Targeting the Achilles’ Heel of Multidrug-Resistant Staphylococcus aureus by the Endocannabinoid Anandamide
Abstract
1. Introduction
2. Results
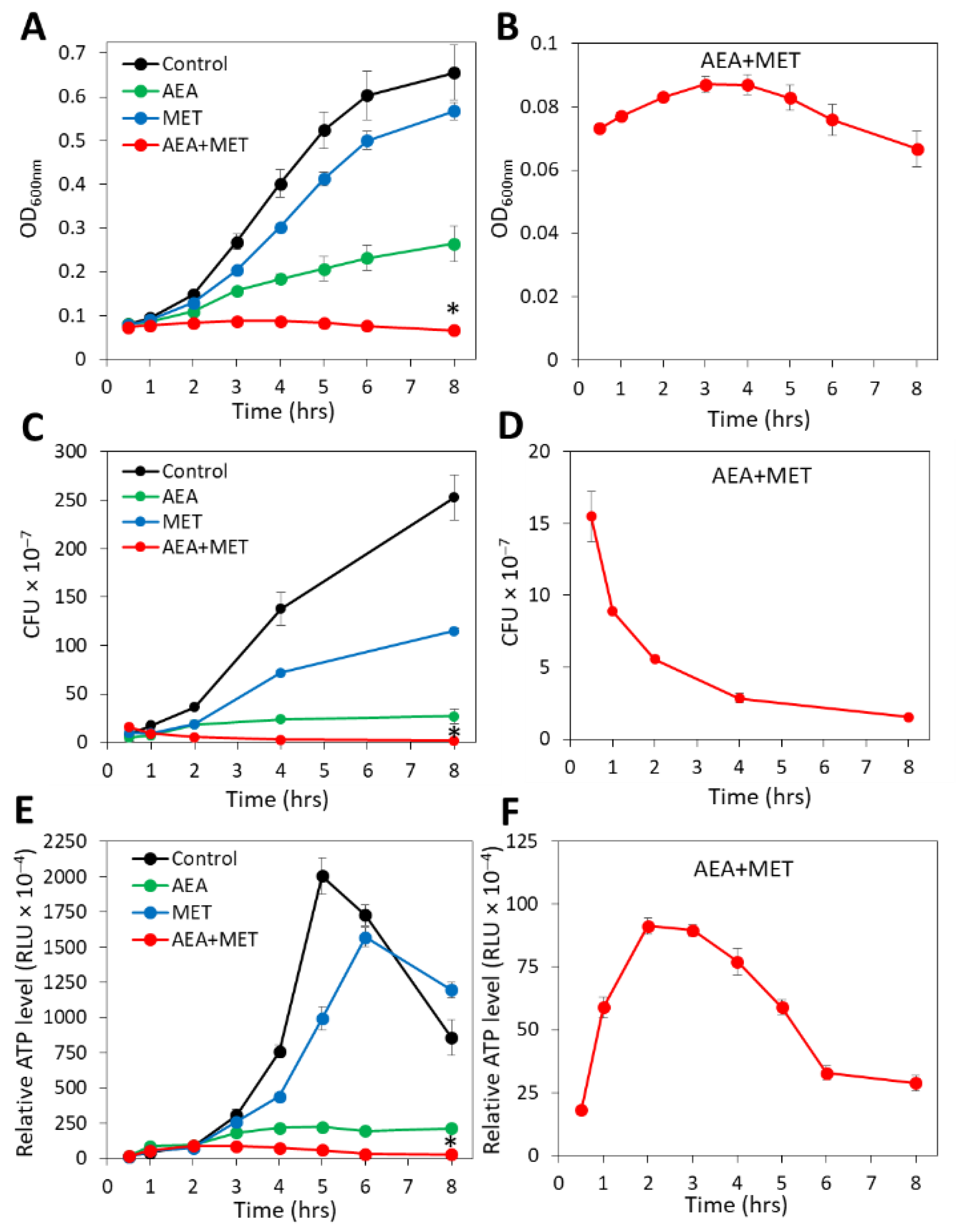
2.1. The Combined Treatment of Anandamide and Methicillin Reduced the Viability of MDRSA
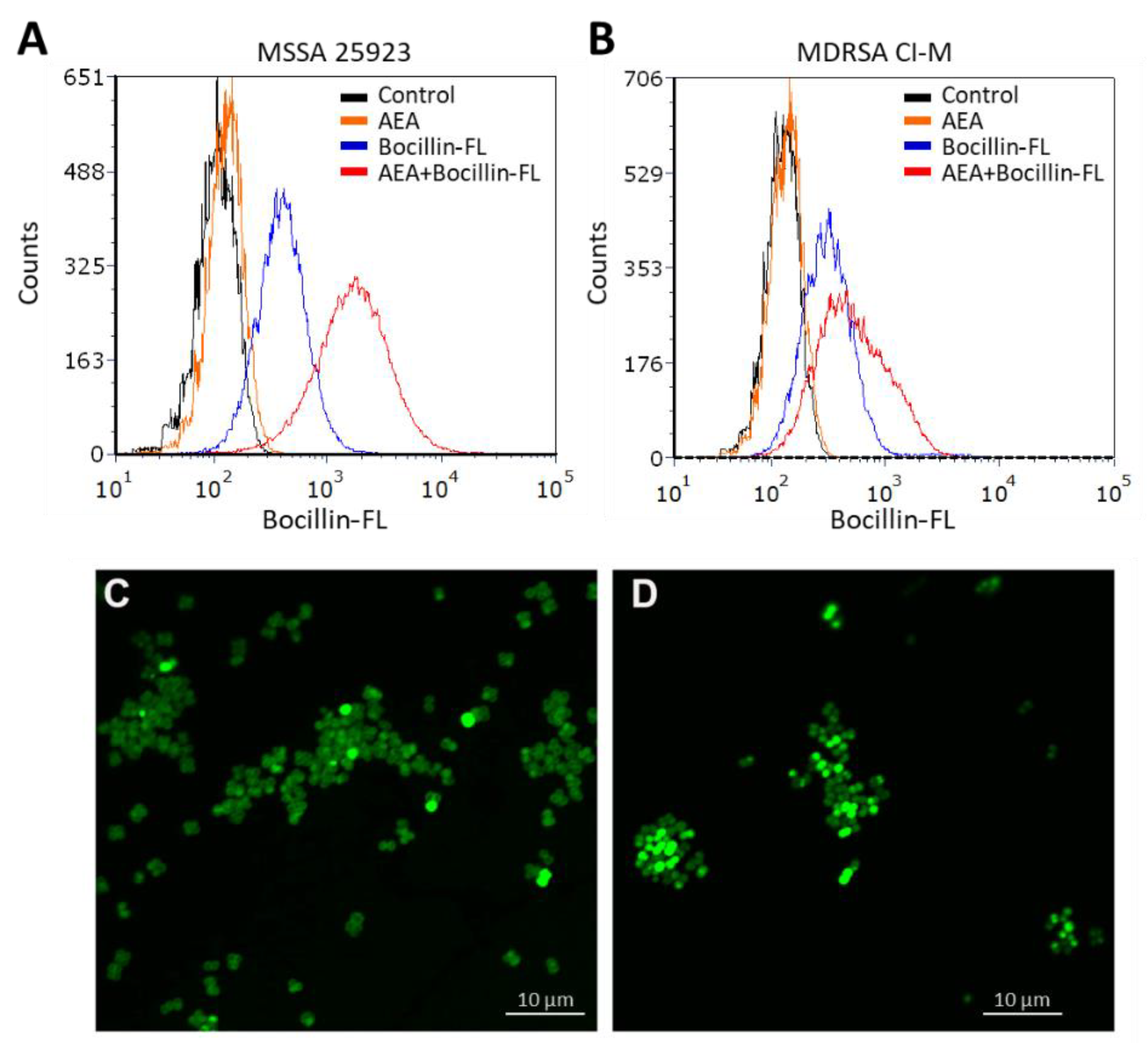
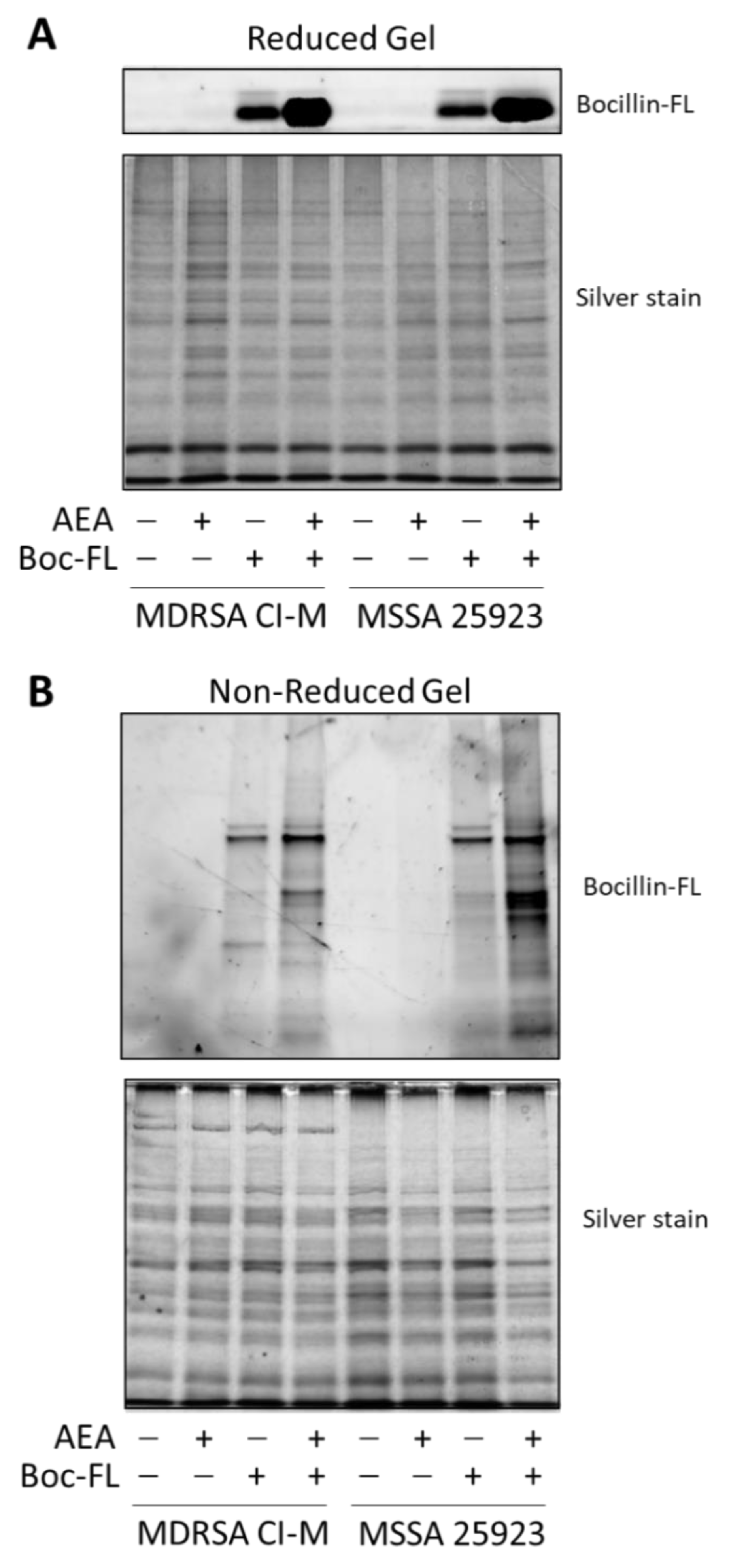
2.2. Anandamide Increases the Intracellular Level of Bocillin-FL
2.3. Anandamide Causes an Intracellular Accumulation of Its Own
2.4. Anandamide Prevents the Secretion of Exopolysaccharides (EPS)
2.5. Anandamide Increases the Extracellular DNA (eDNA) Level
2.6. AEA Does Not Alter the Surface Binding of WGA, ConA, or Dextran to MDRSA CI-M
2.7. AEA Reduces the Cell Wall Teichoic Acid (WTA) Content
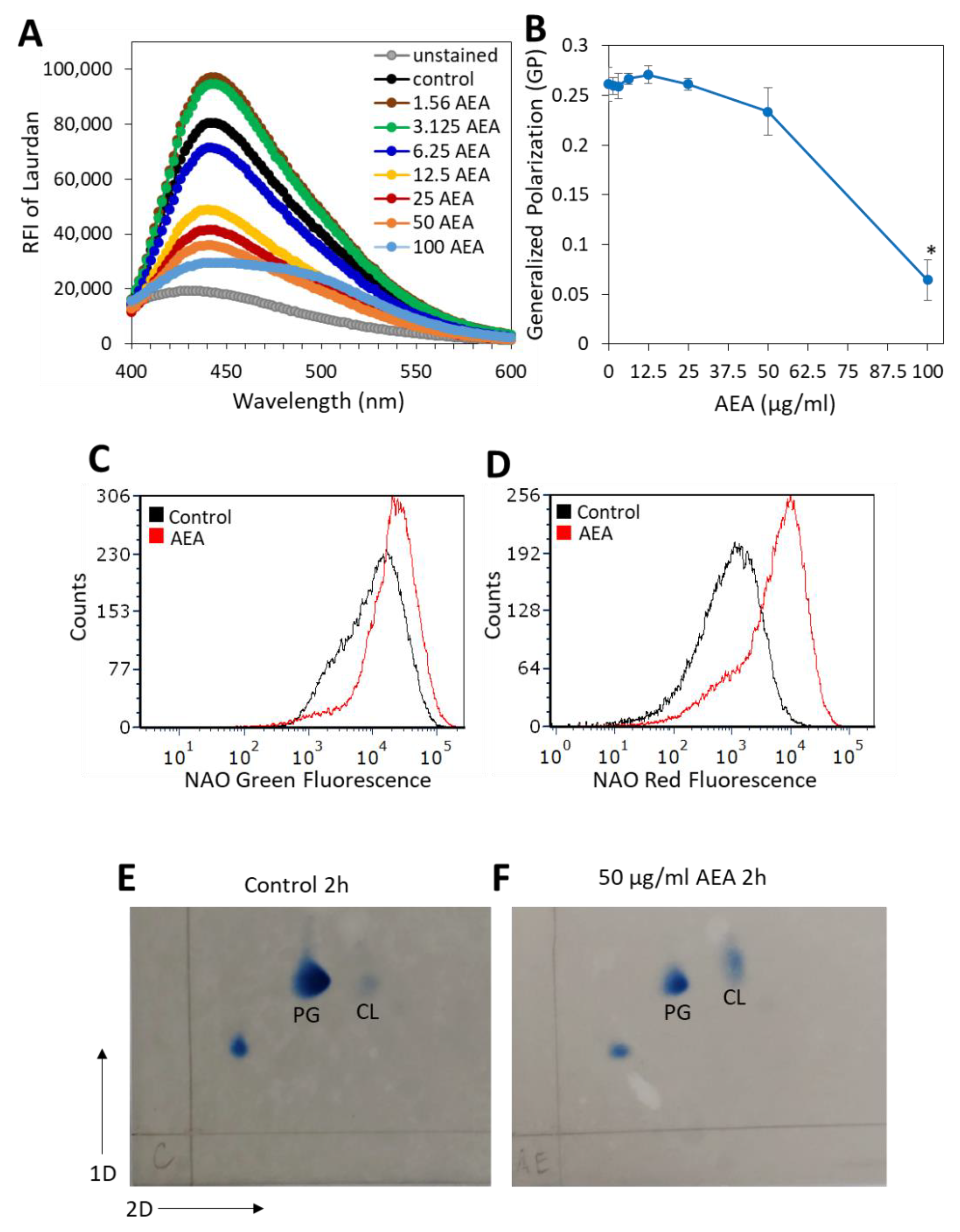
2.8. AEA Alters the Membrane Fluidity and Composition of MDRSA CI-M
2.9. Anandamide Inhibits the Membrane ATPase Activity, but Not the Membrane GTPase Activity
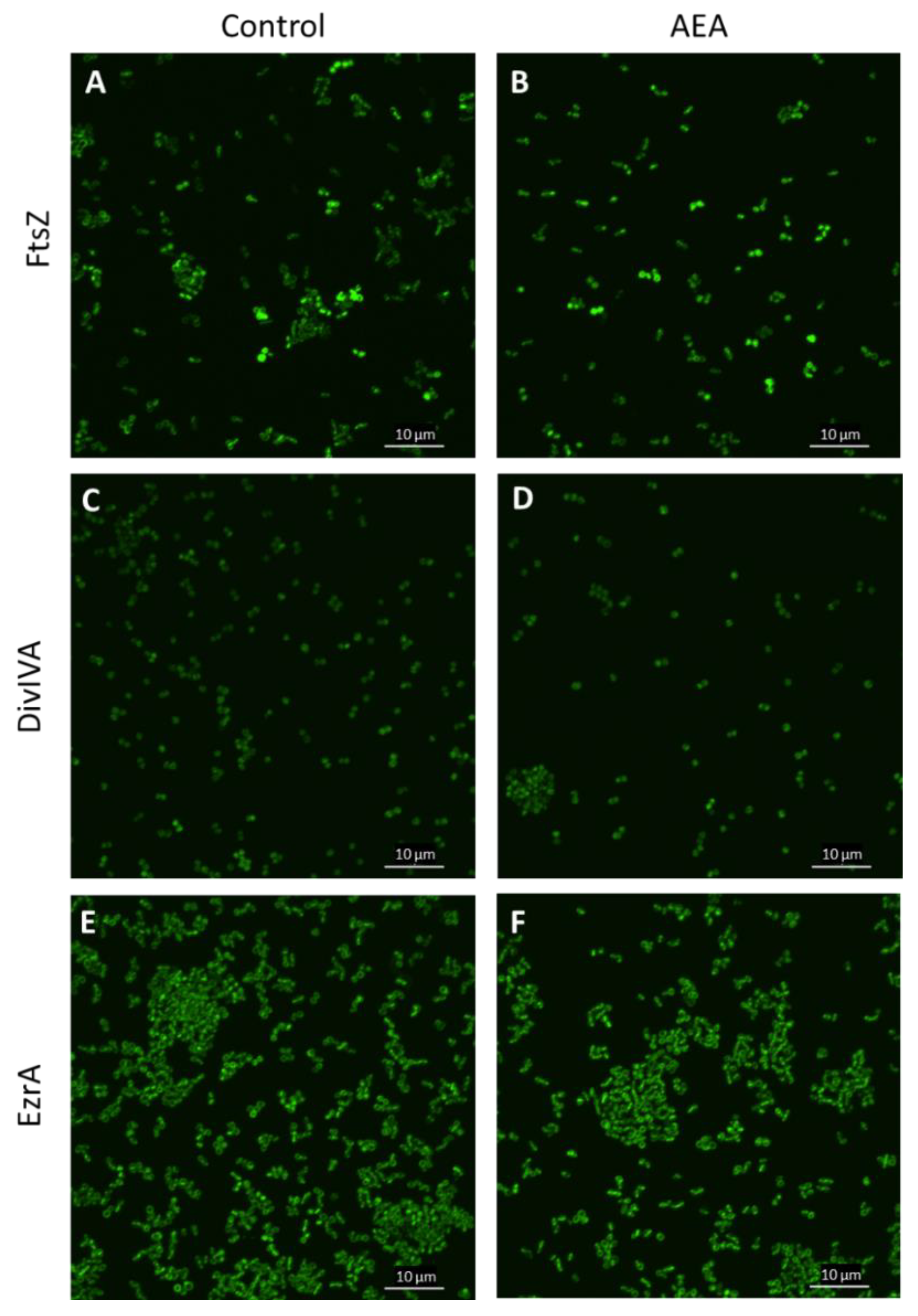
2.10. The Divisome Z-Ring Is Formed in the Presence of AEA
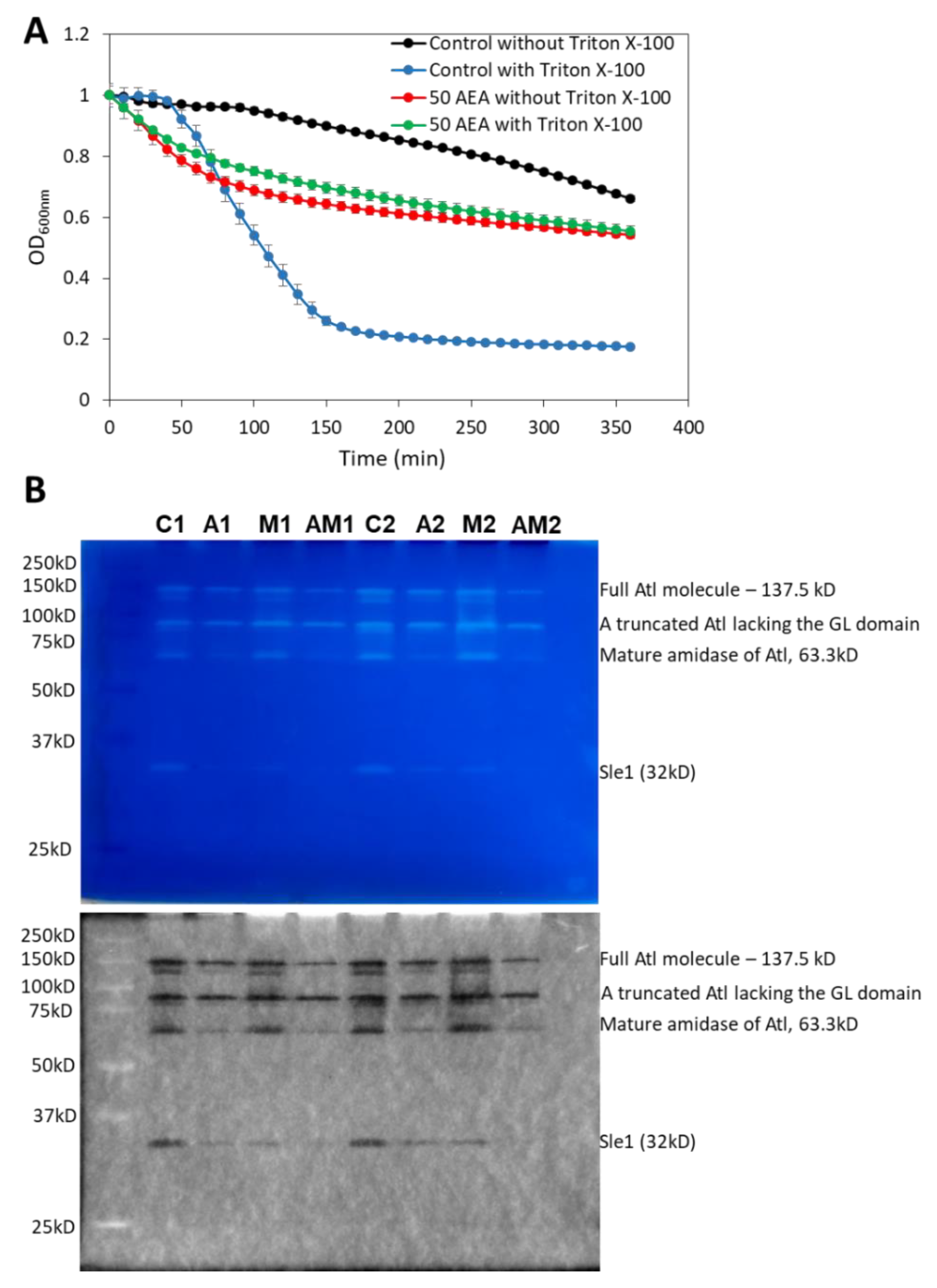
2.11. AEA Induces Spontaneous Cell Lysis despite the Reduced Activity of Autolysins
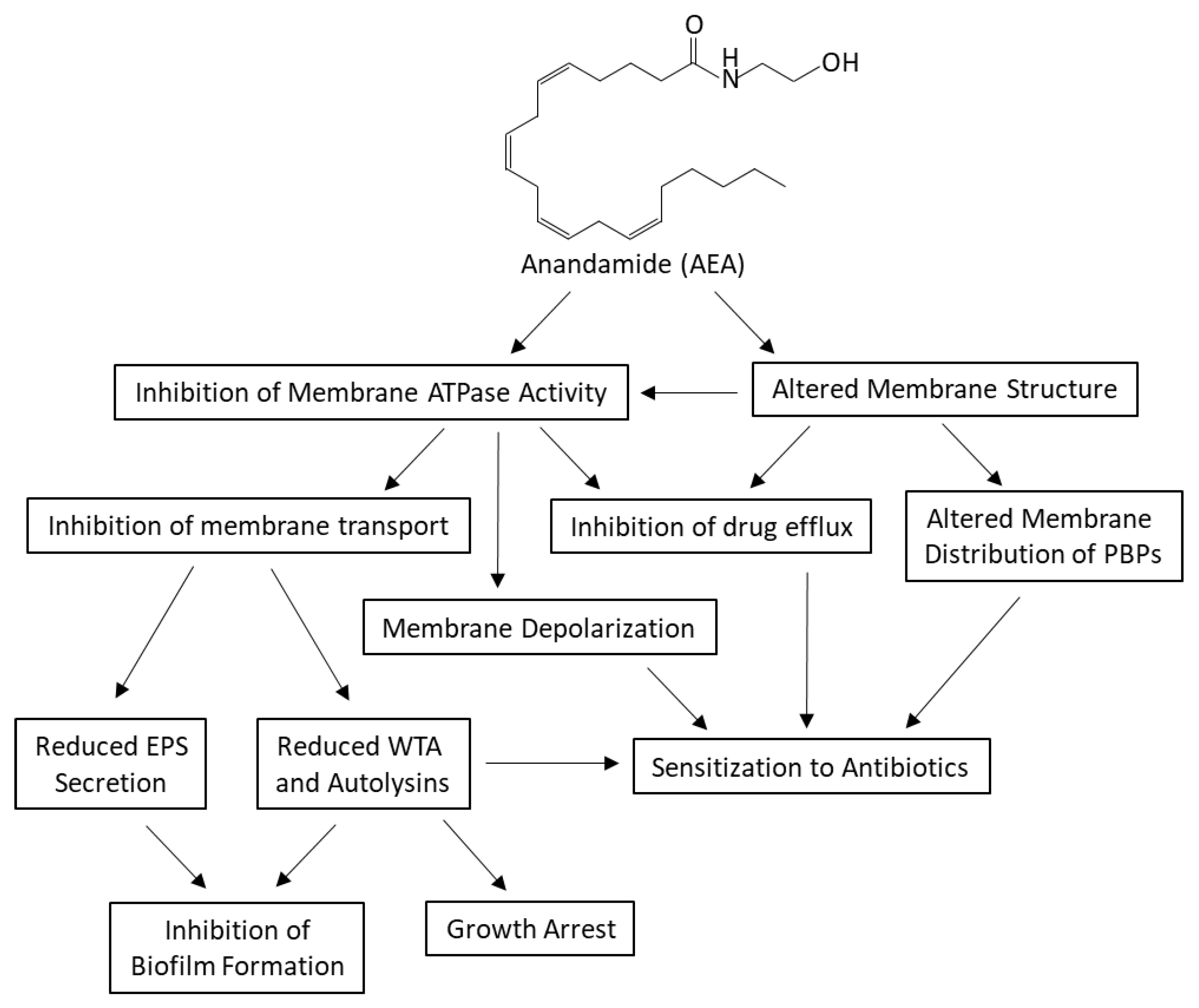
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Bacterial Strains and Culture Conditions
4.3. Determination of Bacterial Growth
4.4. Bocillin-FL Accumulation Assay Using Flow Cytometry and Spinning Disk Confocal Microscopy
4.5. Analysis of Bocillin-FL-Bound Proteins on SDS-PAGE
4.6. Fluorescent AEA Accumulation Assays
4.7. Detection of Exopolysaccharides (EPS) Production by Congo Red
4.8. Detection of Extracellular DNA (eDNA) by Methyl Green Agar Plates
4.9. Bacterial DNA Staining with TOTO-1, SYTO 9, and PI
4.10. Surface Binding of WGA, ConA, and Dextran 10,000
4.11. Determination of the Cell Wall Teichoic Acid (WTA) Content
4.12. Laurdan Membrane Fluidity Assay
4.13. 10-N-Nonyl-Acridine Orange (NAO) Staining of Cardiolipin
4.14. Two-Dimensional Thin-Layer Chromatography of Membrane Lipids
4.15. Membrane ATPase and GTPase Assay
4.16. Autolysis Assay
4.17. Autolysin Zymogram
4.18. Gelatin and Casein Zymogram
4.19. Protease Assay on Gelatin Agar Plates
4.20. RNA and cDNA preparation
4.21. Quantitative Real-Time PCR
4.22. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A. Staphylococcus aureus biofilm: Morphology, genetics, pathogenesis and treatment strategies. Int. J. Environ. Res. Public Health 2021, 18, 7602. [Google Scholar] [CrossRef] [PubMed]
- Valaperta, R.; Tejada, M.R.; Frigerio, M.; Moroni, A.; Ciulla, E.; Cioffi, S.; Capelli, P.; Costa, E. Staphylococcus aureus nosocomial infections: The role of a rapid and low-cost characterization for the establishment of a surveillance system. New Microbiol. 2010, 33, 223–232. [Google Scholar] [PubMed]
- Lee, A.S.; De Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef] [PubMed]
- Kwiecinski, J.M.; Horswill, A.R. Staphylococcus aureus bloodstream infections: Pathogenesis and regulatory mechanisms. Curr. Opin. Microbiol. 2020, 53, 51–60. [Google Scholar] [CrossRef]
- Kwiecinski, J.; Kahlmeter, G.; Jin, T. Biofilm formation by Staphylococcus aureus isolates from skin and soft tissue infections. Curr. Microbiol. 2015, 70, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Schilcher, K.; Horswill, A.R. Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef]
- Ricciardi, B.F.; Muthukrishnan, G.; Masters, E.; Ninomiya, M.; Lee, C.C.; Schwarz, E.M. Staphylococcus aureus evasion of host immunity in the setting of prosthetic joint infection: Biofilm and beyond. Curr. Rev. Musculoskelet. Med. 2018, 11, 389–400. [Google Scholar] [CrossRef]
- de Oliveira, A.; Pinheiro-Hubinger, L.; Pereira, V.C.; Riboli, D.F.M.; Martins, K.B.; Romero, L.C.; Cunha, M. Staphylococcal biofilm on the surface of catheters: Electron microscopy evaluation of the inhibition of biofilm growth by RNAIII inhibiting peptide. Antibiotics 2021, 10, 879. [Google Scholar] [CrossRef]
- Zheng, Y.; He, L.; Asiamah, T.K.; Otto, M. Colonization of medical devices by staphylococci. Environ. Microbiol. 2018, 20, 3141–3153. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal biofilms. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef]
- Guo, Y.; Song, G.; Sun, M.; Wang, J.; Wang, Y. Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Kranjec, C.; Morales Angeles, D.; Torrissen Mårli, M.; Fernández, L.; García, P.; Kjos, M.; Diep, D.B. Staphylococcal biofilms: Challenges and novel therapeutic perspectives. Antibiotics 2021, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Peacock, S.J.; Paterson, G.K. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.S.; Viveiros, M.; Amaral, L.; Couto, I. Multidrug efflux pumps in Staphylococcus aureus: An update. Open Microbiol. J. 2013, 7, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Hassanzadeh, S.; Ganjloo, S.; Pourmand, M.R.; Mashhadi, R.; Ghazvini, K. Epidemiology of efflux pumps genes mediating resistance among Staphylococcus aureus; A systematic review. Microb. Pathog. 2019, 139, 103850. [Google Scholar] [CrossRef]
- Fuda, C.C.; Fisher, J.F.; Mobashery, S. β-Lactam resistance in Staphylococcus aureus: The adaptive resistance of a plastic genome. Cell. Mol. Life Sci. 2005, 62, 2617–2633. [Google Scholar] [CrossRef]
