Abstract
One of the commonly accepted approaches to estimate protein–protein interactions (PPI) in aqueous solutions is the analysis of their translational diffusion. The present review article observes a phenomenological approach to analyze PPI effects via concentration dependencies of self- and collective translational diffusion coefficient for several spheroidal proteins derived from the pulsed field gradient NMR (PFG NMR) and dynamic light scattering (DLS), respectively. These proteins are rigid globular α-chymotrypsin (ChTr) and human serum albumin (HSA), and partly disordered α-casein (α-CN) and β-lactoglobulin (β-Lg). The PPI analysis enabled us to reveal the dominance of intermolecular repulsion at low ionic strength of solution (0.003–0.01 M) for all studied proteins. The increase in the ionic strength to 0.1–1.0 M leads to the screening of protein charges, resulting in the decrease of the protein electrostatic potential. The increase of the van der Waals potential for ChTr and α-CN characterizes their propensity towards unstable weak attractive interactions. The decrease of van der Waals interactions for β-Lg is probably associated with the formation of stable oligomers by this protein. The PPI, estimated with the help of interaction potential and idealized spherical molecular geometry, are in good agreement with experimental data.
1. Introduction
Diffusion is one of the fundamental physical phenomena characterizing functional properties of molecules and their interaction with environment [,,,,,,]. Molecular diffusion is the inevitable component of specific recognition of cell community []. Translational diffusion is the main way of molecular transport in organisms that defines numerous vital activities of the living systems. Previously, the knowledge of protein diffusion was mainly utilized for estimation of the protein hydrodynamic dimensions under different conditions or for evaluation of molecule association. In such systems, the diffusional process is well described by the classical Stokes–Einstein model [,,,]:
where D is the diffusion coefficient, kB is the Boltzmann constant, T is the temperature, η is the solution dynamic viscosity, and Rh is the hydrodynamic radius of a particle approximated as a sphere.
Although the Stokes–Einstein relation was designed for rigid spheres in isotropic medium, it is often used to estimate the size of complicated biological molecules. Unfortunately, frequently, the deviation from the strict spherical shape is substantial, and the use of Stokes–Einstein relation can lead to large inaccuracies and/or misleading conclusions. Additional parameters were introduced to take into account the deviation from the protein spherical shape, i.e., ellipsoid, polymer, rod, or disk [,,,]. However, the related empirical formulas work adequately only in very limited cases. For objects of unknown shape, the size obtained from the Stokes–Einstein equation should be considered as the effective hydrodynamic radius Rh (Figure 1).
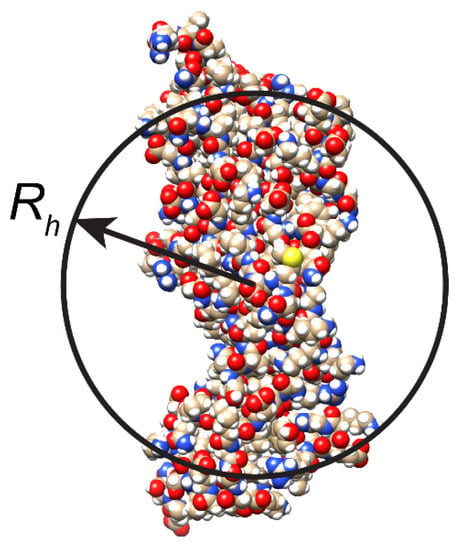
Figure 1.
Schematic representation of effective hydrodynamic radius Rh of non-spherical protein molecule.
In real living systems, translational diffusion of macromolecules significantly deviates from the classical representation of the diffusion in diluted aqueous solutions [,,,,]. The internal environment of a living cell is densely crowded with macromolecules, which create steric barriers for diffusing particles. Living systems contain various types of biological macromolecules, such as DNA, RNA, proteins, and polysaccharides, all engaged in a multitude of specific and non-specific intermolecular interactions []. Thermodynamic heterogeneity and many other factors that exist in the cell affect protein diffusion, providing ambiguous diffusion coefficients [,]. In fact, due to the existence of such macromolecular obstacles (or other cellular components) that represent the so-called “cell” effect, the diffusion coefficient of molecules can exhibit anomalous behavior [,]. These deviations from the classical view (diluted solutions) cause the limitations of common theoretical and experimental approaches, which are mainly used to study protein dynamics [,]. There were many attempts to characterize non-specific intermolecular interactions in terms of excluded volume and restricted motion of studied molecules [,,,,,]. However, the experimental diffusion data show significant deviation of protein diffusion under crowding conditions from the phenomenological predictions [,].
In concentrated/crowded solutions, the non-linear behavior of the diffusion coefficient under the conditions of increased protein concentration was shown [,,]. Such a trend indicates the presence of a significant deviation of protein diffusion from the Stokes–Einstein behavior in concentrated/crowded systems. It was suggested that translational diffusion of macromolecules in crowded environment differs significantly from dilute solution due to the huge number of intermolecular contacts [,,,]. The physical consequence of macromolecular crowding declares itself mainly in the hard-core repulsions and the so-called “soft” interactions []. The hard-core repulsion represents a steric effect arising from the impenetrable nature of atoms, which reduces the available free volume for their diffusive motion. The “soft” interactions include hydrogen bonding, charge–charge, solute–protein, van der Waals, and hydrophobic interactions []. Of these, only the strong electrostatic repulsion of similarly charged molecules prevents their convergence. Other “soft” interactions are attractive and destabilizing, because they favor the expanded conformations that allow the access to attractive surfaces. The effect of intracellular environment modulating protein–protein interactions (PPI) is important because the totality of weak interactions in the cells forms the crowded cellular interior [,,]. The simplest systems for modeling the intermolecular interactions of proteins in cells are solutions with one type of macromolecules at different concentrations []. Therefore, one of the well-known ways for estimation of the intermolecular interactions of proteins in aqueous solutions is the analysis of their translational diffusion in a wide concentration range [,].
Earlier, we have shown that the PPI estimation can be based on comparative analysis of self- and collective translational diffusion [,]. The technique of pulsed field gradient nuclear magnetic resonance (PFG NMR) operates on the experimental time scales exceeding those of the intermolecular collisions. The long-time self-diffusion coefficient Ds is observed as the averaged result of protein diffusivity over a long observation time [,,,,,]. The value of Ds can be characterized by the Stokes–Einstein equation via the coefficient of protein hydrodynamic friction f12 with solvent molecules []:
This relationship is correct only for spherical proteins in dilute homogeneous solution. The Stokes–Einstein equation was found to well-describe the variety of different systems, such as hard sphere dispersions [,], microemulsions [], micellar solutions [], and protein dispersions []. However, for solutions of charged particles in semi-diluted and concentrated solutions, the strong deviation from the Stokes–Einstein relation was experimentally observed [,]. When the protein volume fraction φ increases, the PPI growth results in an additional friction term f22(φ) in the Stokes–Einstein relation. f22(φ) is a phenomenological parameter introduced to describe the self-diffusion slowing down due to the increase in the “local viscosity”. Thus, one can write []:
Such an interpretation of the Stokes–Einstein relation successfully describes the behavior of proteins self-diffusion coefficient in semi-dilute and concentrated solutions [,,].
In dilute solutions, the molecules move independently of each other, whereas in the semi-dilute solutions, the intermolecular interactions result in appearance of new class of motion-collective modes. It is described by the Fick law [,]:
where Dc is the collective diffusion coefficient, j is the diffusion flux vector, ρ is the instantaneous number density (number of molecules per unit volume) at position r and time t.
The collective diffusion coefficient Dc takes into account the local small-displacement mobility of a tracer particle in the medium at equilibrium []. It depends on the microscopic fluctuations in the local concentration of particles and the corresponding local inhomogeneity in the refractive index of medium []. The technique of dynamic light scattering (DLS) is sensitive to the local fluctuations of particle concentration and provides the means for measurements of the short time collective diffusion coefficient Dc.
For dilute systems, where inter-molecular interactions and resulting deviations from the average density are absent, the diffusion coefficient is a constant. In the case of dilute solutions, the self- and collective diffusion coefficients are identical Dc = Ds = D0, where the subscript “0” indicates that interactions between diffusing species are absent. In practice, in most of systems, the concentration of diffusing molecules is noticeable, and the inter-molecular interactions affect the translational diffusion []. At the intermediate concentrations, Dc and Ds differ from D0 and strongly depend on the inter-molecular interactions (Figure 2) [,]. It was shown [,,] that the collective diffusion coefficient Dc tends to increase with the growth of the repulsive interactions between molecules and decrease with the prevalence of attractive ones (Figure 2).
