E.R. BUMPy: A Model-Independent Tool for Constructing Lipid Bilayers of Varying Curvature and Composition. J. Chem. Theory Comput. 2018, 14, 6642–6652. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2007, 29, 1859–1865. [Google Scholar] [CrossRef]
- Qi, Y.; Cheng, X.; Han, W.; Jo, S.; Schulten, K.; Im, W. CHARMM-GUI PACE CG Builder for Solution, Micelle, and Bilayer Coarse-Grained Simulations. J. Chem. Inf. Model. 2014, 54, 1003–1009. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef]
- Han, W.; Schulten, K. Further Optimization of a Hybrid United-Atom and Coarse-Grained Force Field for Folding Simulations: Improved Backbone Hydration and Interactions between Charged Side Chains. J. Chem. Theory Comput. 2012, 8, 4413–4424. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Durrant, J.D.; Amaro, R.E. LipidWrapper: An Algorithm for Generating Large-Scale Membrane Models of Arbitrary Geometry. PLoS Comput. Biol. 2014, 10, e1003720. [Google Scholar] [CrossRef] [PubMed]
- Pezeshkian, W.; Konig, M. TS2CG Tutorial. Available online: http://cgmartini.nl/index.php/2021-martini-online-workshop/tutorials/558-9-ts2cg (accessed on 5 May 2022).
- Noguchi, H. Membrane Simulation Models from Nanometer to Micrometer Scale. J. Phys. Soc. Jpn. 2009, 78, 041007. [Google Scholar] [CrossRef]
- Noguchi, H. Formation of Polyhedral Vesicles and Polygonal Membrane Tubes Induced by Banana-Shaped Proteins. J. Chem. Phys. 2015, 124, 243109. [Google Scholar] [CrossRef]
- Yesylevskyy, S.O.; Rivel, T.; Ramseyer, C. The Influence of Curvature on the Properties of the Plasma Membrane. Insights from Atomistic Molecular Dynamics Simulations. Sci. Rep. 2017, 7, 16078. [Google Scholar] [CrossRef]
- Yesylevskyy, S.; Rivel, T.; Ramseyer, C. Curvature Increases Permeability of the Plasma Membrane for Ions, Water and the Anti-Cancer Drugs Cisplatin and Gemcitabine. Sci. Rep. 2019, 9, 17214. [Google Scholar] [CrossRef]
- Yesylevskyy, S.; Khandelia, H. EnCurv: Simple Technique of Maintaining Global Membrane Curvature in Molecular Dynamics Simulations. J. Chem. Theory Comput. 2021, 17, 1181–1193. [Google Scholar] [CrossRef]
- De Jong, D.H.; Heuer, A. The Influence of Solid Scaffolds on Flat and Curved Lipid Membranes. AIP Adv. 2017, 7, 075007. [Google Scholar] [CrossRef]
- Belessiotis-Richards, A.; Higgins, S.G.; Butterworth, B.; Stevens, M.M.; Alexander-katz, A. Single-Nanometer Changes in Nanopore Geometry Influence Curvature, Local Properties, and Protein Localization in Membrane Simulations. Nano Lett. 2019, 19, 4770–4778. [Google Scholar] [CrossRef]
- Belessiotis-Richards, A.; Higgins, S.G.; Sansom, M.S.P.; Alexander-Katz, A.; Stevens, M.M. Coarse-Grained Simulations Suggest the Epsin N-Terminal Homology Domain Can Sense Membrane Curvature without Its Terminal Amphipathic Helix. ACS Nano 2020, 14, 16919–16928. [Google Scholar] [CrossRef]
- Belessiotis-Richards, A.; Larsen, A.H.; Higgins, S.G.; Stevens, M.M.; Alexander-Katz, A. Coarse-Grained Simulations Suggest Potential Competing Roles of Phosphoinositides and Amphipathic Helix Structures in Membrane Curvature Sensing of the AP180 N-Terminal Homology Domain. J. Phys. Chem. B 2022, 126, 2789–2797. [Google Scholar] [CrossRef] [PubMed]