- Pantosti, A.; Sanchini, A.; Monaco, M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Futur. Microbiol. 2007, 2, 323–334. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Campoccia, D.; Mirzaei, R.; Montanaro, L.; Arciola, C.R. Hijacking of immune defences by biofilms: A multifront strategy. Biofouling 2019, 35, 1055–1074. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 2010, 65, 1955–1958. [Google Scholar] [CrossRef]
- Albayaty, Y.N.; Thomas, N.; Hasan, S.; Prestidgeac, C.A. Penetration of topically used antimicrobials through Staphylococcus aureus biofilms: A comparative study using different models. J. Drug Deliv. Sci. Technol. 2018, 48, 429–436. [Google Scholar] [CrossRef]
- Martínez, J.L.; Rojo, F. Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 768–789. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.R.; Rozen, D.E. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 2006, 4, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef]
- He, X.; Ahn, J. Differential gene expression in planktonic and biofilm cells of multiple antibiotic-resistant Salmonella Typhimurium and Staphylococcus aureus. FEMS Microbiol. Lett. 2011, 325, 180–188. [Google Scholar] [CrossRef]
- Karaman, R.; Jubeh, B.; Breijyeh, Z. Resistance of Gram-positive bacteria to current antibacterial agents and overcoming approaches. Molecules 2020, 25, 2888. [Google Scholar] [CrossRef]
- Singh, A.; Amod, A.; Pandey, P.; Bose, P.; Pingali, M.S.; Shivalkar, S.; Varadwaj, P.K.; Sahoo, A.K.; Samanta, S.K. Bacterial biofilm infections, their resistance to antibiotics therapy and current treatment strategies. Biomed. Mater. 2022, 17, 022003. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Tehrani, K.; Martin, N.I. β-lactam/β-lactamase inhibitor combinations: An update. MedChemComm 2018, 9, 1439–1456. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, T.M.; De Oliveira, C.R.; Chambers, H.F.; Chatterjee, S.S. PBP4: A New Perspective on Staphylococcus aureus β-Lactam Resistance. Microorganisms 2018, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, E.; Koch, G.; Wagner, R.M.; Fekete, A.; Stengel, S.T.; Schneider, J.; Mielich-Süß, B.; Geibel, S.; Markert, S.; Stigloher, C.; et al. Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 2017, 171, 1354–1367.e20. [Google Scholar] [CrossRef] [PubMed]
- Acebrón, I.; Chang, M.; Mobashery, S.; Hermoso, J.A. The Allosteric site for the nascent cell wall in Penicillin-Binding Protein 2a: An Achilles’ heel of methicillin-resistant Staphylococcus aureus. Curr. Med. Chem. 2015, 22, 1678–1686. [Google Scholar] [CrossRef]
- Ferrer-González, E.; Huh, H.; Al-Tameemi, H.M.; Boyd, J.M.; Lee, S.-H.; Pilch, D.S. Impact of FtsZ inhibition on the localization of the Penicillin Binding Proteins in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 2021, 203, e0020421. [Google Scholar] [CrossRef]
- Lee, S.H.; Wang, H.; Labroli, M.; Koseoglu, S.; Zuck, P.; Mayhood, T.; Gill, C.; Mann, P.; Sher, X.; Ha, S.; et al. TarO-specific inhibitors of wall teichoic acid biosynthesis restore β-lactam efficacy against methicillin-resistant staphylococci. Sci. Transl. Med. 2016, 8, 329ra32. [Google Scholar] [CrossRef]
- Wang, H.; Gill, C.J.; Lee, S.H.; Mann, P.; Zuck, P.; Meredith, T.C.; Murgolo, N.; She, X.; Kales, S.; Liang, L.; et al. Discovery of wall teichoic acid inhibitors as potential anti-MRSA β-lactam combination agents. Chem. Biol. 2013, 20, 272–284. [Google Scholar] [CrossRef]
- Campbell, J.; Singh, A.K.; Santa Maria, J.J.P., Jr.; Kim, Y.; Brown, S.; Swoboda, J.G.; Mylonakis, E.; Wilkinson, B.J.; Walker, S. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 2010, 6, 106–116. [Google Scholar] [CrossRef]
- Farha, M.A.; Leung, A.; Sewell, E.W.; D’Elia, M.A.; Allison, S.E.; Ejim, L.; Pereira, P.M.; Pinho, M.G.; Wright, G.D.; Brown, E.D. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem. Biol. 2012, 8, 226–233. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall teichoic acids of Gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [PubMed]
- Winstel, V.; Kühner, P.; Salomon, F.; Larsen, J.; Skov, R.; Hoffmann, W.; Peschel, A.; Weidenmaier, C. Wall teichoic acid glycosylation governs Staphylococcus aureus nasal colonization. mBio 2015, 6, e00632. [Google Scholar] [CrossRef] [PubMed]
- van Dalen, R.; Peschel, A.; van Sorge, N.M. Wall teichoic acid in Staphylococcus aureus host interaction. Trends Microbiol. 2020, 28, 985–998. [Google Scholar] [CrossRef]
- Yeo, W.-S.; Jeong, B.; Ullah, N.; Shah, M.A.; Ali, A.; Kim, K.K.; Bae, T. FtsH sensitizes methicillin-resistant Staphylococcus aureus to β-lactam antibiotics by degrading YpfP, a lipoteichoic acid synthesis enzyme. Antibiotics 2021, 10, 1198. [Google Scholar] [CrossRef] [PubMed]
- Hesser, A.R.; Matano, L.M.; Vickery, C.R.; Wood, B.M.; Santiago, A.G.; Morris, H.G.; Do, T.; Losick, R.; Walker, S. The length of lipoteichoic acid polymers controls Staphylococcus aureus cell size and envelope integrity. J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef] [PubMed]
- Suginaka, H.; Shimatani, M.; Ogawa, M.; Kotani, S. Prevention of penicillin-induced lysis of Staphylococcus aureus by cellular lipoteichoic acid. J. Antibiot. 1979, 32, 73–77. [Google Scholar] [CrossRef]
