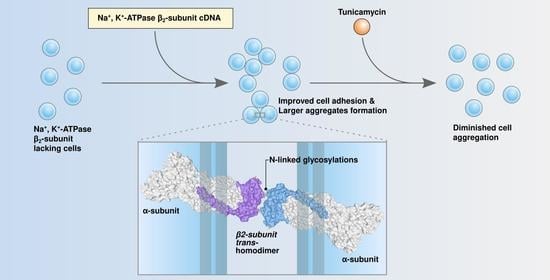
The β2-Subunit (AMOG) of Human Na+, K+-ATPase Is a Homophilic Adhesion Molecule
Abstract
1. Introduction
2. Results
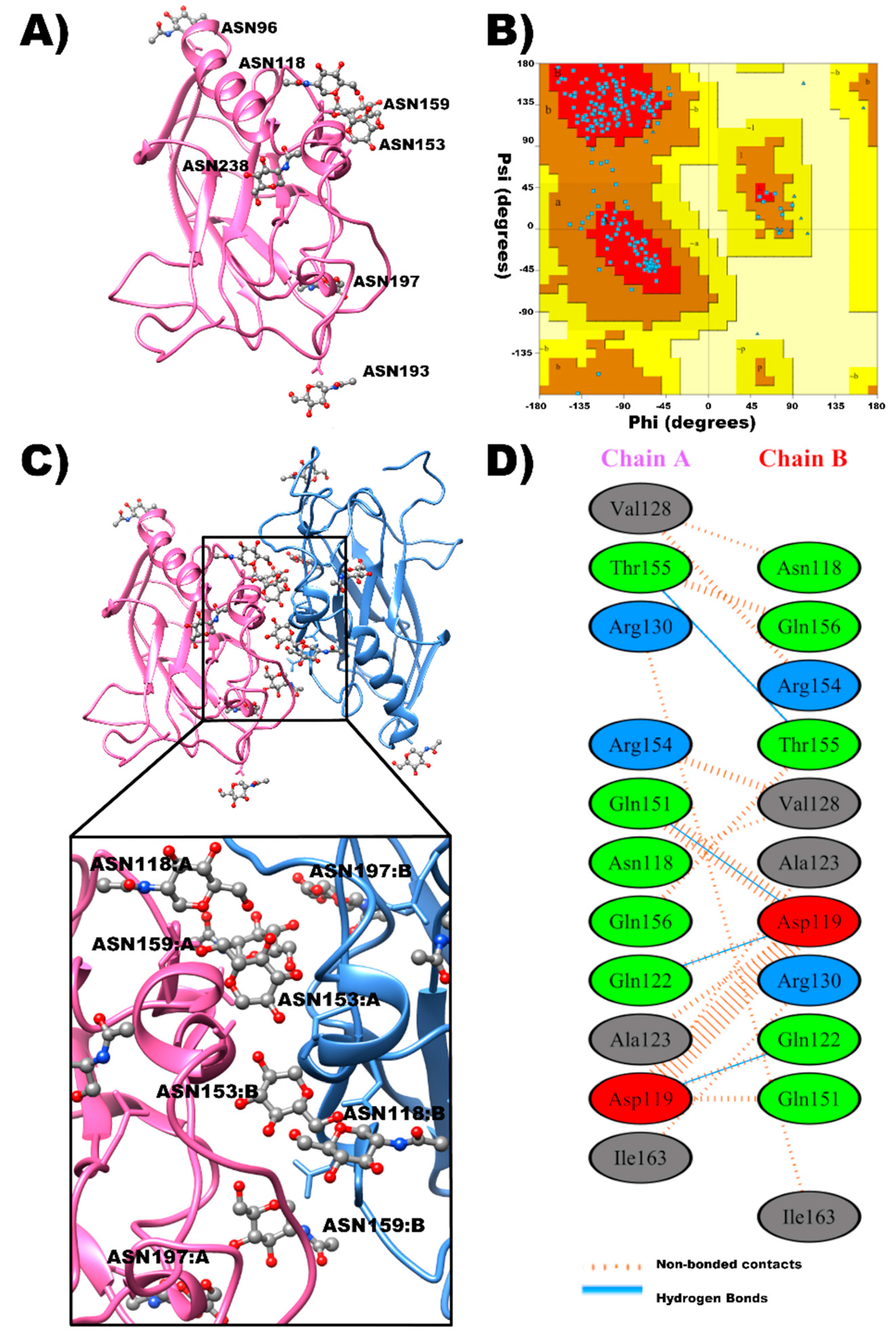
2.1. In Silico Prediction of Glycosylated β2–β2 Trans-Interaction
2.2. Generation of Experimental Tools
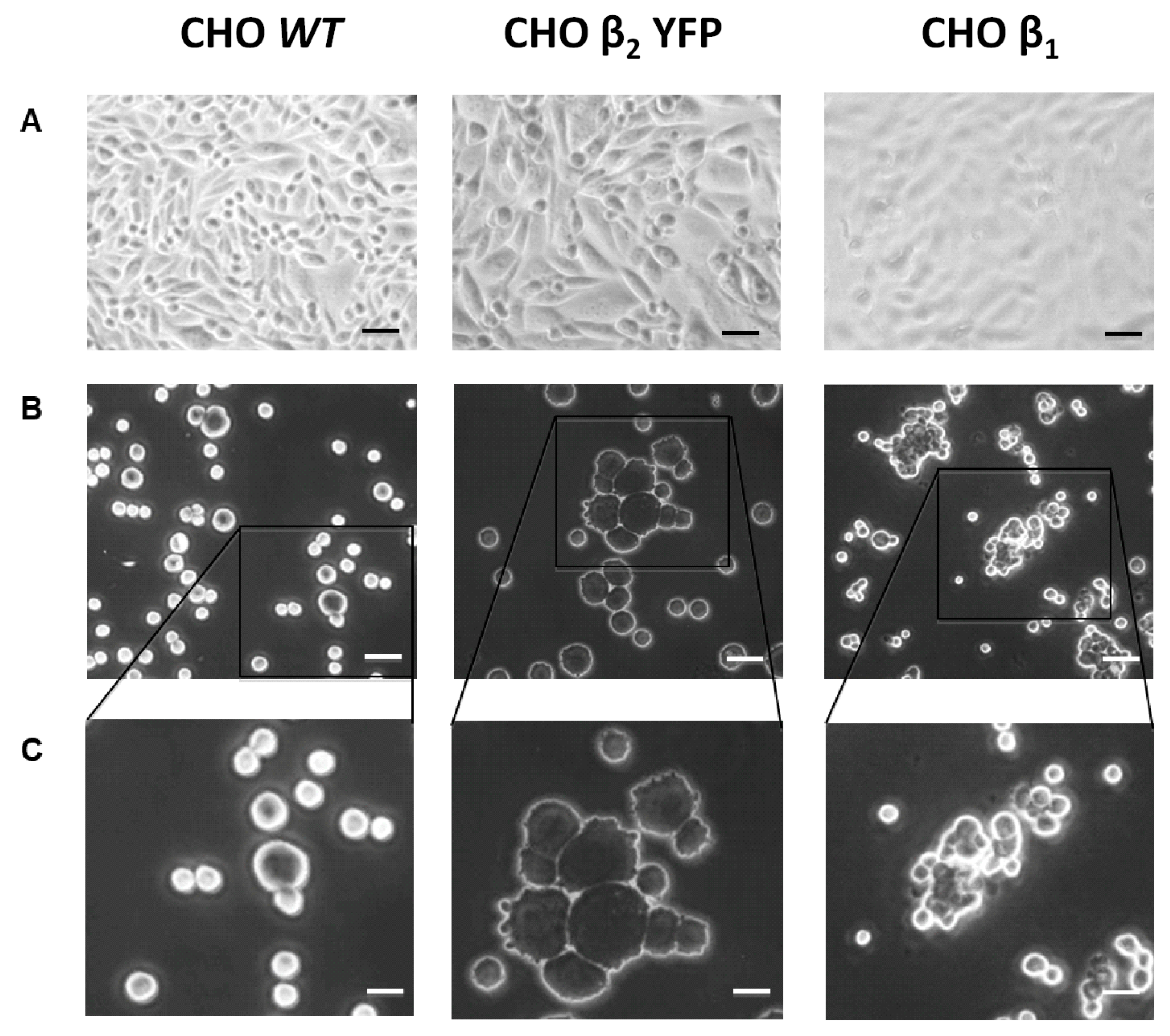
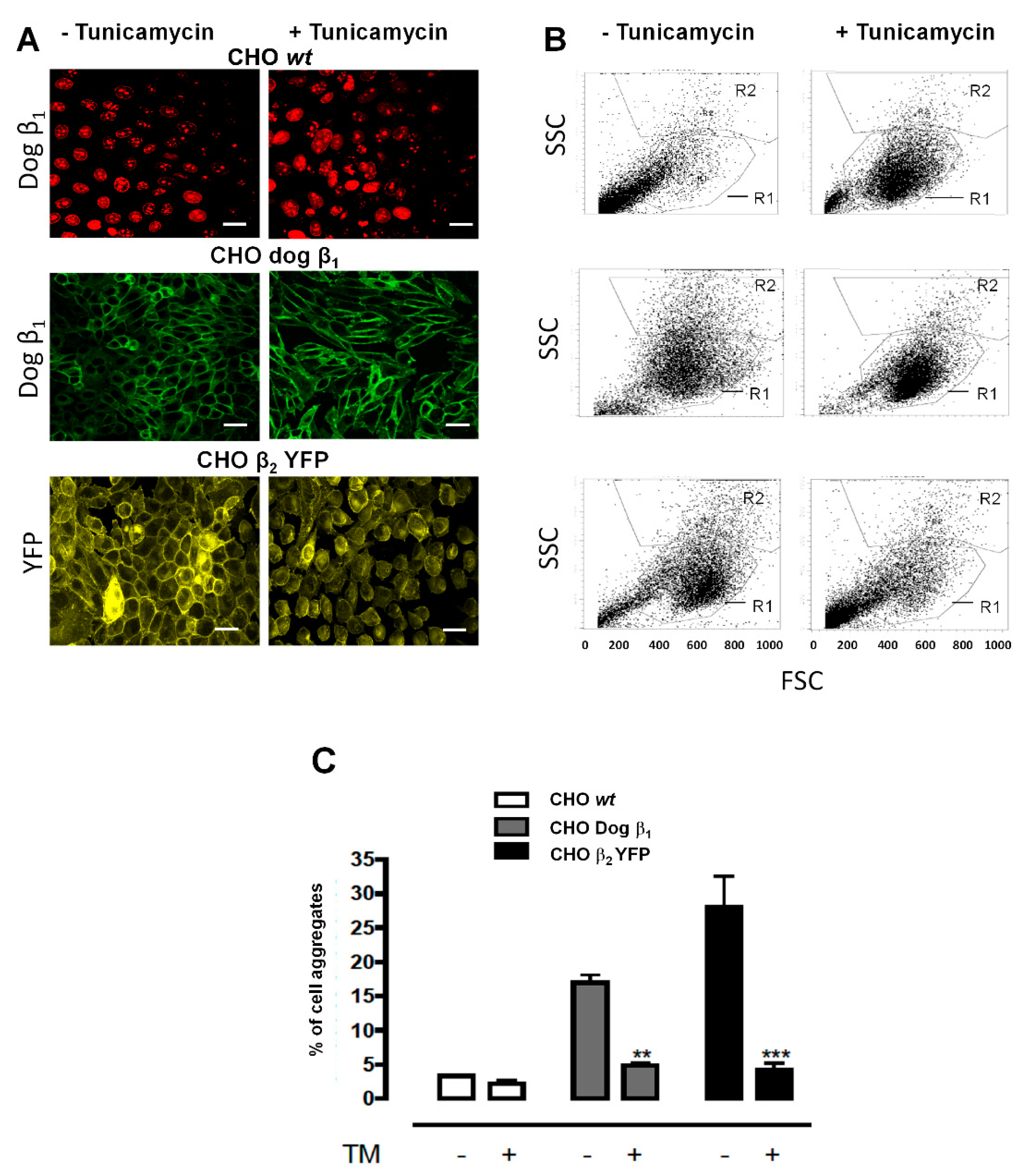
2.3. Over-Expression of β2 YFP Increases Cell Aggregation in a Glycosylation-Dependent Manner
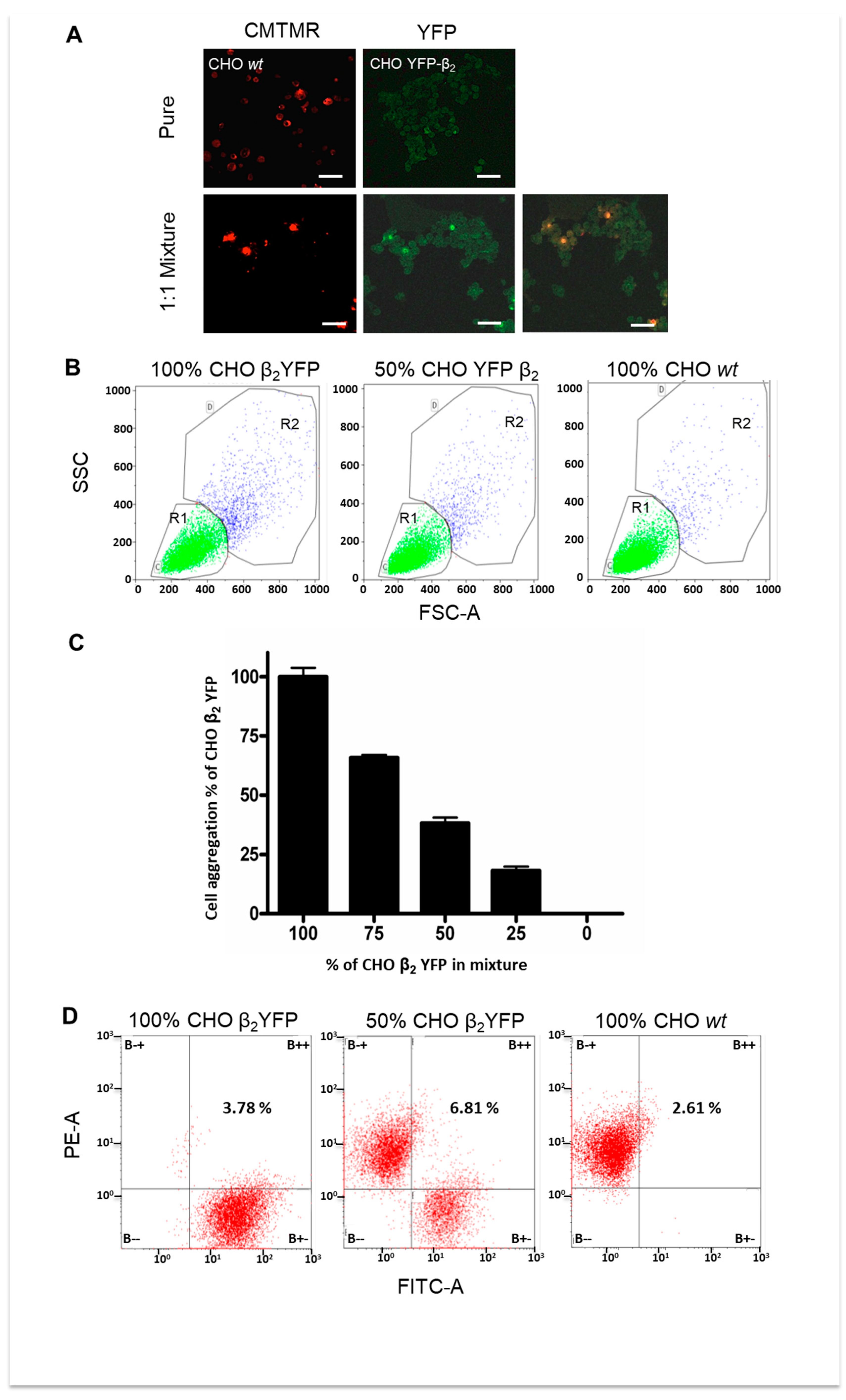
2.4. Cell Aggregation of CHO β2 YFP Decreases When Cocultured with CHO wt Cells
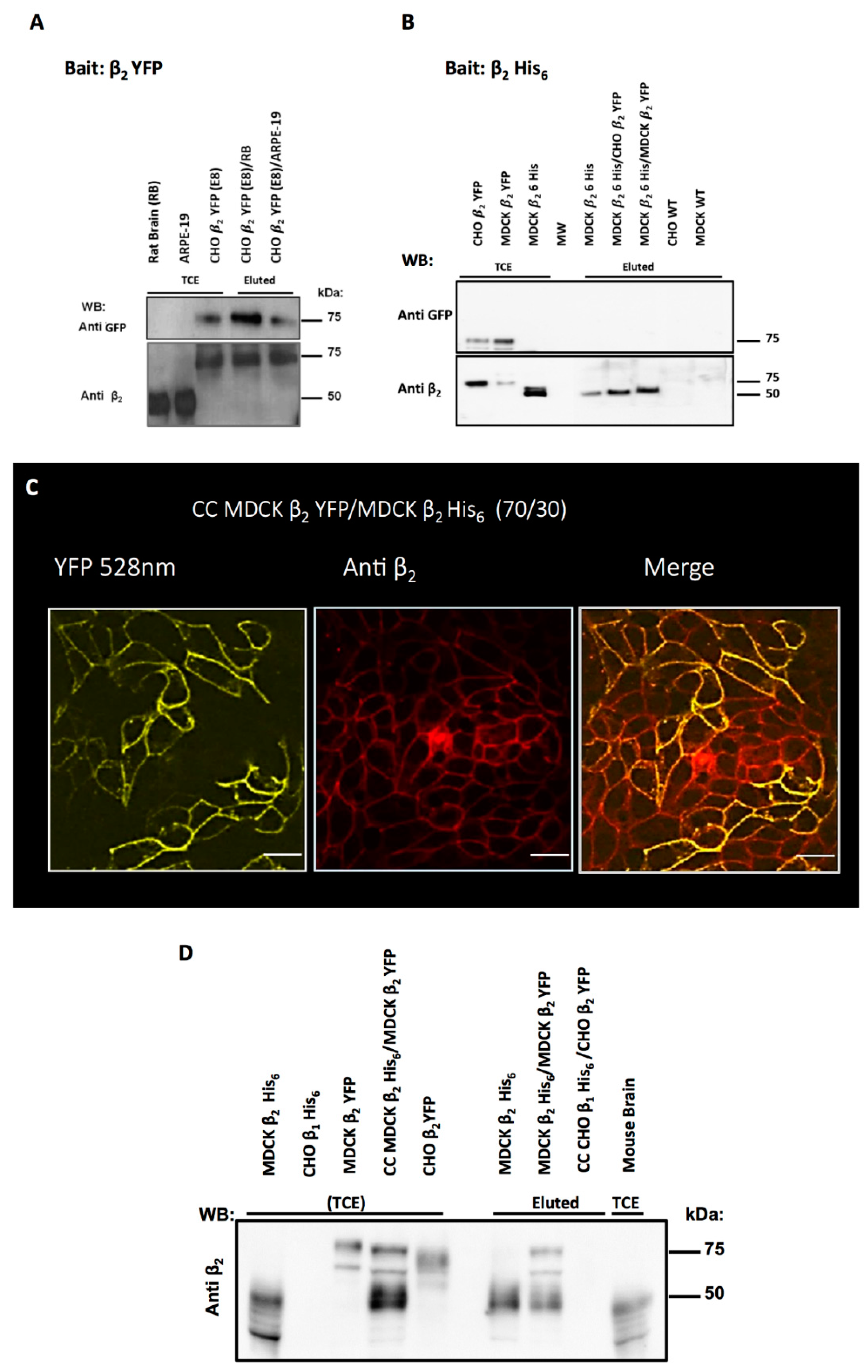
2.5. Cell Aggregation of CHO β2 YFP Is Due to β2–β2 Interactions In Vivo
3. Discussion
3.1. The Role of N-Glycosylation
3.2. β2/AMOG Is a Heterophilic Adhesion Molecule in Astrocytes
3.3. β2/AMOG Is a Homophilic Cell-Adhesion Molecule
4. Materials and Methods
4.1. Cell Culture, Transfection, and Establishment of Stable Cell Line
4.2. Immunofluorescence Imaging
4.3. Primary and Secondary Antibodies
4.4. Cell Aggregation Assay
4.5. Cell Adhesion Assay (Dispase Assay)
4.6. Mixed Monolayers (Co-Cultures)
4.7. Pull-Down Assay
4.8. Co-Affinity Precipitation (Co-AP)
4.9. Molecular Modeling
4.10. Molecular Docking Studies