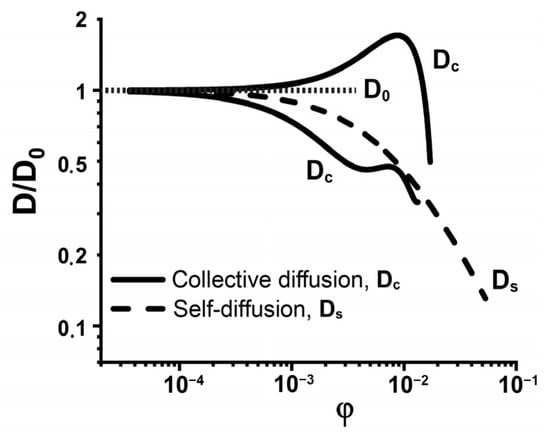
Figure 2.
Schematic representation of protein concentration dependence of collective (Dc) and self-diffusion (Ds) coefficients, Dc in the cases of the repulsive and attractive inter-molecular interactions, D0 is diffusion coefficient in dilute solution. The original data were obtained from [].
The concentration dependencies of the protein self- and collective diffusion coefficients contain information about the contributions from various intermolecular interactions [,,]. Weak PPI are commonly characterized in terms of virial coefficients [,,,] and the friction formalism [,], providing the linkage between solvent- and solute-mediated interactions. To date, the studies of PPI in dilute solutions are limited by the second (paired) virial coefficient A2 [,], which is inter-related to the paired interaction potential determined by the Deryaguin–Landau–Verwey–Overbeek (DLVO) theory [,]. The DLVO theory was applied to describe interactions between biological colloids, such as cells, vesicles, and micelles [,]. Recently, we have proposed a complex approach to study the interactions of protein molecules via the analysis of their diffusive mobility over a wide concentration range []. The next subsection overviews the main points of the Vink theory, which provides an explicit expression for the self- and collective diffusion coefficients in terms of the basic principles of non-equilibrium thermodynamics. The PPI were estimated using the interaction potentials in the frame of DLVO theory and idealized molecular geometry. We have shown that they are in good agreement with the experimental data, thereby indicating the adequacy of this approach for modeling protein interactions in dilute and semi-dilute solutions [,,].
In the present article, PPI were estimated using the examples of spheroidal proteins with various structural organization. They were two rigid globular proteins, α-chymotrypsin (ChTr) and human serum albumin (HSA), with spherical and ellipsoidal form, respectively; another two were spheroidal proteins α-casein (α-CN) and β-lactoglobulin (β-Lg) containing disordered fragments in their structure (see Figure 3). At low solution salinity, the dominance of electrostatic repulsion was shown for all studied proteins. However, α-CN and β-Lg exhibited the significant impact of van der Waals attraction in the total PPI potential, which was related to the tendency of these proteins to form associates. The increase of solution ionic strength resulted in the strong screening of the protein charges leading to decrease of electrostatic inter-protein repulsion for all proteins. The increase in the van der Waals attraction observed for the α-CN and ChTr was responsible for the ability of these proteins to form the short-living protein oligomers. At the same time, the increase in the ionic strength in the β-Lg solution caused the formation of stable oligomers, leading to the decrease in non-specific interactions.
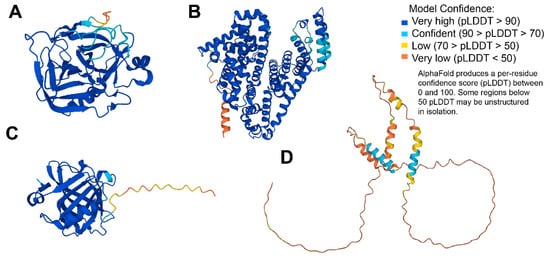
Figure 3.
3D structures modeled for the discussed proteins by AlphaFold []: (A) α–chymotrypsin (UniProt ID: P00766); (B) human serum albumin (UniProt ID: P02768); (C) β-lactoglobulin (UniProt ID: P02754); (D) α-casein (UniProt ID: P02662).
2. Existing Theoretical Descriptions of Protein Translational Diffusion
The study of protein translational diffusion provides the unique way to reveal the intricacies of their inter-molecular interactions. The theory used for interpretation of experimental data is a key step for extracting such information. In the theoretical descriptions of the diffusion process, one can distinguish four levels:
1. Purely phenomenological models with accent on (a) hydrodynamics; (b) free volume theory; (c) effect of steric hindrances, etc. []. Such theories are constructed in an ad hoc manner for description of particular experiments. They usually stress only one, supposedly dominant type of particle interaction and neglect the others. The motivation of such approaches is mainly in the agreement of the corresponding fits with the experimental data rather than in the logical self-consistency of physical principles, which lie in their foundations. This highly superficial level of description was totally exhausted in ideas by the 2000s [].
2. Semi-phenomenological approach based on the standard Stokes–Einstein formalism. It links the particle self-diffusion coefficient Ds with the solute-solvent friction coefficient f12 (Equation (2)). This friction coefficient, in turn, is the function of solution viscosity η as well as the particle size and shape []. In this approach, the random collisions of Brownian particle with solvent molecules define diffusive character of the motion (i.e., the surrounding liquid medium provokes random particle displacements). In this approach, the friction coefficient f12 is a semi-phenomenological function of solution viscosity η and protein size. The dependence of η on molar concentration of the solute C is usually taken as a relationship []:
where ηs is viscosity of pure solvent, [η] is the so-called intrinsic protein viscosity, and kH is the phenomenological parameter known as the Huggins coefficient (named after Maurice L. Huggins (1897–1981)), which is an indicator of the strength of a solvent that typically ranges from about 0.3 (for strong solvents) to 0.5 (for poor solvents).
3. Purely hydrodynamic models based on the stringent solution of corresponding Navier–Stokes equations for hard spheres [], rod-like particles [,], etc. The case of hard spheres seems to be pertinent for spheroidal globular proteins. In this case, the self-diffusion coefficient Ds is obtained as []:
where φ0 ≈ 0.5718 is the critical concentration of dense packing for hard spheres,
The hard-sphere model yields quite reliable results (see, e.g., []). However, its applicability is extremely limited by spheroidal shape of particles and does not take into account various intermolecular interaction. Additional approaches are necessary for taking into account the effects of solute-solute and solute-solvent interactions on protein translational diffusion [].
4. Finally, there is an approach proposed by Hans Vink []. It is based on the frictional formalism of non-equilibrium thermodynamics and provides a fundamental level of description for molecular diffusion. By now, it has become a well-established formalism for both self- and collective (or otherwise mutual) diffusion of various particles. The profound physical principles of non-equilibrium thermodynamics go back to the famous reciprocal relations for kinetic coefficients discovered by Lars Onsager (1903–1976) (see, e.g., []). On the one hand, the Vink’s approach makes use of the phenomenological solute–solute and solvent–solute hydrodynamic friction coefficients, and, on the other hand, it relates collective diffusion coefficient Ds and solute virial coefficients, which include interaction potentials. As a result, the frictional formalism deals with the phenomenological coefficients, and their origin can be justified and revealed in molecular theories of more profound character. A notable characteristic of Vink’s approach is a clear-cut distinction between the self- and collective diffusion. The self-diffusion coefficient Ds refers to the motion of a single particle in solution and depends both on the particle-particle and particle-solvent friction coefficients. As a result, Ds characterizes the movement of solute particles relative to each other. In contrast, collective diffusion coefficient Dc characterizes the flow of solvent molecules relatively to solute particles and, as a result, depends only on the particle-solvent friction coefficient. Therefore, the collective diffusion describes the movement of solute molecules past the solvent ones (the molecules of another type) while the self-diffusion describes the movement of solute molecules past themselves (i.e., actually a displacement of single molecule).
The Vink theory was successfully applied to several systems, such as non-associative fluorinated amphiphile [], water solutions of non-ionic surfactants [], charged block copolymers [], wormlike micelles of non-ionic surfactants [], amylopectin (homopolymer of D-glucose) [], polysterene [], rod-like polymers [] in different solvents, and some other systems [,,]. The dependence of Ds on the solute concentration was measured and interpreted within the framework of Vink theory for a number of globular proteins, including β-lactoglobulin [], hemoglobin [], serum albumin [], ovalbumin [,], and lysozyme []. Additionally, it was applied to monoclonal antibodies [,], α-chymotrypsinogen [], and proteins of various shape and size, such as chymotrypsin, α-casein, and fibrinogen [,,].