- Bæk, K.T.; Bowman, L.; Millership, C.; Dupont Søgaard, M.; Kaever, V.; Siljamäki, P.; Savijoki, K.; Varmanen, P.; Nyman, T.; Gründling, A.; et al. The cell wall polymer lipoteichoic acid becomes nonessential in Staphylococcus aureus cells lacking the ClpX chaperone. mBio 2016, 7, e01228-16. [Google Scholar] [CrossRef] [PubMed]
- Di Scala, C.; Fantini, J.; Yahi, N.; Barrantes, F.J.; Chahinian, H. Anandamide revisited: How Cholesterol and ceramides control receptor-dependent and receptor-independent signal transmission pathways of a lipid neurotransmitter. Biomolecules 2018, 8, 31. [Google Scholar] [CrossRef]
- Banerjee, S.; Sionov, R.V.; Feldman, M.; Smoum, R.; Mechoulam, R.; Steinberg, D. Anandamide alters the membrane properties, halts the cell division and prevents drug efflux in multidrug resistant Staphylococcus aureus. Sci. Rep. 2021, 11, 8690. [Google Scholar] [CrossRef]
- Feldman, M.; Smoum, R.; Mechoulam, R.; Steinberg, D. Potential combinations of endocannabinoid/endocannabinoid-like compounds and antibiotics against methicillin-resistant Staphylococcus aureus. PLoS ONE 2020, 15, e0231583. [Google Scholar] [CrossRef]
- Feldman, M.; Smoum, R.; Mechoulam, R.; Steinberg, D. Antimicrobial potential of endocannabinoid and endocannabinoid-like compounds against methicillin-resistant Staphylococcus aureus. Sci. Rep. 2018, 8, 17696. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.S.; Sobkowiak, B.; Parreira, R.; Edgeworth, J.D.; Viveiros, M.; Clark, T.G.; Couto, I. Genetic Diversity of norA, coding for a main efflux pump of Staphylococcus aureus. Front. Genet. 2019, 9, 710. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; de Lencastre, H.; Tomasz, A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 2001, 98, 10886–10891. [Google Scholar] [CrossRef] [PubMed]
- Truong-Bolduc, Q.C.; Hooper, D.C. The transcriptional regulators NorG and MgrA modulate resistance to both quinolones and β-lactams in Staphylococcus aureus. J. Bacteriol. 2007, 189, 2996–3005. [Google Scholar] [CrossRef]
- Muthian, S.; Nithipatikom, K.; Campbell, W.B.; Hillard, C.J. Synthesis and characterization of a fluorescent substrate for the N-arachidonoylethanolamine (anandamide) transmembrane carrier. J. Pharmacol. Exp. Ther. 2000, 293, 289–295. [Google Scholar]
- DeFrancesco, A.S.; Masloboeva, N.; Syed, A.K.; DeLoughery, A.; Bradshaw, N.; Li, G.-W.; Gilmore, M.S.; Walker, S.; Losick, R. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2017, 114, E5969–E5978. [Google Scholar] [CrossRef]
- Das, T.; Sharma, P.K.; Busscher, H.J.; van der Mei, H.C.; Krom, B.P. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 2010, 76, 3405–3408. [Google Scholar] [CrossRef]
- Rosenberg, M.; Azevedo, N.F.; Ivask, A. Propidium iodide staining underestimates viability of adherent bacterial cells. Sci. Rep. 2019, 9, 6483. [Google Scholar] [CrossRef]
- Okshevsky, M.; Meyer, R.L. Evaluation of fluorescent stains for visualizing extracellular DNA in biofilms. J. Microbiol. Methods 2014, 105, 102–104. [Google Scholar] [CrossRef]
- Holland, L.M.; Conlon, B.; O’Gara, J.P. Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology 2011, 157, 408–418. [Google Scholar] [CrossRef]
- Brown, S.; Xia, G.; Luhachack, L.G.; Campbell, J.; Meredith, T.C.; Chen, C.; Winstel, V.; Gekeler, C.; Irazoqui, J.E.; Peschel, A.; et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. USA 2012, 109, 18909–18914. [Google Scholar] [CrossRef] [PubMed]
- Ulluwishewa, D.; Wang, L.; Pereira, C.; Flynn, S.; Cain, E.; Stick, S.; Reen, F.J.; Ramsay, J.P.; O’Gara, F. Dissecting the regulation of bile-induced biofilm formation in Staphylococcus aureus. Microbiology 2016, 162, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Wanner, S.; Schade, J.; Keinhörster, D.; Weller, N.; George, S.E.; Kull, L.; Bauer, J.; Grau, T.; Winstel, V.; Stoy, H.; et al. Wall teichoic acids mediate increased virulence in Staphylococcus aureus. Nat. Microbiol. 2017, 2, 16257. [Google Scholar] [CrossRef]
- Swoboda, J.G.; Campbell, J.; Meredith, T.C.; Walker, S. Wall teichoic acid function, biosynthesis, and inhibition. ChemBioChem 2009, 11, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-M.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Genet. 2008, 6, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, M.; Vischer, N.O.E.; Strahl, H.; Hamoen, L.W. Assessing membrane fluidity and visualizing fluid membrane domains in bacteria using fluorescent membrane dyes. Bio-Protocol. 2018, 8, e3063. [Google Scholar] [CrossRef]
- Jay, A.G.; Hamilton, J.A. Disorder amidst membrane order: Standardizing Laurdan generalized polarization and membrane fluidity terms. J. Fluoresc. 2016, 27, 243–249. [Google Scholar] [CrossRef]
- Vanounou, S.; Pines, D.; Pines, E.; Parola, A.H.; Fishov, I. Coexistence of domains with distinct order and polarity in fluid bacterial membranes. Photochem. Photobiol. 2002, 76, 1–11. [Google Scholar] [CrossRef]
- El Khoury, M.; Swain, J.; Sautrey, G.; Zimmermann, L.; Van Der Smissen, P.; Décout, J.-L.; Mingeot-Leclercq, M.-P. Targeting bacterial cardiolipin enriched microdomains: An antimicrobial strategy used by amphiphilic aminoglycoside antibiotics. Sci. Rep. 2017, 7, 10697. [Google Scholar] [CrossRef]
- Hernández-Villa, L.; Manrique-Moreno, M.; Leidy, C.; Jemioła-Rzemińska, M.; Ortíz, C.; Strzałka, K. Biophysical evaluation of cardiolipin content as a regulator of the membrane lytic effect of antimicrobial peptides. Biophys. Chem. 2018, 238, 8–15. [Google Scholar] [CrossRef]