- John, L.H.; Preston, G.M.; Sansom, M.S.P.; Clifton, L.A. Large Scale Model Lipid Membrane Movement Induced by a Cation Switch. J. Colloid Interface Sci. 2021, 596, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Meyer, G.R.; Gullingsrud, J.; Schulten, K.; Martinac, B. Molecular Dynamics Study of MscL Interactions with a Curved Lipid Bilayer. Biophys. J. 2006, 91, 1630–1637. [Google Scholar] [CrossRef] [PubMed]
- Martyna, A.; Gómez-Llobregat, J.; Lindén, M.; Rossman, J.S. Curvature Sensing by a Viral Scission Protein. Biochemistry 2016, 55, 3493–3496. [Google Scholar] [CrossRef] [PubMed]
- Stroh, K.S.; Risselada, H.J. Quantifying Membrane Curvature Sensing of Peripheral Proteins by Simulated Buckling and Umbrella Sampling. J. Chem. Theory Comput. 2021, 17, 5276–5286. [Google Scholar] [CrossRef]
- Kamal, M.M.; Mills, D.; Grzybek, M.; Howard, J. Measurement of the Membrane Curvature Preference of Phospholipids Reveals Only Weak Coupling between Lipid Shape and Leaflet Curvature. Proc. Natl. Acad. Sci. USA 2009, 106, 22245–22250. [Google Scholar] [CrossRef]
- Yesylevskyy, S.O.; Ramseyer, C. Determination of Mean and Gaussian Curvatures of Highly Curved Asymmetric Lipid Bilayers: The Case Study of the Influence of Cholesterol on the Membrane Shape. Phys. Chem. Chem. Phys. 2014, 16, 17052–17061. [Google Scholar] [CrossRef]
- Shi, H.; Quint, D.A.; Grason, G.M.; Gopinathan, A.; Huang, K.C. Chiral Twisting in a Bacterial Cytoskeletal Polymer Affects Filament Size and Orientation. Nat. Commun. 2020, 11, 1408. [Google Scholar] [CrossRef]
- Pinot, M.; Vanni, S.; Pagnotta, S.; Lacas-Gervais, S.; Payet, L.A.; Ferreira, T.; Gautier, R.; Goud, B.; Antonny, B.; Barelli, H. Polyunsaturated Phospholipids Facilitate Membrane Deformation and Fission by Endocytic Proteins. Science 2014, 345, 693–697. [Google Scholar] [CrossRef]
- Baoukina, S.; Marrink, S.J.; Tieleman, D.P. Molecular Structure of Membrane Tethers. Biophys. J. 2012, 102, 1866–1871. [Google Scholar] [CrossRef]
- Baoukina, S.; Ingólfsson, H.I.; Marrink, S.J.; Tieleman, D.P. Curvature-Induced Sorting of Lipids in Plasma Membrane Tethers. Adv. Theory Simul. 2018, 1, 1800034. [Google Scholar] [CrossRef]
- Masone, D.; Uhart, M.; Bustos, D.M. Bending Lipid Bilayers: A Closed-Form Collective Variable for Effective Free-Energy Landscapes in Quantitative Biology. J. Chem. Theory Comput. 2018, 14, 2240–2245. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, B. Curvature as a Collective Coordinate in Enhanced Sampling Membrane Simulations. J. Chem. Theory Comput. 2019, 15, 6551–6561. [Google Scholar] [CrossRef]
- Fiorin, G.; Marinelli, F.; Faraldo-Gómez, J.D. Direct Derivation of Free Energies of Membrane Deformation and Other Solvent Density Variations from Enhanced Sampling Molecular Dynamics. J. Comput. Chem. 2020, 41, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Tribello, G.A.; Bonomi, M.; Branduardi, D.; Camilloni, C.; Bussi, G. PLUMED 2: New Feathers for an Old Bird. Comput. Phys. Commun. 2014, 185, 604–613. [Google Scholar] [CrossRef]
- The PLUMED Consortium. Promoting Transparency and Reproducibility in Enhanced Molecular Simulations. Nat. Methods 2019, 16, 670–673. [Google Scholar] [CrossRef]
- Boyd, K.J.; Alder, N.N.; May, E.R. Buckling under Pressure: Curvature-Based Lipid Segregation and Stability Modulation in Cardiolipin-Containing Bilayers. Langmuir 2017, 33, 6937–6946. [Google Scholar] [CrossRef]