For the self-diffusion coefficient of particle Ds, the Vink theory yields:
where f22 and f12 are referred to the solute–solute and solvent–solute molar hydrodynamic friction coefficients, respectively, and c and c1 are the solute and solvent molar concentrations, respectively. For the normalized Ds, one has
where D0 is the protein diffusion coefficient at infinite dilution.
Partial volume of the solvent molecule is denoted as υ1, and that of the solute as υ2. Then, the volume fraction of solute is φ = cυ2 and the analogous expression can be written for the solvent. The sum of solvent and solute volume fractions equals to 1:
If we denote:
Equation (10) can be rewritten as:
On the other hand, for the collective diffusion coefficient Dc, Vink’s formalism yields:
where
Here A2, A3, … are the second, third, etc., solute virial coefficients, respectively, in molar concentration units. They characterize the solute-solute (protein–protein, in our case) interactions. The second virial coefficient A2 is a valuation of pairwise interactions. However, if the solute concentration increases, there is inevitable need to introduce the multi-particle interactions, which are characterized by the higher-order virial coefficients.
In the course of our investigation attempts, we tried the hydrodynamic model of Michio Tokuyama and Irwin Oppenheim (type 3) in [], the semi-phenomenological approach (type2) [], and the Vink theory (type 4) [,]. In our opinion, the Vink’s approach gives the most profound and fundamental microscopic level of the description of diffusion coefficients for proteins with both regular (globular and cylindrical) or partially disordered structure.
3. Paired PPI Potential
The Vink’s formalism relates Dc to the second virial coefficient A2, which is one of the most important PPI characteristics. Its value is determined by the combined action of various inter-molecule interactions, manifesting themselves in the potential of mean force W. The William G. McMillan–Joseph E. Mayer solution theory provides the relationship between A2 and W []:
where is the activity of pure solvent, is the hard-sphere contribution to A2, d is the diameter of protein molecule, and r is the radial coordinate. The lower limit of integration in (12) is chosen as d + 3Å to take into account a layer of water bound to the protein [].
For protein molecules in solution, the pair interaction potential, W(r), is usually modeled within the framework of classical DLVO theory of the colloid suspension stability [,]. According to the DLVO theory, the total interaction potential is mainly determined by the sum of a long-ranged Coulomb potential and the van der Waals interactions [,]:
where Wel(r)—electrostatic interaction potential, WvdW(r)—van der Waals interaction potential.
In our previous studies, several models of colloidal particles have been successfully applied for estimation of the interaction potential of proteins with different mass, shape, and structural rigidity [,]. We distinguished the model of porous colloid particle [,] as the most suitable in all cases, where protein molecules do not form associates in a rather wide concentration range []. The “porous” model represents the most complete description of protein charge shell including the surface charge distributions and the counter-ion layer Figure S1 (for calculation details see supplementary material). However, when we deal with the probable protein association, the “porous” model fails in description of experimental data []. For these cases, it is better to use the Yukawa electrostatic potential [], which considers the effective ζ-potential as a charge characteristic of a protein molecule and may be used to obtain the satisfactory interpretation of the experimental results [].
4. Self-and Collective Diffusion of Spheroidal Proteins
The concentration dependencies of the self-diffusion coefficients obtained with PFG NMR for ChTr, HSA, β-Lg, and α-CN are presented in Figure 4 [,,,]. The initial near-horizontal parts of the curves (Figure 4) characterize the region of the dilute solutions with the diffusion coefficients D0 of 15.2 · 10−10 m2/s, 9.63 · 10−11 m2/s, 8.65 · 10−11 m2/s, and 7.82 · 10−11 m2/s for ChTr, β-Lg, α-CN and has, respectively. We have estimated the protein hydrodynamic radii using the Stokes–Einstein equation (Equation (1)). These evaluations revealed that the Rh values of the diffusing particles are 1.8 nm, 3.5 nm, 2.9 nm, and 3.2 nm for ChThasHSA, β-Lg, and α-CN respectively.
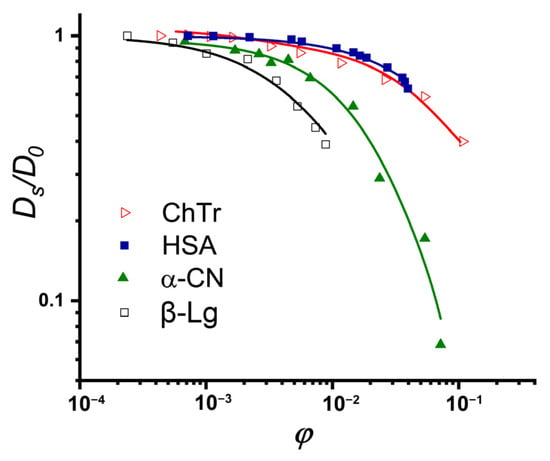
Figure 4.
Normalized concentration dependencies of self- diffusion coefficients of ChTr (pH = 3.5, I = 0.01 M), HSA (pH = 7.2, I = 0.01 M), α-CN (pH = 7.0, I = 0.01 M), and β-Lg (pH = 7.0, I = 0.003 M) The original data were obtained from [,,].
The preliminary analysis of the obtained concentration dependencies for protein self–diffusion coefficient shows that the diffusive mobilities of β-Lg and αS-CN are lower than those of the HSA and ChTr. The sharper, in comparison with the globular HSA and ChTr, concentration-dependent decrease in the α-CN self-diffusion is probably caused by the mostly disordered structure of α-casein molecule [] (see Figure 3C). It is striking that β-Lg has the most precocious decrease of the diffusive mobility against its smallest molecular weight (18 kDa), whereas the presence of protein associates has not been proven. The reason for such early decrease of β-Lg self-diffusion is probably related with the partly disordered structure of β-Lg (see Figure 3B) and the presence of significant attractive PPI of β-Lg at low protein concentration [,,].
We compared the translational diffusion coefficients obtained using the DLS and PFG NMR methods to get the information about weak intermolecular interactions. Figure 5 shows the concentration dependencies of ChTr, HSA, β-Lg, and αS-CN obtained by these two independent experimental methods, which observe different diffusion effects characterized by the self-diffusion coefficient Ds for NMR and the collective diffusion coefficient Dc for DLS. The estimation of the protein intermolecular interactions involves analysis of the translational diffusion using the methods that are sensitive to various molecular effects [].
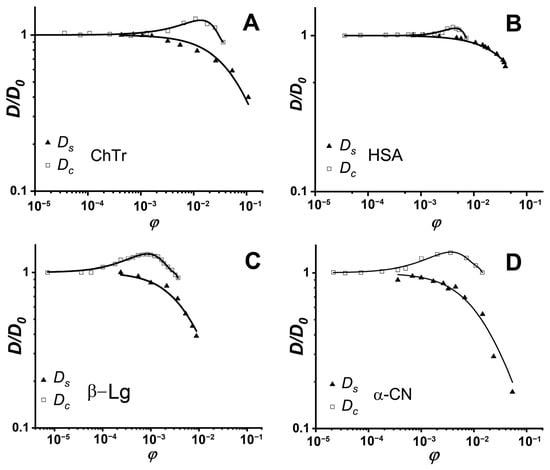
Figure 5.
Normalized concentration dependencies of protein self- (triangles) and collective (squares) diffusion coefficients: (A) ChTr (pH = 3.5, I = 0.01 M); (B) HSA (pH = 7.2, I = 0.01 M); (C) β-Lg (pH = 7.0, I = 0.003 M); (D) α-CN (pH = 7.0, I = 0.01 M). Solid lines denote fits of experimental data by the Vink’s algorithm. The original data were obtained from [,,,].
Earlier, Vink theory was successfully applied to the approximation of the experimental data on the self- and collective diffusion of proteins. It was shown that Vink theory well-described the experimental data obtained in the studied concentration range for spheroidal ChTr, HSA, β-Lg, and α-CN [,,,]. The numerical fitting of the experimental self- and collective diffusion data gives the friction and virial coefficients, respectively. According to the Vink’s approach, the hydrodynamic interactions are taken into account by introduction of the solvent–solute (f12) and the solute–solute (f22) friction coefficients. For dilute protein solutions, solvent–solute friction coefficient f12 can be determined by the Stokes–Einstein relation (Equation (2)). Using the thus retrieved f12 values and the fitting parameter ρ, the f22 values were calculated. The f22 value characterizes the influence of the direct and hydrodynamic interactions between the protein molecules on the protein self-diffusion [,]. Figure 6 shows f22 for ChTr, HSA, β-Lg, and α-CN in dilute solution (φ = 0.003). The f22 values for β-Lg and α-CN were found to be higher than those for ChTr and HSA. It can be associated with the influence of disordered fragments in β-Lg and α-CN structure, which can provide the steric PPI prior to the associates formation [,].