- Jiang, J.H.; Bhuiyan, M.S.; Shen, H.H.; Cameron, D.R.; Rupasinghe, T.W.T.; Wu, C.M.; Le Brun, A.P.; Kostoulias, X.; Domene, C.; Fulcher, A.J.; et al. Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 3722–3727. [Google Scholar] [CrossRef] [PubMed]
- Mileykovskaya, E.; Dowhan, W. Visualization of Phospholipid Domains in Escherichia coli by Using the cardiolipin-specific fluorescent dye 10- N -Nonyl Acridine Orange. J. Bacteriol. 2000, 182, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Benarroch, J.M.; Asally, M. The Microbiologist’s Guide to membrane potential dynamics. Trends Microbiol. 2020, 28, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Silber, N.; de Opitz, C.L.M.; Mayer, C.; Sass, P. Cell division protein FtsZ: From structure and mechanism to antibiotic target. Future Microbiol. 2020, 15, 801–831. [Google Scholar] [CrossRef] [PubMed]
- Bottomley, A.; Liew, A.T.F.; Kusuma, K.D.; Peterson, E.; Seidel, L.; Foster, S.J.; Harry, E. Coordination of chromosome segregation and cell division in Staphylococcus aureus. Front. Microbiol. 2017, 8, 1575. [Google Scholar] [CrossRef]
- Eswara, P.J.; Brzozowski, R.S.; Viola, M.G.; Graham, G.; Spanoudis, C.; Trebino, C.; Jha, J.; Aubee, J.I.; Thompson, K.M.; Camberg, J.L.; et al. An essential Staphylococcus aureus cell division protein directly regulates FtsZ dynamics. eLife 2018, 7, e38856. [Google Scholar] [CrossRef]
- Mukherjee, A.; Saez, C.; Lutkenhaus, J. Assembly of an FtsZ mutant deficient in GTPase activity has implications for FtsZ assembly and the role of the Z Ring in cell division. J. Bacteriol. 2001, 183, 7190–7197. [Google Scholar] [CrossRef]
- Lyu, Z.; Coltharp, C.; Yang, X.; Xiao, J. Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional Superresolution imaging. Biopolymers 2016, 105, 725–734. [Google Scholar] [CrossRef]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell wall hydrolases in bacteria: Insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol. 2019, 10, 331. [Google Scholar] [CrossRef]
- Forsberg, C.W.; Rogers, H.J. Autolytic enzymes in growth of bacteria. Nature 1971, 229, 272–273. [Google Scholar] [CrossRef]
- Nega, M.; Tribelli, P.M.; Hipp, K.; Stahl, M.; Götz, F. New insights in the coordinated amidase and glucosaminidase activity of the major autolysin (Atl) in Staphylococcus aureus. Commun. Biol. 2020, 3, 695. [Google Scholar] [CrossRef] [PubMed]
- Do, T.; Schaefer, K.; Santiago, A.G.; Coe, K.A.; Fernandes, P.B.; Kahne, D.; Pinho, M.G.; Walker, S. Staphylococcus aureus cell growth and division are regulated by an amidase that trims peptides from uncrosslinked peptidoglycan. Nat. Microbiol. 2020, 5, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, J.; Fujiwara, T.; Yamada, S.; Suzawa, Y.; Nishida, T.; Oyamada, Y.; Hayashi, I.; Yamagishi, J.; Komatsuzawa, H.; Sugai, M. Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 2005, 58, 1087–1101. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, K.B.; Gatto, C.; Walker, S.; Wilkinson, B.J. Exposure of Staphylococcus aureus to Targocil blocks translocation of the major autolysin Atl across the membrane, resulting in a significant decrease in autolysis. Antimicrob. Agents Chemother. 2018, 62, e00323-18. [Google Scholar] [CrossRef]
- Ranjit, D.K.; Endres, J.L.; Bayles, K.W. Staphylococcus aureus CidA and LrgA proteins exhibit Holin-like properties. J. Bacteriol. 2011, 193, 2468–2476. [Google Scholar] [CrossRef]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E.; Smeltzer, M.S.; Bayles, K.W. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef]
- Buchad, H.; Nair, M. The small RNA SprX regulates the autolysin regulator WalR in Staphylococcus aureus. Microbiol. Res. 2021, 250, 126785. [Google Scholar] [CrossRef]
- Groicher, K.H.; Firek, B.A.; Fujimoto, D.F.; Bayles, K.W. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 2000, 182, 1794–1801. [Google Scholar] [CrossRef]
- Vaz, F.; Filipe, S.R. Preparation and analysis of crude autolytic enzyme extracts from Staphylococcus aureus. Bio-Protoc. 2015, 5, e1678. [Google Scholar] [CrossRef]
- Thalsø-Madsen, I.; Torrubia, F.R.; Xu, L.; Petersen, A.; Jensen, C.; Frees, D. The Sle1 cell wall amidase is essential for β-lactam resistance in community-acquired methicillin-resistant Staphylococcus aureus USA300. Antimicrob. Agents Chemother. 2019, 64, e01931-19. [Google Scholar] [CrossRef]
- Bose, J.L.; Lehman, M.K.; Fey, P.D.; Bayles, K.W. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS ONE 2012, 7, e42244. [Google Scholar] [CrossRef] [PubMed]
- Brötz-Oesterhelt, H.; Vorbach, A. Reprogramming of the caseinolytic protease by ADEP antibiotics: Molecular mechanism, cellular consequences, therapeutic potential. Front. Mol. Biosci. 2021, 8, 690902. [Google Scholar] [CrossRef] [PubMed]
- Donegan, N.P.; Marvin, J.S.; Cheung, A.L. Role of Adaptor TrfA and ClpPC in controlling levels of SsrA-tagged proteins and antitoxins in Staphylococcus aureus. J. Bacteriol. 2014, 196, 4140–4151. [Google Scholar] [CrossRef] [PubMed]