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jorgensen, P.L.; Skou, J.C. Preparation of highly active (Na+ + K+)-ATPase from the outer medulla of rabbit kidney. Biochem. Biophys. Res. Commun. 1969, 37, 39–46. [Google Scholar] [CrossRef]
- Skou, J.C. The (Na+, K+)-ATPase: Coupling of the reaction with ATP to the reaction with Na+ and K+. Ann. N. Y. Acad. Sci. 1982, 402, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.P.; Farley, R.A. All three potential N-glycosylation sites of the dog kidney (Na+, K+)-ATPase beta-subunit contain oligosaccharide. Biochim. Biophys. Acta 1988, 954, 50–57. [Google Scholar] [CrossRef]
- Reinhard, L.; Tidow, H.; Clausen, M.J.; Nissen, P. Na+,K+-ATPase as a docking station: Protein-protein complexes of the Na+,K+-ATPase. Cell Mol. Life Sci. 2013, 70, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Contreras, R.G.; Lázaro, A.; Bolivar, J.J.; Flores-Maldonado, C.; Sánchez, S.H.; González-Mariscal, L.; García-Villegas, M.R.; Valdés, J.; Cereijido, M. A novel type of cell-cell cooperation between epithelial cells. J. Membr. Biol. 1995, 145, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Shoshani, L.; Contreras, R.G.; Roldan, M.L.; Moreno, J.; Lazaro, A.; Balda, M.S.; Matter, K.; Cereijido, M. The polarized expression of Na+,K+-ATPase in epithelia depends on the association between beta-subunits located in neighboring cells. Mol. Biol. Cell 2005, 16, 1071–1081. [Google Scholar] [CrossRef]