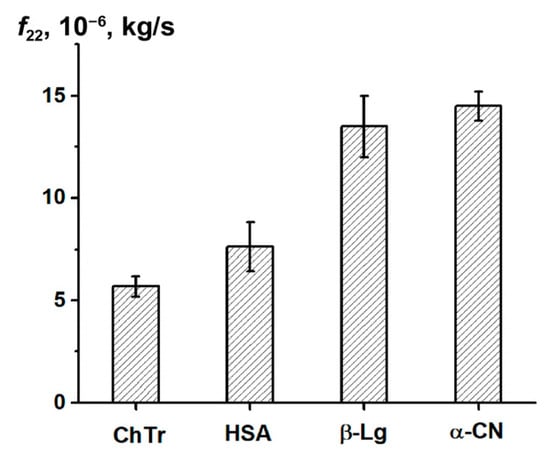
Figure 6.
Protein–protein friction coefficient f22 for diluted protein solutions.
The collective diffusion coefficient and its approximation by the Vink’s algorithm (squares on Figure 5) made it possible to obtain the sets of virial coefficients (Table S1) [,]. Intermolecular interactions of participating partners manifest themselves in the number and values of virial coefficients. An analytical relationship between the experimental and theoretical values exists only for the second virial coefficient A2. This value contains information about pairwise intermolecular interactions, which are possible in dilute solutions. In the case of the semi-diluted and concentrated solutions, A2 cannot provide all information about protein interactions, since it is necessary to take into account the influence of the many-body interactions via the higher order virial coefficients.
The second virial coefficient A2 can be obtained using different experimental methods, such as dynamic light scattering (DLS) [], static light scattering (SLS) [], gas-chromatographic elution [], and membrane osmometry [] measurements. In previous works, the A2 values for ChTr, HSA, β-Lg, and α-CN were determined independently with DLS and SLS techniques [,,,,]. Table 1 shows that the difference in A2 values of ChTr, has, and β-Lg obtained by two light-scattering methods is rather significant, which can be explained by the different protein environment in corresponding experiments. Furthermore, the difference of A2 values for α-CN by a factor of approximately 40 may be a result of the α-CN associate formation detected in the DLS experiment [,]. Therefore, the subsequent analysis of PPI for α-CN was based on the SLS data for α-CN monomers, and corresponding A2 values were obtained by Dickinson et al. []. For other proteins, the PPI estimations were based on the diffusion data from the DLS data combined with the Vink’s algorithm.

Table 1.
Second virial coefficients of proteins obtained by the light scattering [,,,,,].
5. Paired PPI Potential of Spheroidal Proteins
The second virial coefficient A2 is sensitive to the nature of “soft” PPI. The McMillan–Mayer theory (Equation (14)) is usually used for the quantifying of PPI, providing the relationship between the A2 and the total paired interaction potential W []. The effective interaction potential in the framework of the DLVO theory is represented by the attraction–repulsion balance between two molecules in solution and is determined by the contributions of electrostatic and van der Waals interactions (Equation (15)). The calculation of PPI potentials of spheroidal proteins ChTr, HSA, β-Lg, and α-CN was based on the model of spherical porous colloidal particle [,] (for calculation details, see Supporting Information). The corresponding data on the PPI potentials are presented in Figure 7. It was found that the main contribution to intermolecular interactions of all studied spheroidal proteins is made by electrostatic repulsion potential Wel(r). However, in the cases of β-Lg and α-CN, the contribution of the van der Waals interaction was more noticeable. The stronger van der Waals potentials of β-Lg and α-CN are probably associated with the propensity of these proteins to self-associate [,,,,]. The flexible disordered domains of β-Lg and α-CN can provide the attractive PPI potential during the associate formation. To create the favorable conditions for attractive interactions of proteins resulting in their association, it is necessary to reduce electrostatic repulsion. As a rule, for alteration of the electrostatic interactions, one can use the change in the ionic strength (i.e., the changes in the concentration of the free ions in solution).
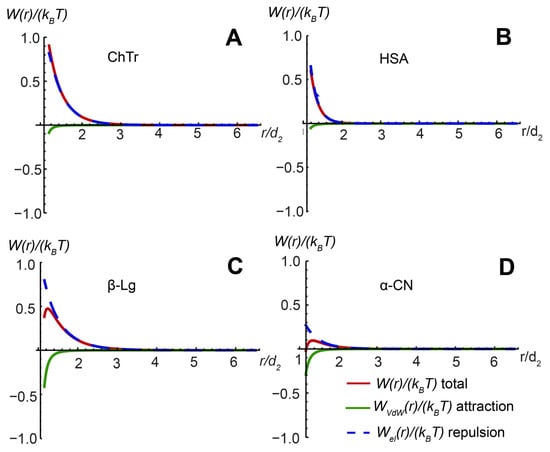
Figure 7.
Total paired PPI potential and its constituent elements for (A) ChTr (pH = 3.5, I = 0.01 M); (B) has (pH = 7.2, I = 0.01 M); (C) β-Lg (pH = 7.0, I = 0.003 M); (D) α-CN (pH = 7.0, I = 0.01 M). The original data for ChTr ahasHSA were obtained from [,].
6. Ionic Strength Influence on Repulsion–Attraction Balance in PPI
At low ionic strength (0.003 M–0.01 M), all spheroidal proteins have positive A2 values indicating the prevalence of the paired repulsive potential. The increase in the ionic strength (0.01 M–1.0 M) shows the strong charge screening reflected in the decrease in the Debye (screening) length κ−1 and negative value of A2 for ChTr, β-Lg, and α-CN (Table 2) [,,,]. It should be noted that for the rigid ChTr, a negative A2 is observed at a sufficiently high ionic strength (1.0 M). A negative value of A2 in the framework of DLVO theory characterizes the dominance of the van der Waals attractions [,,,]. Other factors affecting PPI, such as the steric ones, hydrogen bonding, and short-range hydration forces, are not included in the DLVO representation. These attractive effects can be considered as a correction to the van der Waals term by adjusting the Hamaker coefficient H []. A strong screening of protein charges leads to a significant probability of the neighboring protein molecules to stick and self-assemble, which is expressed in the increasing values of Hamaker constant (Table 2).

Table 2.
Second virial coefficient A2, Debye screening length κ−1, Hamaker constant H of ChTr, β-Lg, and α-CN at various ionic strength I values [,,,].
Finally, using the protein–protein second virial coefficient A2 at increasing ionic strength, we estimated the contributions of the electrostatic and van der Waals interactions to the total paired PPI potential WI (for calculation details, see Supplementary Information). Our results show that the increase in the salinity of the protein solutions associated with a strong screening of protein charges results in the significant decrease of the electrostatic repulsion and the dominance of the protein–protein attraction (Figure 8).
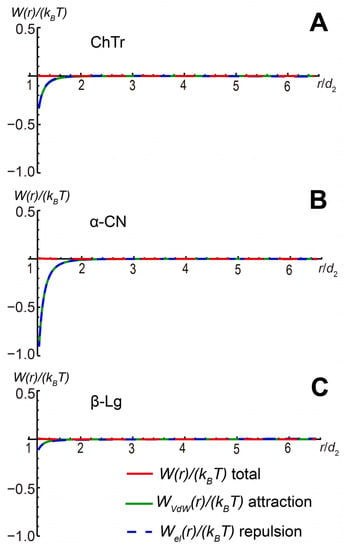
Figure 8.
Total paired PPI potential and its contributing parts for (A) ChTr (pH = 3.5, I = 1.0 M); (B) α-CN (pH = 7.0, I = 0.1 M); (C) β-Lg (pH = 7.0, I = 0.1 M).
The β-Lg and α-CN self-association is highly dependent on the ionic strength (I) of the solution [,,,,]. Furthermore, β-Lg at I = 0.1 M could form stable oligomers, leading to a decrease in the resulting weak non-specific PPI. In our opinion, the van der Waals attraction of protein molecules contributes to the further self-association of proteins. This effect is especially pronounced for α-CN. The main reason for this behavior is likely the non-electrostatic interactions between disordered fragments of its molecules. However, these intra-molecular interactions are relatively weak and unstable in solution. With a further increase in the salinity or due to other favorable factors, these attractive interactions lead to the formation of stable protein self-associates, as was observed for β-Lg [,].