- Cino, E.A.; Tieleman, D.P. Curvature-Based Sorting of Eight Lipid Types in Asymmetric Buckled Plasma Membrane Models. Biophys. J. 2022, 121, 2060–2068. [Google Scholar] [CrossRef]
- Mandal, T.; Spagnolie, S.E.; Audhya, A.; Cui, Q. Protein-Induced Membrane Curvature in Coarse-Grained Simulations. Biophys. J. 2021, 120, 3211–3221. [Google Scholar] [CrossRef]
- Gómez-Llobregat, J.; Elías-Wolff, F.; Lindén, M. Anisotropic Membrane Curvature Sensing by Amphipathic Peptides. Biophys. J. 2016, 110, 197–204. [Google Scholar] [CrossRef]
- Elías-Wolff, F.; Lindén, M.; Lyubartsev, A.P.; Brandt, E.G. Curvature Sensing by Cardiolipin in Simulated Buckled Membranes. Soft Matter 2019, 15, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Elías-Wolff, F.; Lindén, M.; Lyubartsev, A.P.; Brandt, E.G. Computing Curvature Sensitivity of Biomolecules in Membranes by Simulated Buckling. J. Chem. Theory Comput. 2018, 14, 1643–1655. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.H.; John, L.H.; Sansom, M.S.P.; Corey, R.A. Specific Interactions of Peripheral Membrane Proteins with Lipids: What Can Molecular Simulations Show Us? Biosci. Rep. 2022, 42, BSR20211406. [Google Scholar] [CrossRef] [PubMed]
- Peter, B.J.; Kent, H.M.; Mills, I.G.; Vallis, Y.; Butler, P.J.G.; Evans, P.R.; McMahon, H.T. BAR Domains as Sensors of Membrane Curvature: The Amphiphysin BAR Structure. Science 2004, 303, 495–499. [Google Scholar] [CrossRef]
- Callan-Jones, A.; Bassereau, P. Curvature-Driven Membrane Lipid and Protein Distribution. Curr. Opin. Solid State Mater. Sci. 2013, 17, 143–150. [Google Scholar] [CrossRef]
- Simunovic, M.; Prévost, C.; Andrew, C.J.; Bassereau, P. Physical Basis of Some Membrane Shaping Mechanisms. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20160034. [Google Scholar] [CrossRef]
- Bhatia, V.K.; Madsen, K.L.; Bolinger, P.Y.; Kunding, A.; Hedegard, P.; Gether, U.; Stamou, D. Amphipathic Motifs in BAR Domains Are Essential for Membrane Curvature Sensing. EMBO J. 2009, 28, 3303–3314. [Google Scholar] [CrossRef]
- Aimon, S.; Callan-Jones, A.; Berthaud, A.; Pinot, M.; Toombes, G.E.S.; Bassereau, P. Membrane Shape Modulates Transmembrane Protein Distribution. Dev. Cell 2014, 28, 212–218. [Google Scholar] [CrossRef]
- Bhatia, V.K.; Hatzakis, N.S.; Stamou, D. A Unifying Mechanism Accounts for Sensing of Membrane Curvature by BAR Domains, Amphipathic Helices and Membrane-Anchored Proteins. Semin. Cell Dev. Biol. 2010, 21, 381–390. [Google Scholar] [CrossRef]
- Wichmann, N.; Lund, P.M.; Hansen, M.B.; Hjørringgaard, C.U.; Larsen, J.B.; Kristensen, K.; Andresen, T.L.; Simonsen, J.B. Applying Flow Cytometry to Identify the Modes of Action of Membrane-Active Peptides in a Label-Free and High-Throughput Fashion. Biochim. Biophys. Acta Biomembr. 2022, 1864, 183820. [Google Scholar] [CrossRef]
- Hoover, B.M.; Shen, Z.; Gahan, C.G.; Lynn, D.M.; Van Lehn, R.C.; Murphy, R.M. Membrane Remodeling and Stimulation of Aggregation Following α-Synuclein Adsorption to Phosphotidylserine Vesicles. J. Phys. Chem. B 2021, 125, 1582–1594. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.C.; Capraro, B.R.; Tian, A.; Isas, J.M.; Langen, R.; Baumgart, T. Quantifying Membrane Curvature Generation of Drosophila Amphiphysin N-BAR Domains. J. Phys. Chem. Lett. 2010, 1, 3401–3406. [Google Scholar] [CrossRef] [PubMed]