- Padilla-Benavides, T.; Roldan, M.L.; Larre, I.; Flores-Benitez, D.; Villegas-Sepulveda, N.; Contreras, R.G.; Cereijido, M.; Shoshani, L. The polarized distribution of Na+, K+-ATPase: Role of the interaction between beta subunits. Mol. Biol. Cell 2010, 21, 2217–2225. [Google Scholar] [CrossRef]
- Dunbar, L.A.; Roush, D.L.; Courtois-Coutry, N.; Muth, T.R.; Gottardi, C.J.; Rajendran, V.; Geibel, J.; Kashgarian, M.; Caplan, M.J. Sorting of ion pumps in polarized epithelial cells. Ann. N. Y. Acad. Sci. 1997, 834, 514–523. [Google Scholar] [CrossRef]
- Lian, W.N.; Wu, T.W.; Dao, R.L.; Chen, Y.J.; Lin, C.H. Deglycosylation of Na+,K+-ATPase causes the basolateral protein to undergo apical targeting in polarized hepatic cells. J. Cell Sci. 2006, 119, 11–22. [Google Scholar] [CrossRef][Green Version]
- Vagin, O.; Turdikulova, S.; Sachs, G. Recombinant addition of N-glycosylation sites to the basolateral Na+,K+-ATPase beta1 subunit results in its clustering in caveolae and apical sorting in HGT-1 cells. J. Biol. Chem. 2005, 280, 43159–43167. [Google Scholar] [CrossRef]