- Jousselin, A.; Kelley, W.L.; Barras, C.; Lew, D.P.; Renzoni, A. The Staphylococcus aureus thiol/oxidative stress global regulator Spx controls trfA, a gene implicated in cell wall antibiotic resistance. Antimicrob. Agents Chemother. 2013, 57, 3283–3292. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase inhibitors in the 21st century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Lekshmi, M.; Ammini, P.; Adjei, J.; Sanford, L.M.; Shrestha, U.; Kumar, S.; Varela, M.F. Modulation of antimicrobial efflux pumps of the major facilitator superfamily in Staphylococcus aureus. AIMS Microbiol. 2018, 4, 1–18. [Google Scholar] [CrossRef]
- Kvist, M.; Hancock, V.; Klemm, P. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 2008, 74, 7376–7382. [Google Scholar] [CrossRef]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef]
- Zimmermann, S.; Klinger-Strobel, M.; Bohnert, J.A.; Wendler, S.; Rodel, J.; Pletz, M.W.; Loffler, B.; Tuchscherr, L. Clinically approved drugs inhibit the Staphylococcus aureus multidrug NorA efflux pump and reduce biofilm formation. Front. Microbiol. 2019, 10, 2762. [Google Scholar] [CrossRef]
- Sionov, R.V.; Steinberg, D. Anti-microbial activity of phytocannabinoids and endocannabinoids in the light of their physiological and pathophysiological roles. Biomedicines 2022, 10, 631. [Google Scholar] [CrossRef]
- Chung, H.S.; Yao, Z.; Goehring, N.W.; Kishony, R.; Beckwith, J.; Kahne, D. Rapid β-lactam-induced lysis requires successful assembly of the cell division machinery. Proc. Natl. Acad. Sci. USA 2009, 106, 21872–21877. [Google Scholar] [CrossRef] [PubMed]
- Lui, H.K.; Gao, W.; Cheung, K.C.; Jin, W.B.; Sun, N.; Kan, J.W.Y.; Wong, I.L.K.; Chiou, J.; Lin, D.; Chan, E.W.C.; et al. Boosting the efficacy of anti-MRSA β-lactam antibiotics via an easily accessible, non-cytotoxic and orally bioavailable FtsZ inhibitor. Eur. J. Med. Chem. 2019, 163, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-González, E.; Kaul, M.; Parhi, A.K.; LaVoie, E.J.; Pilch, D.S. β-lactam antibiotics with a high affinity for PBP2 act synergistically with the FtsZ-targeting agent TXA707 against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2017, 61, e00863-17. [Google Scholar] [CrossRef]
- Tan, C.M.; Therien, A.G.; Lu, J.; Lee, S.H.; Caron, A.; Gill, C.J.; Lebeau-Jacob, C.; Benton-Perdomo, L.; Monteiro, J.M.; Pereira, P.M.; et al. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci. Transl. Med. 2012, 4, 126ra35. [Google Scholar] [CrossRef] [PubMed]
- Barrows, J.M.; Goley, E.D. FtsZ dynamics in bacterial division: What, how, and why? Curr. Opin. Cell Biol. 2021, 68, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Michalik, S.; Varming, A.N.; Andersen, J.H.; Albrecht, D.; Jelsbak, L.; Krieger, S.; Ohlsen, K.; Hecker, M.; Gerth, U.; et al. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J. Proteome Res. 2013, 12, 547–558. [Google Scholar] [CrossRef]
- Bæk, K.T.; Gründling, A.; Mogensen, R.G.; Thøgersen, L.; Petersen, A.; Paulander, W.; Frees, D. β-Lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease. Antimicrob. Agents Chemother. 2014, 58, 4593–4603. [Google Scholar] [CrossRef]
- Bai, J.; Zhu, X.; Zhao, K.; Yan, Y.; Xu, T.; Wang, J.; Zheng, J.; Huang, W.; Shi, L.; Shang, Y.; et al. The role of ArlRS in regulating oxacillin susceptibility in methicillin-resistant Staphylococcus aureus indicates it is a potential target for antimicrobial resistance breakers. Emerg. Microbes Infect. 2019, 8, 503–515. [Google Scholar] [CrossRef]
- Lau, J.; Hernandez-Alicea, L.; Vass, R.H.; Chien, P. A Phosphosignaling adaptor primes the AAA+ protease ClpXP to drive cell cycle-regulated proteolysis. Mol. Cell 2015, 59, 104–116. [Google Scholar] [CrossRef]
- Collinson, I. The Dynamic ATP-Driven Mechanism of bacterial protein translocation and the critical role of phospholipids. Front. Microbiol. 2019, 10, 1217. [Google Scholar] [CrossRef]
- Strahl, H.; Hamoen, L.W. Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. USA 2010, 107, 12281–12286. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Shiau, C.W.; Chang, Y.C.; Kung, H.N.; Wu, J.C.; Lim, C.H.; Yeo, H.H.; Chang, H.C.; Chien, H.S.; Huang, S.H.; et al. SC5005 dissipates the membrane potential to kill Staphylococcus aureus persisters without detectable resistance. J. Antimicrob. Chemother. 2021, 76, 2049–2056. [Google Scholar] [CrossRef] [PubMed]
- Patton, T.G.; Yang, S.-J.; Bayles, K.W. The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol. Microbiol. 2006, 59, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Rice, K.C.; Bayles, K.W. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 2008, 72, 85–109. [Google Scholar] [CrossRef]
- Sadykov, M.R.; Windham, I.H.; Widhelm, T.J.; Yajjala, V.K.; Watson, S.M.; Endres, J.L.; Bavari, A.I.; Thomas, V.C.; Bose, J.L.; Bayles, K.W. CidR and CcpA synergistically regulate Staphylococcus aureus cidABC expression. J. Bacteriol. 2019, 201, e00371-19. [Google Scholar] [CrossRef]
- Salamaga, B.; Kong, L.; Pasquina-Lemonche, L.; Lafage, L.; von Und Zur Muhlen, M.; Gibson, J.F.; Grybchuk, D.; Tooke, A.K.; Panchal, V.; Culp, E.J.; et al. Demonstration of the role of cell wall homeostasis in Staphylococcus aureus growth and the action of bactericidal antibiotics. Proc. Natl. Acad. Sci. USA 2021, 118, e2106022118. [Google Scholar] [CrossRef]
- Delaune, A.; Poupel, O.; Mallet, A.; Coic, Y.-M.; Msadek, T.; Dubrac, S. Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS ONE 2011, 6, e17054. [Google Scholar] [CrossRef]
- Mediati, D.G.; Wu, S.; Wu, W.; Tree, J.J. Networks of resistance: Small RNA control of antibiotic resistance. Trends Genet. 2021, 37, 35–45. [Google Scholar] [CrossRef]