- Galic, M.; Jeong, S.; Tsai, F.C.; Joubert, L.M.; Wu, Y.I.; Hahn, K.M.; Cui, Y.; Meyer, T. External Push and Internal Pull Forces Recruit Curvature-Sensing N-BAR Domain Proteins to the Plasma Membrane. Nat. Cell Biol. 2012, 14, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Agudo-Canalejo, J.; Discher, D.E. Biomembrane Adhesion to Substrates Topographically Patterned with Nanopits. Biophys. J. 2018, 115, 1292–1306. [Google Scholar] [CrossRef]
- Zhao, W.; Hanson, L.; Lou, H.Y.; Akamatsu, M.; Chowdary, P.D.; Santoro, F.; Marks, J.R.; Grassart, A.; Drubin, D.G.; Cui, Y.; et al. Nanoscale Manipulation of Membrane Curvature for Probing Endocytosis in Live Cells. Nat. Nanotechnol. 2017, 12, 750–756. [Google Scholar] [CrossRef]
- Rzepiela, A.J.; Louhivuori, M.; Peter, C.; Marrink, S.J. Hybrid Simulations: Combining Atomistic and Coarse-Grained Force Fields Using Virtual Sites. Phys. Chem. Chem. Phys. 2011, 13, 10437–10448. [Google Scholar] [CrossRef]
- Senn, H.M.; Thiel, W. QM/MM Methods for Biomolecular Systems. Angew. Chem. Int. Ed. 2009, 48, 1198–1229. [Google Scholar] [CrossRef]
- Orsi, M.; Noro, M.G.; Essex, J.W. Dual-Resolution Molecular Dynamics Simulation of Antimicrobials in Biomembranes. J. R. Soc. Interface 2011, 8, 826–841. [Google Scholar] [CrossRef]
- Liu, Y.; De Vries, A.H.; Barnoud, J.; Pezeshkian, W.; Melcr, J.; Marrink, S.J. Dual Resolution Membrane Simulations Using Virtual Sites. J. Phys. Chem. B 2020, 124, 3944–3953. [Google Scholar] [CrossRef]
- Larsen, A.H.; Sansom, M.S.P. Binding of Ca2+-Independent C2 Domains to Lipid Membranes: A Multi-Scale Molecular Dynamics Study. Structure 2021, 29, 1200–1213. [Google Scholar] [CrossRef]
- Vickery, O.N.; Stansfeld, P.J. CG2AT2: An Enhanced Fragment-Based Approach for Serial Multi-Scale Molecular Dynamics Simulations. J. Chem. Theory Comput. 2021, 17, 6472–6482. [Google Scholar] [CrossRef] [PubMed]
- Wassenaar, T.A.; Pluhackova, K.; Böckmann, R.A.; Marrink, S.J.; Tieleman, D.P. Going Backward: A Flexible Geometric Approach to Reverse Transformation from Coarse Grained to Atomistic Models. J. Chem. Theory Comput. 2014, 10, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.; Lorenz, C.D. LiPyphilic: A Python Toolkit for the Analysis of Lipid Membrane Simulations. J. Chem. Theory Comput. 2021, 17, 5907–5919. [Google Scholar] [CrossRef]
- Bhatia, H.; Ingólfsson, H.I.; Carpenter, T.S.; Lightstone, F.C.; Bremer, P.T. MemSurfer: A Tool for Robust Computation and Characterization of Curved Membranes. J. Chem. Theory Comput. 2019, 15, 6411–6421. [Google Scholar] [CrossRef]
- Fábián, B.; Javanainen, M. CurD: A Tool for Diffusion Analyses on Curved Membranes. ChemRxiv 2021. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larsen, A.H. Molecular Dynamics Simulations of Curved Lipid Membranes. Int. J. Mol. Sci. 2022, 23, 8098. https://doi.org/10.3390/ijms23158098
Larsen AH. Molecular Dynamics Simulations of Curved Lipid Membranes. International Journal of Molecular Sciences. 2022; 23(15):8098. https://doi.org/10.3390/ijms23158098
Chicago/Turabian StyleLarsen, Andreas Haahr. 2022. "Molecular Dynamics Simulations of Curved Lipid Membranes" International Journal of Molecular Sciences 23, no. 15: 8098. https://doi.org/10.3390/ijms23158098
APA StyleLarsen, A. H. (2022). Molecular Dynamics Simulations of Curved Lipid Membranes. International Journal of Molecular Sciences, 23(15), 8098. https://doi.org/10.3390/ijms23158098