- Vagin, O.; Turdikulova, S.; Tokhtaeva, E. Polarized membrane distribution of potassium-dependent ion pumps in epithelial cells: Different roles of the N-glycans of their beta subunits. Cell Biochem. Biophys. 2007, 47, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Antonicek, H.; Persohn, E.; Schachner, M. Biochemical and functional characterization of a novel neuron-glia adhesion molecule that is involved in neuronal migration. J. Cell Biol. 1987, 104, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Antonicek, H.; Schachner, M. The Adhesion Molecule on Glia (AMOG) Incorporated into lipid vesicles binds to subpopulations of neurons. J. Neu. 1988, 8, 2961–2966. [Google Scholar] [CrossRef]
- Gloor, S.; Antonicek, H.; Sweadner, J.K.; Pagliusi, S.; Frank, R.; Moos, M.; Schachner, M. The adhesion molecule on glia (AMOG) is a homologue of the beta subunit of the Na+,K+-ATPase. J. Cell Biol. 1990, 110, 165–174. [Google Scholar] [CrossRef]
- Morth, J.P.; Pedersen, B.P.; Toustrup-Jensen, M.S.; Sørensen, T.L.; Petersen, J.; Andersen, J.P.; Vilsen, B.; Nissen, P. Crystal structure of the sodium–potassium pump. Nature 2007, 450, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, T.; Ogawa, H.; Cornelius, F.; Toyoshima, C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature 2009, 459, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Bab-Dinitz, E.; Albeck, S.; Peleg, Y.; Brumfeld, V.; Gottschalk, K.E.; Karlish, S.J. A C-terminal lobe of the beta subunit of Na+,K+-ATPase and H+,K+-ATPase resembles cell adhesion molecules. Biochemistry 2009, 48, 8684–8691. [Google Scholar] [CrossRef]