- Eyraud, A.; Tattevin, P.; Chabelskaya, S.; Felden, B. A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res. 2014, 42, 4892–4905. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, S.; Sun, B. SpoVG regulates cell wall metabolism and oxacillin resistance in methicillin-Resistant Staphylococcus aureus strain N315. Antimicrob. Agents Chemother. 2016, 60, 3455–3461. [Google Scholar] [CrossRef]
- Beltrame, C.O.; Côrtes, M.F.; Bonelli, R.R.; Côrrea, A.B.; Botelho, A.M.; Américo, M.A.; Fracalanzza, S.E.; Figueiredo, A.M. Inactivation of the autolysis-related genes lrgB and yycI in Staphylococcus aureus increases cell lysis-dependent eDNA release and enhances biofilm development in vitro and in vivo. PLoS ONE 2015, 10, e0138924. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, X.; Qin, J.; Cheng, S.; Yeo, W.-S.; He, L.; Ma, X.; Liu, X.; Li, M.; Bae, T. The ATP-dependent protease ClpP inhibits biofilm formation by regulating Agr and cell wall hydrolase Sle1 in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2017, 7, 181. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, M.; Walker, S. Envelope structures of Gram-positive bacteria. Curr. Top Microbiol. Immunol. 2017, 404, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.; Singh, A.K.; Swoboda, J.G.; Gilmore, M.S.; Wilkinson, B.J.; Walker, S. An antibiotic that inhibits a late step in wall teichoic acid biosynthesis induces the cell wall stress stimulon in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Liew, A.T.F.; Theis, T.; Jensen, S.O.; Garcia-Lara, J.; Foster, S.J.; Firth, N.; Lewis, P.J.; Harry, E.J. A simple plasmid-based system that allows rapid generation of tightly controlled gene expression in Staphylococcus aureus. Microbiology 2011, 157, 666–676. [Google Scholar] [CrossRef]
- Sionov, R.V.; Tsavdaridou, D.; Aqawi, M.; Zaks, B.; Steinberg, D.; Shalish, M. Tooth mousse containing casein phosphopeptide-amorphous calcium phosphate prevents biofilm formation of Streptococcus mutans. BMC Oral Health 2021, 21, 1–10. [Google Scholar] [CrossRef]
- Zhao, G.; Meier, T.I.; Kahl, S.D.; Gee, K.R.; Blaszczak, L.C. BOCILLIN FL, a sensitive and commercially available reagent for detection of Penicillin-Binding Proteins. Antimicrob. Agents Chemother. 1999, 43, 1124–1128. [Google Scholar] [CrossRef]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef]
- Umemura, S.; Itoh, J.; Takekoshi, S.; Hasegawa, H.; Yasuda, M.; Osamura, R.Y.; Watanabe, K. Methyl Green staining and DNA strands in vitro: High affinity of Methyl Green dye to cytosine and guanine. Acta Histochem. ET Cytochem. 2003, 36, 361–366. [Google Scholar] [CrossRef][Green Version]
- Ron-Doitch, S.; Frušić-Zlotkin, M.; Soroka, Y.; Duanis-Assaf, D.; Amar, D.; Kohen, R.; Steinberg, D. eDNA-mediated cutaneous protection against UVB damage conferred by Staphylococcal epidermal colonization. Microorganisms 2021, 9, 788. [Google Scholar] [CrossRef]
- Schneider-Rayman, M.; Steinberg, D.; Sionov, R.V.; Friedman, M.; Shalish, M. Effect of epigallocatechin gallate on dental biofilm of Streptococcus mutans: An in vitro study. BMC Oral Health 2021, 21, 447. [Google Scholar] [CrossRef] [PubMed]
- Meredith, T.C.; Swoboda, J.G.; Walker, S. Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus. J. Bacteriol. 2008, 190, 3046–3056. [Google Scholar] [CrossRef] [PubMed]
- Covas, G.; Vaz, F.; Henriques, G.; Pinho, M.G.; Filipe, S.R. Analysis of cell wall teichoic acids in Staphylococcus aureus. Methods Mol. Biol. 2016, 1440, 201–213. [Google Scholar] [CrossRef]
- Strahl, H.; Bürmann, F.; Hamoen, L.W. The actin homologue MreB organizes the bacterial cell membrane. Nat. Commun. 2014, 5, 3442. [Google Scholar] [CrossRef]
- Sündermann, A.; Eggers, L.F.; Schwudke, D. Liquid extraction: Bligh and Dyer. In Encyclopedia of Lipidomics; Wenk, M.R., Ed.; Springer: Dordrecht, The Netherlands, 2016. [Google Scholar]
- Hoelscher, G.L.; Hudson, M.C. Characterization of an unusual fluoride-resistant Streptococcus mutans isolate. Curr. Microbiol. 1996, 32, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Duanis-Assaf, D.; Kenan, E.; Sionov, R.; Steinberg, D.; Shemesh, M. Proteolytic activity of Bacillus subtilis upon κ-Casein undermines its “Caries-Safe” effect. Microorganisms 2020, 8, 221. [Google Scholar] [CrossRef]
- Villet, R.A.; Truong-Bolduc, Q.C.; Wang, Y.; Estabrooks, Z.; Medeiros, H.; Hooper, D.C. Regulation of expression of abcA and its response to environmental conditions. J. Bacteriol. 2014, 196, 1532–1539. [Google Scholar] [CrossRef]
- Batte, J.L.; Sahukhal, G.S.; Elasri, M.O. MsaB and CodY interact to regulate Staphylococcus aureus capsule in a nutrient-dependent manner. J. Bacteriol. 2018, 200, e00294-18. [Google Scholar] [CrossRef]
- Chung, P.Y.; Chung, L.Y.; Navaratnam, P. Transcriptional profiles of the response of methicillin-resistant Staphylococcus aureus to pentacyclic triterpenoids. PLoS ONE 2013, 8, e56687. [Google Scholar] [CrossRef]
- Neumann, Y.; Ohlsen, K.; Donat, S.; Engelmann, S.; Kusch, H.; Albrecht, D.; Cartron, M.; Hurd, A.; Foster, S.J. The effect of skin fatty acids on Staphylococcus aureus. Arch. Microbiol. 2015, 197, 245–267. [Google Scholar] [CrossRef]