7. Conclusions
PPIs have a pivotal role in biological processes in living systems, controlling and modulating the direction protein functioning, such as, for example, signal transduction, associated with various diseases, including cancer, infections, and neurodegenerative diseases [].
In the present article, we analyzed the uniform approach to study intermolecular interactions of proteins in solutions. This approach is based on the analysis of the translational diffusion data. It was applied to a set of the spheroidal proteins differing in degree of structural (dis)order. The reviewed approach carries out the inter-complementary analysis of the protein self- and collective diffusion coefficients obtained by the experimental methods of nuclear magnetic resonance with pulsed gradient of magnetic field (PFG NMR) and spectroscopy of dynamic light scattering (DLS). The combination of concentration dependencies for coefficients of self- and collective diffusion with the Vink theory (phenomenological approach based on the formalism of non-equilibrium thermodynamics) enables one to obtain the sets of friction and virial coefficients for proteins studied. The second and higher virial coefficients were obtained for estimation of pair and multi-particle intermolecular interactions in solutions with low values of the ionic strength (0.003–0.01 M) for ChTr, HSA, α-CN, and β-Lg. The McMillan–Mayer theory can be used for quantitative estimation of the non-specific PPI. This theory provides the relationship between the second virial coefficient A2 and the effective potential of paired interactions W(r) within the framework of DLVO theory. In this theory, the balance of attraction-repulsion interactions between the two protein molecules in solution depends on the electrostatic and van der Waals potentials. The positive value of the second virial coefficient A2 for spheroidal ChTr, HSA, α-CN, and β-Lg at low ionic strengths (0.003–0.01 M) means the dominance of the intermolecular repulsion. The increase in ionic strength (0.1–1.0 M) led to the screening of the protein charges and, as a result, to the decrease in the electrostatic potential. The increase in the van der Waals potential for ChTr and α-CN can explain the propensity of these proteins to weak unstable attractive interactions. The decrease in the strength of the van der Waals interaction for β-Lg is probably associated with oligomers formation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23169240/s1.
Author Contributions
Conceptualization, Y.F.Z. and V.N.U.; investigation A.M.K. and A.E.S.; writing—original draft preparation, A.M.K. and A.E.S.; writing—review and editing, Y.F.Z. and V.N.U.; visualization, A.M.K. and V.N.U.; project administration, Y.F.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the government assignment for Federal Research Center Kazan Scientific Center of Russian Academy of Sciences (122011800137-0).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data in this study are available on reasonable request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bouchaud, J.-P.; Georges, A. Anomalous diffusion in disordered media: Statistical mechanisms, models and physical applications. Phys. Rep. 1990, 195, 127–293. [Google Scholar] [CrossRef]
- Metzler, R.; Klafter, J. Anomalous stochastic processes in the fractional dynamics framework: Fokker-Planck equation, dispersive transport, and non-exponential relaxation. In Advances in Chemical Physicsc; Prigogine, I., Rice, A.S., Eds.; John wiley and Sons, Inc.: Hoboken, NJ, USA, 2001; Volume 116, pp. 223–264. [Google Scholar]
- Price, W.S. Pulsed-field gradient nuclear magnetic resonance as a tool for studying translational diffusion: Part 1. Basic theory. Concepts Magn. Reson. Educ. J. 1997, 9, 299–336. [Google Scholar] [CrossRef]
- Sarhangi, S.M.; Matyushov, D.V. Driving forces of protein diffusion. J. Phys. Chem. Lett. 2020, 11, 10137–10143. [Google Scholar] [CrossRef] [PubMed]
- Karzar-Jeddi, M.; Romero-Vargas Castrillón, S. Dynamics of water monolayers confined by chemically heterogeneous surfaces: Observation of surface-induced anisotropic diffusion. J. Phys. Chem. B. 2017, 121, 9666–9675. [Google Scholar] [CrossRef] [PubMed]
- Arrio-Dupont, M.; Foucault, G.; Vacher, M.; Devaux, P.F.; Cribier, S. Translational diffusion of globular proteins in the cytoplasm of cultured muscle cells. Biophys. J. 2000, 78, 901–907. [Google Scholar] [CrossRef]
- Bashardanesh, Z.; Elf, J.; Zhang, H.; Van der Spoel, D. Rotational and translational diffusion of proteins as a function of concentration. ACS Omega 2019, 4, 20654–20664. [Google Scholar] [CrossRef]
- Halford, S.E.; Marko, J.F. How do site-specific DNA-binding proteins and their targets? Nucleic Acids Res. 2004, 32, 3040–3052. [Google Scholar] [CrossRef]
- Muramatsu, N.; Minton, A.P. Tracer diffusion of globular proteins in concentrated protein solutions. Proc. Natl. Acad. Sci. USA 1988, 85, 2984–2988. [Google Scholar] [CrossRef]
- Roosen-Runge, F.; Hennig, M.; Zhang, F.; Jacobs, R.M.J.; Sztucki, M.; Schober, H.; Seydel, T.; Schreibe, F. Protein self-diffusion in crowded solutions. Proc. Natl. Acad. Sci. USA 2011, 108, 11815–11820. [Google Scholar] [CrossRef]
- Balakrishnan, G.; Durand, D.; Nicolai, T. Particle diffusion in globular protein gels in relation to the gel structure. Biomacromolecules 2011, 12, 450–456. [Google Scholar] [CrossRef]
- de la Torre, J.; Huertas, M.L.; Carrasco, B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000, 78, 719–730. [Google Scholar] [CrossRef]
- Koenderink, G.H.; Zhang, H.; Aarts, D.G.; Lettinga, M.P.; Philipse, A.P.; Nägele, G. On the validity of Stokes–Einstein–Debye relations for rotational diffusion in colloidal suspensions. Faraday Discuss. 2003, 123, 335–354. [Google Scholar] [CrossRef] [PubMed]
- Balboa Usabiaga, F.; Xie, X.; Delgado-Buscalioni1, R.; Donev, A. The Stokes-Einstein relation at moderate Schmidt number. J. Chem. Phys. 2013, 139, 214113. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, D.; Zhang, L. Molecular dynamics study on nanoparticle diffusion in polymer melts: A test of the Stokes−Einstein law. J. Phys. Chem. C 2008, 112, 6653–6661. [Google Scholar] [CrossRef]
- Kok, C.M.; Rudin, A. Relationship between the hydrodynamic radius and the radius of gyration of a polymer in solution. Die Makromol. 1981, 2, 655–659. [Google Scholar] [CrossRef]
- Chebotareva, N.A.; Kurganov, B.I.; Livanova, N.B. Biochemical effects of molecular crowding. Biochemistry 2004, 69, 1239–1251. [Google Scholar] [CrossRef]
- Rivas, G.; Minton, A.P. Macromolecular crowding in vitro, in vivo, and in between. Trends Biochem. Sci. 2016, 41, 970–981. [Google Scholar] [CrossRef]
- Monteith, W.B.; Cohen, R.D.; Smith, A.E.; Guzman-Cisneros, E.; Pielak, G.J. Quinary structure modulates protein stability in cells. Proc. Natl. Acad. Sci. USA 2015, 112, 1739–1742. [Google Scholar] [CrossRef]
- Danielsson, J.; Mu, X.; Lang, L.; Wang, H.; Binolfi, A.; Theillet, F.-X.; Bekei, B.; Logan, D.T.; Selenko, P.; Wennerström, H.; et al. Thermodynamics of protein destabilization in live cells. Proc. Natl. Acad. Sci. USA 2015, 112, 12402–12407. [Google Scholar] [CrossRef]
- Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. What macromolecular crowding can do to a protein. Int. J. Mol. Sci. 2014, 15, 23090–23140. [Google Scholar] [CrossRef]