- Tokhtaeva, E.; Sachs, G.; Sun, H.; Dada, L.A.; Sznajder, J.I.; Vagin, O. Identification of the amino acid region involved in the intercellular interaction between the β1 subunits of Na+/K+-ATPase. J. Cell Sci. 2012, 125, 1605–1616. [Google Scholar] [CrossRef]
- Páez, O.; Martínez-Archundia, M.; Villegas-Sepúlveda, N.; Roldan, M.L.; Correa-Basurto, J.; Shoshani, L. A Model for the Homotypic Interaction between Na+,K+-ATPase β1 Subunits Reveals the Role of Extracellular Residues 221–229 in Its Ig-Like Domain. Int. J. Mol. Sci. 2019, 20, 4538. [Google Scholar] [CrossRef]
- Müller-Husmann, G.; Gloor, S.; Schachner, M. Functional characterization of β isoforms of murine Na+,K+-ATPase the adhesion molecule on glia (AMOG/β2), but not β1, promotes neurite outgrowth. J. Biol. Chem. 1993, 268, 26260–26267. [Google Scholar] [CrossRef]
- Senner, V.; Schmidtpeter, S.; Braune, S.; Puttmann, S.; Thanos, S.; Bartsch, U.; Schachner, M.; Paulus, W. AMOG/β2 and glioma invasion: Does loss of AMOG make tumour cells run amok? Neuropathol. Appl. Neurobiol. 2003, 29, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Kleene, R.; Loers, G.; Langer, J.; Frobert, Y.; Buck, F.; Schachner, M. Prion protein regulates glutamate-dependent lactate transport of astrocytes. J. Neu. 2007, 27, 12331–12340. [Google Scholar] [CrossRef] [PubMed]