- Ishii, K.; Adachi, T.; Yasukawa, J.; Suzuki, Y.; Hamamoto, H.; Sekimizu, K. Induction of virulence gene expression in Staphylococcus aureus by pulmonary surfactant. Infect. Immun. 2014, 82, 1500–1510. [Google Scholar] [CrossRef]
- Liang, X.; Zheng, L.; Landwehr, C.; Lunsford, D.; Holmes, D.; Ji, Y. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 2005, 187, 5486–5492. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Shang, Y.; Wu, Y.; Wu, J.; Chen, J.; Wang, Z.; Sun, X.; Xu, G.; Deng, Q.; Qu, D.; et al. Diclazuril inhibits biofilm formation and hemolysis of Staphylococcus aureus. ACS Infect. Dis. 2021, 7, 1690–1701. [Google Scholar] [CrossRef] [PubMed]
- Jousselin, A.; Manzano, C.; Biette, A.; Reed, P.; Pinho, M.G.; Rosato, A.E.; Kelley, W.L.; Renzoni, A. The Staphylococcus aureus chaperone PrsA is a new auxiliary factor of oxacillin resistance affecting Penicillin-Binding Protein 2A. Antimicrob. Agents Chemother. 2015, 60, 1656–1666. [Google Scholar] [CrossRef] [PubMed]
- Salaheen, S.; Peng, M.; Joo, J.; Teramoto, H.; Biswas, D. Eradication and sensitization of methicillin resistant Staphylococcus aureus to methicillin with bioactive extracts of berry pomace. Front. Microbiol. 2017, 8, 253. [Google Scholar] [CrossRef]
- Sethupathy, S.; Vigneshwari, L.; Valliammai, A.; Balamurugan, K.; Pandian, S.K. L-Ascorbyl 2,6-Dipalmitate inhibits biofilm and virulence in methicillin-resistant Staphylococcus aureus and prevents triacylglyceride accumulation in Caenorhabditis elegans. RSC Adv. 2017, 7, 23392–23406. [Google Scholar] [CrossRef]
- Kannappan, A.; Srinivasan, R.; Nivetha, A.; Annapoorani, A.; Pandian, S.K.; Ravi, A.V. Anti-virulence potential of 2-hydroxy-4-methoxybenzaldehyde against methicillin-resistant Staphylococcus aureus and its clinical isolates. Appl. Microbiol. Biotechnol. 2019, 103, 6747–6758. [Google Scholar] [CrossRef]
- Wassmann, C.S.; Højrup, P.; Klitgaard, J.K. Cannabidiol is an effective helper compound in combination with bacitracin to kill Gram-positive bacteria. Sci. Rep. 2020, 10, 4112. [Google Scholar] [CrossRef]
- Hall, J.W.; Yang, J.; Guo, H.; Ji, Y. The Staphylococcus aureus AirSR Two-component system mediates reactive oxygen species resistance via transcriptional regulation of Staphyloxanthin production. Infect. Immun. 2017, 85, e00838-16. [Google Scholar] [CrossRef]
- Sun, H.; Yang, Y.; Xue, T.; Sun, B. Modulation of cell wall synthesis and susceptibility to vancomycin by the two-component system AirSR in Staphylococcus aureus NCTC8325. BMC Microbiol. 2013, 13, 286. [Google Scholar] [CrossRef]
- Crosby, H.A.; Tiwari, N.; Kwiecinski, J.M.; Xu, Z.; Dykstra, A.; Jenul, C.; Fuentes, E.J.; Horswill, A.R. The Staphylococcus aureus ArlRS two-component system regulates virulence factor expression through MgrA. Mol. Microbiol. 2020, 113, 103–122. [Google Scholar] [CrossRef] [PubMed]
- Fournier, B.; Klier, A.; Rapoport, G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 2001, 41, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, C.; Hussain, M.; Peters, G.; Götz, F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 1997, 24, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Houston, P.; Rowe, S.E.; Pozzi, C.; Waters, E.M.; O’Gara, J.P. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 2011, 79, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Shoji, M.; Cui, L.; Iizuka, R.; Komoto, A.; Neoh, H.M.; Watanabe, Y.; Hishinuma, T.; Hiramatsu, K. walK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 3870–3881. [Google Scholar] [CrossRef]
- Alqarzaee, A.A.; Chaudhari, S.S.; Islam, M.M.; Kumar, V.; Zimmerman, M.C.; Saha, R.; Bayles, K.W.; Frees, D.; Thomas, V.C. Staphylococcal ClpXP protease targets the cellular antioxidant system to eliminate fitness-compromised cells in stationary phase. Proc. Natl. Acad. Sci. USA 2021, 118, e2109671118. [Google Scholar] [CrossRef]
- Fletcher, J.R.; Villareal, A.R.; Penningroth, M.R.; Hunter, R.C. Staphylococcus aureus overcomes anaerobe-derived short-chain fatty acid stress via FadX and the CodY regulon. J. Bacteriol. 2022, e0006422. [Google Scholar] [CrossRef]
- Pohl, K.; Francois, P.; Stenz, L.; Schlink, F.; Geiger, T.; Herbert, S.; Goerke, C.; Schrenzel, J.; Wolz, C. CodY in Staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J. Bacteriol. 2009, 191, 2953–2963. [Google Scholar] [CrossRef]
- Majerczyk, C.D.; Sadykov, M.R.; Luong, T.T.; Lee, C.; Somerville, G.A.; Sonenshein, A.L. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 2008, 190, 2257–2265. [Google Scholar] [CrossRef]
- Waters, N.R.; Samuels, D.J.; Behera, R.K.; Livny, J.; Rhee, K.Y.; Sadykov, M.R.; Brinsmade, S.R. A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 2016, 101, 495–514. [Google Scholar] [CrossRef]
- Majerczyk, C.D.; Dunman, P.M.; Luong, T.T.; Lee, C.Y.; Sadykov, M.R.; Somerville, G.A.; Bodi, K.; Sonenshein, A.L. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 2010, 192, 2861–2877. [Google Scholar] [CrossRef] [PubMed]
- Bowman, L.; Palmer, T. The Type VII Secretion System of Staphylococcus. Annu. Rev. Microbiol. 2021, 75, 471–494. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, S.; Ehlert, K.; Tschierske, M.; Labischinski, H.; Berger-Bächi, B. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. USA 1999, 96, 9351–9356. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.A.; Yoshii, Y.; Yamada, S.; Nagakura, M.; Kinjo, Y.; Mizunoe, Y.; Okuda, K.I. Roles of lytic transglycosylases in biofilm formation and β-lactam resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63, e01277-19. [Google Scholar] [CrossRef]