- Goodsell, D.S. A look inside the living cell. Am. Sci. 1992, 80, 457–465. [Google Scholar]
- Banks, D.S.; Fradin, C. Anomalous diffusion of proteins due to molecular crowding. Biophys. J. 2005, 89, 2960–2971. [Google Scholar] [CrossRef] [PubMed]
- Długosz, M.; Trylska, J. Diffusion in crowded biological environments: Applications of Brownian dynamics. BMC Biophys. 2011, 4, 3. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Q.; Cheung, M.S. A physics-based approach of coarse-graining the cytoplasm of Escherichia coli (CGCYTO). Biophys. J. 2012, 102, 2353–2361. [Google Scholar] [CrossRef]
- Ritchie, K.; Shan, X.Y.; Kondo, J.; Iwasawa, K.; Fujiwara, T.; Kusumi, A. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys. J. 2005, 88, 2266–2277. [Google Scholar] [CrossRef]
- Gabdoulline, R.R.; Wade, R.C. Brownian dynamics simulation of protein–protein diffusional encounter. Methods 1998, 14, 329–341. [Google Scholar] [CrossRef]
- Mintont, A.P. The effect of volume occupancy upon the thermodynamic activity of proteins: Some biochemical consequences. Mol. Cell. Biochem. 1983, 55, 119–140. [Google Scholar] [CrossRef]
- Kuznetsova, I.M.; Zaslavsky, B.Y.; Breydo, L.; Turoverov, K.K.; Uversky, V.N. Beyond the excluded volume effects: Mechanistic complexity of the crowded milieu. Molecules 2015, 20, 1377–1409. [Google Scholar] [CrossRef]
- Mintont, A.P.; Colclasuret, G.C.; Parkert, J.C. Model for the role of macromolecular crowding in regulation of cellular volume (swelling-activated ion transporters/exduded volume/scaled particle theory). Physiology 1992, 89, 10504–10506. [Google Scholar] [CrossRef]
- Minton, A.P. Influence of excluded volume upon macromolecular structure and associations in ‘crowded” media. Curr. Opin. Biotechnol. 1997, 8, 65–69. [Google Scholar] [CrossRef]
- Fardet, A.; Hoebler, C.; Djelveh, G.; Barry, J.L. Restricted bovine serum albumin diffusion through the protein network of pasta. J. Agric. Food Chem. 1998, 46, 4635–4641. [Google Scholar] [CrossRef][Green Version]
- Boyer, P.M.; Hsu, J.T. Experimental studies of restricted protein diffusion in an agarose matrix. AIChE J. 1992, 38, 259–272. [Google Scholar] [CrossRef]
- Bicout, D.J.; Field, M.J. Stochastic dynamics simulations of macromolecular diffusion in a model of the cytoplasm of Escherichia coli. J. Phys. Chem. 1996, 100, 2489–2497. [Google Scholar] [CrossRef]
- Zhou, X.Z. Calculation of translational friction and intrinsic viscosity. II. Application to globular proteins. Biophys. J. 1995, 69, 2298–2303. [Google Scholar] [CrossRef]
- Tokuyama, M.; Moriki, T.; Kimura, Y. Self-diffusion of biomolecules in solution. Phys. Rev. E 2011, 83, 51402. [Google Scholar] [CrossRef]
- Melnikova, D.L.; Skirda, V.D.; Nesmelova, I.V. Effect of intrinsic disorder and self-association on the translational diffusion of proteins: The case of α-casein. J. Phys. Chem. B 2017, 121, 2980–2988. [Google Scholar] [CrossRef] [PubMed]
- Nesmelova, I.V.; Skirda, V.D.; Fedotov, V.D. Generalized concentration dependence of globular protein self-diffusion coefficients in aqueous solutions. Biopolymers 2002, 63, 132–140. [Google Scholar] [CrossRef]
- Yu, I.; Mori, T.; Ando, T.; Harada, R.; Jung, J.; Sugita, Y.; Feig, M. Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. Elife 2016, 5, e19274. [Google Scholar] [CrossRef]
- Nawrocki, G.; Wang, P.H.; Yu, I.; Sugita, Y.; Feig, M. Slow-down in diffusion in crowded protein solutions correlates with transient cluster formation. J. Phys. Chem. B 2017, 121, 11072–11084. [Google Scholar] [CrossRef]
- Beck, C.; Grimaldo, M.; Roosen-Runge, F.; Braun, M.K.; Zhang, F.; Schreiber, F.; Seydel, T. Nanosecond tracer diffusion as a probe of the solution structure and molecular mobility of protein assemblies: The case of ovalbumin. J. Phys. Chem. B 2018, 122, 8343–8350. [Google Scholar] [CrossRef]
- Braun, M.K.; Grimaldo, M.; Roosen-Runge, F.; Hoffmann, I.; Czakkel, O.; Sztucki, M.; Zhang, F.; Schreiber, F.; Seydel, T. Crowding-controlled cluster size in concentrated aqueous protein solutions: Structure, self-and collective diffusion. J. Phys. Chem. Lett. 2017, 8, 2590–2596. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.; Li, C.; Pielak, G.J. Soft interactions and crowding. Biophys. Rev. 2013, 5, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Qin, L.Y.; Gong, Z.; Qin, S.; Zhou, H.X.; Tang, C. Preferential interactions of a crowder protein with the specific binding site of a native protein complex. J. Phys. Chem. Lett. 2022, 13, 792–800. [Google Scholar] [CrossRef] [PubMed]
- McConkey, E.H. Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc. Natl. Acad. Sci. USA 1982, 79, 3236–3240. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.D.; Pielak, G.J. A cell is more than the sum of its (dilute) parts: A brief history of quinary structure. Protein Sci. 2017, 26, 403–413. [Google Scholar] [CrossRef]
- Pareek, V.; Tian, H.; Winograd, N.; Benkovic, S.J. Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science 2020, 368, 283–290. [Google Scholar] [CrossRef]
- Grobelny, S.; Erlkamp, M.; Möller, J.; Tolan, M.; Winter, R. Intermolecular interactions in highly concentrated protein solutions upon compression and the role of the solvent. J. Chem. Phys. 2014, 141, 22D506. [Google Scholar] [CrossRef]
- Blanco, M.A.; Perevozchikova, T.; Martorana, V.; Manno, M.; Roberts, C.J. Protein–protein interactions in dilute to concentrated solutions: α-Chymotrypsinogen in acidic conditions. J. Phys. Chem. B 2014, 118, 5817–5831. [Google Scholar] [CrossRef]
- Yu, M.; Silva, T.C.; van Opstal, A.; Romeijn, S.; Every, H.A.; Jiskoot, W.; Witkamp, G.-J.; Ottens, M. The investigation of protein diffusion via H-cell microfluidics. Biophys. J. 2019, 116, 595–609. [Google Scholar] [CrossRef]
- Kusova, A.M.; Sitnitsky, A.E.; Faizullin, D.A.; Zuev, Y.F. Protein translational diffusion and intermolecular interactions of globular and intrinsically unstructured proteins. J. Phys. Chem. A 2019, 123, 10190–10196. [Google Scholar] [CrossRef]
- Kusova, A.M.; Sitnitsky, A.E.; Zuev, Y.F. Impact of intermolecular attraction and repulsion on molecular diffusion and virial coefficients of spheroidal and rod-shaped proteins. J. Mol. Liq. 2021, 323, 114927. [Google Scholar] [CrossRef]
- Price, W.S. Pulsed-field gradient nuclear magnetic resonance as a tool for studying translational diffusion: Part II. Experimental aspects. Concepts Magn. Reson. 1998, 10, 197–237. [Google Scholar] [CrossRef]
- Appell, J.; Porte, G.; Buhler, E. Self-diffusion and collective diffusion of charged colloids studied by dynamic light scattering. J. Phys. Chem. B 2005, 109, 13186–13194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Genz, U.; Klein, R. Collective diffusion of charged spheres in the presence of hydrodynamic interaction. Phys. A Stat. Mech. Its Appl. 1991, 171, 26–42. [Google Scholar] [CrossRef]
- Scalettar, B.A.; Hearst, J.E.; Klein, M.P. FRAP and FCS studies of self-diffusion and mutual diffusion in entangled DNA solutions. Macromolecules 1989, 22, 4550–4559. [Google Scholar] [CrossRef]
- Kops-Werkhoven, M.M.; Vrij, A.; Lekkerkerker, H.N.W. On the relation between diffusion, sedimentation, and friction. J. Chem. Phys. 1983, 78, 2760–2763. [Google Scholar] [CrossRef]