- Vagin, O.; Tokhtaeva, E.; Sachs, G. The role of the beta 1 subunit of the Na,K-ATPase and its glycosylation in cell-cell adhesion. J. Biol. Chem. 2006, 281, 39573–39587. [Google Scholar] [CrossRef] [PubMed]
- Vagin, O.; Sachs, G.; Tokhtaeva, E. The roles of Na,K-ATPase beta-1 subunit in pump sorting and epithelial integrity. J. Bioenerg. Biomembr. 2007, 39, 367–372. [Google Scholar] [CrossRef]
- Heller, M.; von der Ohe, M.; Kleene, R.; Mohajeri, M.H.; Schachner, M. The immunoglobulin-superfamily molecule basigin is a binding protein for oligomannosidic carbohydrates: An anti-idiotypic approach. J. Neurochem. 2003, 84, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Yan, Y.; Huang, S.-Y. A new pairwise shape-based scoring function to consider long-range interactions for protein-protein docking. Biophys. J. 2017, 112 (Suppl. 1), 470a. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Huen, A.C.; Park, J.K.; Godsel, L.M.; Chen, X.; Bannon, L.J.; Amargo, E.V.; Hudson, T.Y.; Mongiu, A.K.; Leigh, I.M.; Kelsell, D.P.; et al. Intermediate filament–membrane attachments function synergistically with actin-dependent contacts to regulate intercellular adhesive strength. J. Cell Biol. 2002, 159, 1005–1017. [Google Scholar] [CrossRef]