- Stapleton, M.R.; Horsburgh, M.J.; Hayhurst, E.J.; Wright, L.; Jonsson, I.M.; Tarkowski, A.; Kokai-Kun, J.F.; Mond, J.J.; Foster, S.J. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 2007, 189, 7316–7325. [Google Scholar] [CrossRef]
- Yu, D.; Zhao, L.; Xue, T.; Sun, B. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol. 2012, 12, 288. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Zhao, L.; Sun, B. LuxS/AI-2 system is involved in antibiotic susceptibility and autolysis in Staphylococcus aureus NCTC 8325. Int. J. Antimicrob. Agents 2013, 41, 85–89. [Google Scholar] [CrossRef]
- Ma, R.; Qiu, S.; Jiang, Q.; Sun, H.; Xue, T.; Cai, G.; Sun, B. AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int. J. Med. Microbiol. 2017, 307, 257–267. [Google Scholar] [CrossRef]
- Zhao, L.; Xue, T.; Shang, F.; Sun, H.; Sun, B. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect. Immun. 2010, 78, 3506–3515. [Google Scholar] [CrossRef]
- Yang, S.J.; Xiong, Y.Q.; Yeaman, M.R.; Bayles, K.W.; Abdelhady, W.; Bayer, A.S. Role of the LytSR two-component regulatory system in adaptation to cationic antimicrobial peptides in Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 3875–3882. [Google Scholar] [CrossRef]
- Brunskill, E.W.; Bayles, K.W. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 1996, 178, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.M.; Alexander, J.A.N.; Choo, E.J.; Basuino, L.; da Costa, T.M.; Severin, A.; Chung, M.; Aedo, S.; Strynadka, N.C.J.; Tomasz, A.; et al. High-level resistance of Staphylococcus aureus to β-Lactam antibiotics mediated by Penicillin-Binding Protein 4 (PBP4). Antimicrob. Agents Chemother. 2017, 61, e02727-16. [Google Scholar] [CrossRef] [PubMed]
- Roch, M.; Lelong, E.; Panasenko, O.O.; Sierra, R.; Renzoni, A.; Kelley, W.L. Thermosensitive PBP2a requires extracellular folding factors PrsA and HtrA1 for Staphylococcus aureus MRSA β-lactam resistance. Commun. Biol. 2019, 2, 417. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yeo, W.S.; Bae, T. The SaeRS two-component system of Staphylococcus aureus. Genes 2016, 7, 81. [Google Scholar] [CrossRef]
- Lou, Q.; Ma, Y.; Qu, D. Two-component signal transduction system SaeRS is involved in competence and penicillin susceptibility in Staphylococcus epidermidis. J. Basic Microbiol. 2016, 56, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Lou, Q.; Zhu, T.; Hu, J.; Ben, H.; Yang, J.; Yu, F.; Liu, J.; Wu, Y.; Fischer, A.; Francois, P.; et al. Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiol. 2011, 11, 146. [Google Scholar] [CrossRef]
- Geoghegan, J.A.; Corrigan, R.M.; Gruszka, D.T.; Speziale, P.; O’Gara, J.P.; Potts, J.R.; Foster, T.J. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2010, 192, 5663–5673. [Google Scholar] [CrossRef]
- Formosa-Dague, C.; Speziale, P.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc. Natl. Acad. Sci. USA 2016, 113, 410–415. [Google Scholar] [CrossRef]
- Lauderdale, K.J.; Boles, B.R.; Cheung, A.L.; Horswill, A.R. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 2009, 77, 1623–1635. [Google Scholar] [CrossRef]
- Kathirvel, M.; Buchad, H.; Nair, M. Enhancement of the pathogenicity of Staphylococcus aureus strain Newman by a small noncoding RNA SprX1. Med. Microbiol. Immunol. 2016, 205, 563–574. [Google Scholar] [CrossRef]
- Pamp, S.J.; Frees, D.; Engelmann, S.; Hecker, M.; Ingmer, H. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2006, 188, 4861–4870. [Google Scholar] [CrossRef] [PubMed]
- Panasenko, O.O.; Bezrukov, F.; Komarynets, O.; Renzoni, A. YjbH solubility controls Spx in Staphylococcus aureus: Implication for MazEF Toxin-Antitoxin system regulation. Front. Microbiol. 2020, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Poupel, O.; Proux, C.; Jagla, B.; Msadek, T.; Dubrac, S. SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog. 2018, 14, e1006917. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sionov, R.V.; Banerjee, S.; Bogomolov, S.; Smoum, R.; Mechoulam, R.; Steinberg, D. Targeting the Achilles’ Heel of Multidrug-Resistant Staphylococcus aureus by the Endocannabinoid Anandamide. Int. J. Mol. Sci. 2022, 23, 7798. https://doi.org/10.3390/ijms23147798
Sionov RV, Banerjee S, Bogomolov S, Smoum R, Mechoulam R, Steinberg D. Targeting the Achilles’ Heel of Multidrug-Resistant Staphylococcus aureus by the Endocannabinoid Anandamide. International Journal of Molecular Sciences. 2022; 23(14):7798. https://doi.org/10.3390/ijms23147798
Chicago/Turabian StyleSionov, Ronit Vogt, Shreya Banerjee, Sergei Bogomolov, Reem Smoum, Raphael Mechoulam, and Doron Steinberg. 2022. "Targeting the Achilles’ Heel of Multidrug-Resistant Staphylococcus aureus by the Endocannabinoid Anandamide" International Journal of Molecular Sciences 23, no. 14: 7798. https://doi.org/10.3390/ijms23147798
APA StyleSionov, R. V., Banerjee, S., Bogomolov, S., Smoum, R., Mechoulam, R., & Steinberg, D. (2022). Targeting the Achilles’ Heel of Multidrug-Resistant Staphylococcus aureus by the Endocannabinoid Anandamide. International Journal of Molecular Sciences, 23(14), 7798. https://doi.org/10.3390/ijms23147798