- Cichocki, B.; Felderhof, B.U. Long-time self-diffusion coefficient and zero-frequency viscosity of dilute suspensions of spherical Brownian particles. J. Chem. Phys. 1988, 89, 3705–3709. [Google Scholar] [CrossRef]
- Kops-Werkhoven, M.M.; Fijnaut, H.M. Dynamic behavior of silica dispersions studied near the optical matching point. J. Chem. Phys. 1982, 77, 2242–2253. [Google Scholar] [CrossRef]
- Van Blaaderen, A.; Peetermans, J.; Maret, G.; Dhont, J.K.G. Long-time self-diffusion of spherical colloidal particles measured with fluorescence recovery after photobleaching. J. Chem. Phys. 1992, 96, 4591–4603. [Google Scholar] [CrossRef]
- Cebula, D.J.; Ottewill, R.H.; Ralston, J.; Pusey, P.N. Investigations of microemulsions by light scattering and neutron scattering. J. Chem. Soc. Faraday Trans. 1 1981, 77, 2585–2612. [Google Scholar] [CrossRef]
- Chatenay, D.; Urbach, W.; Messager, R.; Langevin, D. Self-diffusion of interacting micelles: FRAPP study of micelles self-diffusion. J. Chem. Phys. 1987, 86, 2343–2351. [Google Scholar] [CrossRef]
- Licinio, P.; Delaye, M. Mutual and self-diffusion in concentrated α-crystallin protein dispersions. A dynamic light scattering study. J. Phys. 1988, 49, 975–981. [Google Scholar] [CrossRef][Green Version]
- Imhof, A.; Van Blaaderen, A.; Maret, G.; Mellema, J.; Dhont, J.K.G. A comparison between the long-time self-diffusion and low shear viscosity of concentrated dispersions of charged colloidal silica spheres. J. Chem. Phys. 1994, 100, 2170–2181. [Google Scholar] [CrossRef]
- Vink, H. Mutual diffusion and self-diffusion in the frictional formalism of non-equilibrium thermodynamics. J. Chem. Soc. Faraday Trans. 1 1985, 81, 1725–1730. [Google Scholar] [CrossRef]
- Kozer, N.; Kuttner, Y.Y.; Haran, G.; Schreiber, G. Protein-protein association in polymer solutions: From dilute to semidilute to concentrated. Biophys. J. 2007, 92, 2139–2149. [Google Scholar] [CrossRef]
- Dear, B.J.; Chowdhury, A.; Hung, J.J.; Karouta, C.A.; Ramachandran, K.; Nieto, M.P.; Wilks, L.R.; Sharma, A.; Shay, T.Y.; Cheung, J.K.; et al. Relating collective diffusion, protein–protein interactions, and viscosity of highly concentrated monoclonal antibodies through dynamic light scattering. Ind. Eng. Chem. Res. 2019, 58, 22456–22471. [Google Scholar] [CrossRef]
- Kusova, A.M.; Sitnitsky, A.E.; Zuev, Y.F. Effect of structural disorder on hydrodynamic behavior of alpha-casein according to PFG NMR spectroscopy. Appl. Magn. Reson. 2018, 49, 499–509. [Google Scholar] [CrossRef]
- Sobac, B.; Dehaeck, S.; Bouchaudy, A.; Salmon, J.B. Collective diffusion coefficient of a charged colloidal dispersion: Interferometric measurements in a drying drop. Soft Matter 2020, 16, 8213–8225. [Google Scholar] [CrossRef]
- Bowen, W.R.; Mongruel, A. Calculation of the collective diffusion coefficient of electrostatically stabilised colloidal particles. Colloids Surfaces A 1998, 138, 161–172. [Google Scholar] [CrossRef]
- Yuan, G.; Wang, X.; Han, C.C.; Wu, C. Reexamination of slow dynamics in semidilute solutions: From correlated concentration fluctuation to collective diffusion. Macromolecules 2006, 39, 3642–3647. [Google Scholar] [CrossRef]
- Dallari, F.; Jain, A.; Sikorski, M.; Möller, J.; Bean, R.; Boesenberg, U.; Frenzel, L.; Goy, C.; Hallmann, J.; Kim, Y.; et al. Microsecond hydrodynamic interactions in dense colloidal dispersions probed at the European XFEL. IUCrJ 2021, 8, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Le Bon, C.; Nicolai, T.; Kuil, M.E.; Hollander, J.G. Self-diffusion and cooperative diffusion of globular proteins in solution. J. Phys. Chem. B 1999, 103, 10294–10299. [Google Scholar] [CrossRef]
- Galantini, L.; Giampaolo, S.M.; Mannina, L.; Pavel, N.V.; Viel, S. Study of intermicellar interactions and micellar sizes in ionic micelle solutions by comparing collective diffusion and self-diffusion coefficients. J. Phys. Chem. B 2004, 108, 4799–4805. [Google Scholar] [CrossRef]
- Lahtinen, J.M.; Mašın, M.; Laurila, T.; Ala-Nissila, T.; Chvoj, Z. Many-particle diffusion in continuum: Influence of a periodic surface potential. J. Chem. Phys. 2002, 116, 7666–7672. [Google Scholar] [CrossRef]
- Scherer, T.M.; Liu, J.; Shire, S.J.; Minton, A.P. Intermolecular interactions of IgG1 monoclonal antibodies at high concentrations characterized by light scattering. J. Phys. Chem. B 2010, 114, 12948–12957. [Google Scholar] [CrossRef]
- Raut, A.S.; Kalonia, D.S. Opalescence in monoclonal antibody solutions and its correlation with intermolecular interactions in dilute and concentrated solutions. J. Pharm. Sci. 2015, 104, 1263–1274. [Google Scholar] [CrossRef]
- Otting, G.; Wüthrich, K. Heteronuclear filters in two-dimensional [1H, 1H]-NMR spectroscopy: Combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys. 1990, 23, 39–96. [Google Scholar] [CrossRef]
- Coen, C.J.; Blanch, H.W.; Prausnitz, J.M. Salting out of aqueous proteins: Phase equilibria and intermolecular potentials. AIChE J. 1995, 41, 996–1004. [Google Scholar] [CrossRef]
- Haynes, C.A.; Tamura, K.; Korfer, H.R.; Blanch, H.W.; Prausnitz, J.M. Thermodynamic properties of aqueous. alpha.-chymotrypsin solution from membrane osmometry measurements. J. Phys. Chem. 1992, 96, 905–912. [Google Scholar] [CrossRef]
- Vilker, V.L.; Colton, C.K.; Smith, K.A. The osmotic pressure of concentrated protein solutions: Effect of concentration and pH in saline solutions of bovine serum albumin. J. Colloid Interface Sci. 1981, 79, 548–566. [Google Scholar] [CrossRef]
- Jia, D.; Muthukumar, M. Effect of salt on the ordinary–extraordinary transition in solutions of charged macromolecules. J. Am. Chem. Soc. 2019, 141, 5886–5896. [Google Scholar] [CrossRef] [PubMed]
- Felderhof, B.U.; Deutch, J.M. Frictional properties of dilute polymer solutions. I. Rotational friction coefficient. J. Chem. Phys. 1975, 62, 2391–2397. [Google Scholar] [CrossRef]
- Tsapikouni, T.S.; Allen, S.; Missirlisa, Y.F. Measurement of interaction forces between fibrinogen coated probes and mica surface with the atomic force microscope: The pH and ionic strength effect. Biointerphases 2008, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Curtis, R.A.; Ulrich, J.; Montaser, A.; Prausnitz, J.M.; Blanch, H.W. Protein–protein interactions in concentrated electrolyte solutions. Biotechnol. Bioeng. 2002, 79, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Marra, J.; Israelachvili, J. Direct measurements of forces between phosphatidylcholine and phosphatidylethanolamine bilayers in aqueous electrolyte solutions. Biochemistry 1985, 24, 4608–4618. [Google Scholar] [CrossRef]
- McIntosh, T.J.; Magid, A.D.; Simon, S.A. Interactions between charged, uncharged, and zwitterionic bilayers containing phosphatidylglycerol. Biophys. J. 1990, 57, 1187–1197. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, a.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Masaro, L.; Zhu, X.X. Physical models of diffusion for polymer solutions, gels and solids. Prog. Polym. Sci. 1999, 24, 731–775. [Google Scholar] [CrossRef]
- Padding, J.T. Theory of Polymer Dynamics; Advanced Courses in Macroscopic Physical Chemistry; University of Cambridge: Cambridge, UK, 2005; pp. 21–28. [Google Scholar]
- Rubinstein, M.; Colby, R.H. Polymer Physics; Oxford University Press: New York, NY, USA, 2003; Volume 23, p. 259. [Google Scholar]