- Jiang, Q.; Xie, Q.; Hu, C.; Yang, Z.; Huang, P.; Shen, H.; Schachner, M.; Zhao, W. Glioma malignancy is linked to interdependent and inverse AMOG and L1 adhesion molecule expression. BMC Cancer 2019, 19, 911. [Google Scholar] [CrossRef] [PubMed]
- Vagin, O.; Tokhtaeva, E.; Yakubov, I.; Shevchenko, E.; Sachs, G. Inverse correlation between the extent of N-glycan branching and intercellular adhesion in epithelia. Contribution of the Na,K-ATPase beta1 subunit. J. Biol. Chem. 2008, 283, 2192–2202. [Google Scholar] [CrossRef] [PubMed]
- Magyar, J.P.; Bartsch, U.; Wang, Z.Q.; Howells, N.; Aguzzi, A.; Wagner, E.F.; Schachner, M. Degeneration of neural cells in the central nervous system of mice deficient in the gene for the adhesion molecule on Glia, the beta 2 subunit of murine Na,K-ATPase. J. Cell Biol. 1994, 127, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Lobato-Álvarez, J.A.; López-Murillo, T.d.C.; Vilchis-Nestor, C.A.; Roldan-Gutierrez, M.L.R.; Paez-Gómez, O.; Shoshani, L. Epithelial Na+,K+-ATPase—A Sticky Pump; Najman, S., Ed.; Cell Biology—New Insights [Internet]; IntechOpen: London, UK, 2016; Available online: https://www.intechopen.com/chapters/49171 (accessed on 29 May 2022).
- Xie, Z. Molecular mechanisms of Na/K-ATPase-mediated signal transduction. Ann. N. Y. Acad. Sci. 2003, 986, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Scheidenhelm, D.K.; Cresswell, J.; Haipek, C.A.; Fleming, T.P.; Mercer, R.W.; Gutmann, D.H. Akt-dependent cell size regulation by the adhesion molecule on glia occurs independently of phosphatidylinositol 3-kinase and Rheb signaling. Mol. Cell Biol. 2005, 8, 3151–3162. [Google Scholar] [CrossRef]
- Litan, A.; Li, Z.; Tokhtaeva, E.; Kelly, P.; Vagin, O.; Langhans, S.A. A Functional Interaction Between Na,K-ATPase β2-Subunit/AMOG and NF2/Merlin Regulates Growth Factor Signaling in Cerebellar Granule Cells. Mol. Neurobiol. 2019, 56, 7557–7571. [Google Scholar] [CrossRef] [PubMed]
- Dankovich, T.M.; Rizzoli, S.O. The Synaptic Extracellular Matrix: Long-Lived, Stable, and Still Remarkably Dynamic. Front. Synaptic. Neurosci. 2022, 8, 854956. [Google Scholar] [CrossRef]
- Zipursky, S.L.; Grueber, W.B. The molecular basis of self-avoidance. Annu. Rev. Neurosci. 2013, 36, 547–568. [Google Scholar] [CrossRef]
- Rotoli, D.; Cejas, M.M.; Maeso, M.D.; Pérez-Rodríguez, N.D.; Morales, M.; Ávila, J.; Mobasheri, A.; Martín-Vasallo, P. The Na,K-ATPase β-Subunit Isoforms Expression in Glioblastoma Multiforme: Moonlighting Roles. Int. J. Mol. Sci. 2017, 18, 2369. [Google Scholar] [CrossRef]
- Barreto, N.; Caballero, M.; Bonfanti, A.P.; de Mato, F.C.P.; Munhoz, J.; da Rocha-E-Silva, T.A.A.; Sutti, R.; Vitorino-Araujo, J.L.; Verinaud, L.; Rapôso, C. Spider venom components decrease glioblastoma cell migration and invasion through RhoA-ROCK and Na+/K+-ATPase β2: Potential molecular entities to treat invasive brain cancer. Cancer Cell Int. 2020, 20, 576. [Google Scholar] [CrossRef]
- Li, S.; Dai, Z.; Yang, D.; Li, W.; Dai, H.; Sun, B.; Liu, X.; Xie, X.; Xu, R.; Zhao, X. Targeting β2 subunit of Na+/K+-ATPase induces glioblastoma cell apoptosis through elevation of intracellular Ca2. Am. J. Cancer Res. 2019, 9, 1293–1308. [Google Scholar] [PubMed]
- Vilchis-Nestor, C.A.; Roldán, M.L.; Leonardi, A.; Navea, J.G.; Padilla-Benavides, T.; Shoshani, L. Ouabain Enhances Cell-Cell Adhesion Mediated by β1 Subunits of the Na+,K+-ATPase in CHO Fibroblasts. Int. J. Mol. Sci. 2019, 20, 2111. [Google Scholar] [CrossRef] [PubMed]
- Contreras, R.G.; Shoshani, L.; Flores-Maldonado, C.; Lázaro, A.; Monroy, A.O.; Roldán, M.L.; Fiorentino, R.; Cereijido, M. E-Cadherin and tight junctions between epithelial cells of different animal species. Pflug. Arch. 2002, 444, 467–475. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Thornton, J.M. PROCHECK: Validation of protein structure coordinates. In Crystalography of Biological Macromolecules; Rossmann, M.G., Arnold, E., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2006; pp. 722–725. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roldán, M.L.; Ramírez-Salinas, G.L.; Martinez-Archundia, M.; Cuellar-Perez, F.; Vilchis-Nestor, C.A.; Cancino-Diaz, J.C.; Shoshani, L. The β2-Subunit (AMOG) of Human Na+, K+-ATPase Is a Homophilic Adhesion Molecule. Int. J. Mol. Sci. 2022, 23, 7753. https://doi.org/10.3390/ijms23147753
Roldán ML, Ramírez-Salinas GL, Martinez-Archundia M, Cuellar-Perez F, Vilchis-Nestor CA, Cancino-Diaz JC, Shoshani L. The β2-Subunit (AMOG) of Human Na+, K+-ATPase Is a Homophilic Adhesion Molecule. International Journal of Molecular Sciences. 2022; 23(14):7753. https://doi.org/10.3390/ijms23147753
Chicago/Turabian StyleRoldán, María Luisa, Gema Lizbeth Ramírez-Salinas, Marlet Martinez-Archundia, Francisco Cuellar-Perez, Claudia Andrea Vilchis-Nestor, Juan Carlos Cancino-Diaz, and Liora Shoshani. 2022. "The β2-Subunit (AMOG) of Human Na+, K+-ATPase Is a Homophilic Adhesion Molecule" International Journal of Molecular Sciences 23, no. 14: 7753. https://doi.org/10.3390/ijms23147753
APA StyleRoldán, M. L., Ramírez-Salinas, G. L., Martinez-Archundia, M., Cuellar-Perez, F., Vilchis-Nestor, C. A., Cancino-Diaz, J. C., & Shoshani, L. (2022). The β2-Subunit (AMOG) of Human Na+, K+-ATPase Is a Homophilic Adhesion Molecule. International Journal of Molecular Sciences, 23(14), 7753. https://doi.org/10.3390/ijms23147753