- Tokuyama, M.; Oppenheim, I. Dynamics of hard-sphere suspensions. Phys. Rev. E 1994, 50, R16. [Google Scholar] [CrossRef]
- Shaqfeh, E.S.G.; Fredrickson, G.H. The hydrodynamic stress in a suspension of rods. Phys. Fluids A 1990, 2, 7–24. [Google Scholar] [CrossRef]
- Dhont, J.K.G.; Briels, W.J. Viscoelasticity of suspensions of long, rigid rods. Colloids Surfaces A 2003, 213, 131–156. [Google Scholar] [CrossRef]
- Sherwood, J.D. The primary electroviscous effect in a suspension of rods. J. Fluid Mech. 1981, 111, 347–366. [Google Scholar] [CrossRef]
- Tyrrell, H.J.V.; Harris, K.R. Diffusion in Liquids: A Theoretical and Experimental Study; Butterworth-Heinemann: Oxford, UK, 2013. [Google Scholar]
- Taraban, M.B.; Yu, L.; Feng, Y.; Jouravleva, E.V.; Anisimov, M.A.; Jiang, Z.X.; Yu, Y.B. Conformational transition of a non-associative fluorinated amphiphile in aqueous solution. RSC Adv. 2014, 4, 54565–54575. [Google Scholar] [CrossRef]
- Brown, W.; Rymden, R. Static and dynamical properties of a nonionic surfactant (C12E6) in aqueous solution. J. Phys. Chem. 1987, 91, 3565–3571. [Google Scholar] [CrossRef]
- Kjøniksen, A.L.; Zhu, K.; Behrens, M.A.; Pedersen, J.S.; Nyström, B. Effects of temperature and salt concentration on the structural and dynamical features in aqueous solutions of charged triblock copolymers. J. Phys. Chem. B 2011, 115, 2125–2139. [Google Scholar] [CrossRef]
- Einaga, Y. Wormlike micelles of polyoxyethylene alkyl ethers CiEj. Polym. J. 2009, 41, 157–173. [Google Scholar] [CrossRef]
- Wassenius, H.; Löfroth, J.; Nydén, M. NMR diffusometry and dynamic light scattering studies of amylopectin: Effect of shearing and heating on the size distribution and diffusion behaviour. Starch-Stärke 2006, 58, 66–81. [Google Scholar] [CrossRef]
- Rauch, J.; Köhler, W. Collective and thermal diffusion in dilute, semidilute, and concentrated solutions of polystyrene in toluene. J. Chem. Phys. 2003, 119, 11977–11988. [Google Scholar] [CrossRef]
- Bu, Z.; Russo, P.S.; Tipton, D.L.; Negulescu, I.I. Self-diffusion of rodlike polymers in isotropic solutions. Macromolecules 1994, 27, 6871–6882. [Google Scholar] [CrossRef]
- Daivis, P.J.; Pinder, D.N.; Callaghan, P.T. Dynamic light scattering and pulsed gradient spin-echo NMR measurements of diffusion in polystyrene-poly (vinyl methyl ether)-toluene solutions. Macromolecules 1992, 25, 170–178. [Google Scholar] [CrossRef]
- Law, S.J.; Britton, M.M. Sizing of reverse micelles in microemulsions using NMR measurements of diffusion. Langmuir 2012, 28, 11699–11706. [Google Scholar] [CrossRef]
- Petsev, D.N.; Denkov, N.D. Diffusion of charged colloidal particles at low volume fraction: Theoretical model and light scattering experiments. J. Colloid Interface Sci. 1992, 149, 329–344. [Google Scholar] [CrossRef]
- Keller, K.H.; Canales, E.R.; Il Yum, S. Tracer and mutual diffusion coefficients of proteins. J. Phys. Chem. 1971, 75, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Everhart, C.H.; Johnson, C.S., Jr. The determination of tracer diffusion coefficients for proteins by means of pulsed field gradient NMR with applications to hemoglobin. J. Magn. Reson. 1982, 48, 466–474. [Google Scholar] [CrossRef]
- Coffman, J.L.; Lightfoot, E.N.; Root, T.W. Protein diffusion in porous chromatographic media studied by proton and fluorine PFG-NMR. J. Phys. Chem. B 1997, 101, 2218–2223. [Google Scholar] [CrossRef]
- Nicoud, L.; Jagielski, J.; Pfister, D.; Lazzari, S.; Massant, J.; Lattuada, M.; Morbidelli, M. Kinetics of monoclonal antibody aggregation from dilute toward concentrated conditions. J. Phys. Chem. B 2016, 120, 3267–3280. [Google Scholar] [CrossRef]
- Kusova, A.M.; Sitnitsky, A.E.; Idiyatullin, B.Z.; Bakirova, D.R.; Zuev, Y.F. The effect of shape and concentration on translational diffusion of proteins measured by PFG NMR. Appl. Magn. Reson. 2018, 49, 35–51. [Google Scholar] [CrossRef]
- Kusova, A.M.; Sitnitsky, A.E.; Zuev, Y.F. The role of pH and ionic Strength in the attraction–repulsion balance of fibrinogen interactions. Langmuir 2021, 27, 10394–10401. [Google Scholar] [CrossRef]
- Zuev, Y.F.; Litvinov, R.I.; Sitnitsky, A.E.; Idiyatullin, B.Z.; Bakirova, D.R.; Galanakis, D.K.; Zhmurov, A.; Barsegov, V.; Weisel, J.W. Conformational flexibility and self-association of fibrinogen in concentrated solutions. J. Phys. Chem. B 2017, 121, 7833–7843. [Google Scholar] [CrossRef]
- Hill, T.L. An Introduction to Statistical Thermodynamics; Dover Publications, Inc.: New York, NY, USA, 1986. [Google Scholar]
- Leckband, D.; Sivasankar, S. Forces controlling protein interactions: Theory and experiment. Colloids Surf. B 1999, 14, 83–97. [Google Scholar] [CrossRef]
- Israelachvili, J.N. Intermolecular and Surface Forces, 3rd ed.; Academic Press: Cambridge, MA, USA, 2011; pp. 291–338. [Google Scholar]
- Ohshima, H. Biophysical Chemistry of Biointerfaces; John Wiley & Sons: New York, NY, USA, 2011. [Google Scholar]
- Ohshima, H. Theory of Colloid and Interfacial Electric Phenomena; Elsevier: Sheffield, UK, 2006. [Google Scholar]
- Giera, B.; Zepeda-Ruiz, L.A.; Pascall, A.J.; Weisgraber, T.H. Mesoscale particle-based model of electrophoretic deposition. Langmuir 2017, 33, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Kusova, A.M.; Iskhakova, A.K.; Zuev, Y.F. NMR and dynamic light scattering give different diffusion information for short-living protein oligomers. Human serum albumin in water solutions of metal ions. Eur. Biophys. J. 2022, 51, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Nilsson, H.; Roos, H.; Halle, B. Protein self-association in solution: The bovine β-lactoglobulin dimer and octamer. Protein Sci. 2003, 12, 2404–2411. [Google Scholar] [CrossRef]
- Laneuville, S.I.; Turgeon, S.L.; Sanchez, C.; Paquin, P. Gelation of native β-lactoglobulin induced by electrostatic attractive interaction with xanthan gum. Langmuir 2006, 22, 7351–7357. [Google Scholar] [CrossRef] [PubMed]
- Verheul, M.; Pedersen, J.S.; Roefs, S.P.; de Kruif, K.G. Association behavior of native β-lactoglobulin. Biopolym. Orig. Res. Biomol. 1999, 49, 11–20. [Google Scholar] [CrossRef]
- Hung, J.J.; Zeno, W.F.; Chowdhury, A.A.; Dear, B.J.; Ramachandran, K.; Nieto, M.P.; Shay, T.Y.; Karouta, C.A.; Cheung, J.K.; Truskett, T.M.; et al. Self-diffusion of a highly concentrated monoclonal antibody by fluorescence correlation spectroscopy: Insight into protein–protein interactions and self-association. Soft Matter 2019, 15, 6660–6676. [Google Scholar] [CrossRef]
- Kontopidis, G.; Holt, C.; Sawyer, L. Invited review: β-lactoglobulin: Binding properties, structure, and function. J. Dairy Sci. 2004, 87, 785–796. [Google Scholar] [CrossRef]
- Schaink, H.M.; Smit, J.A.M. Determination of the osmotic second virial coefficient and the dimerization of β-lactoglobulin in aqueous solutions with added salt at the isoelectric point. Phys. Chem. Chem. Phys. 2000, 2, 1537–1541. [Google Scholar] [CrossRef]
- Cruickshank, A.J.B.; Gainey, B.W.; Hicks, C.P.; Letcher, T.M.; Moody, R.W.; Young, C.L. Gas-liquid chromatographic determination of cross-term second virial coefficients using glycerol. Benzene+ nitrogen and benzene+ carbon dioxide at 50 C. Trans. Faraday Soc. 1969, 65, 1014–1031. [Google Scholar] [CrossRef]
- Dickinson, E.; Semenova, M.G.; Antipova, A.S. Salt stability of casein emulsions. Food Hydrocoll. 1998, 12, 227–235. [Google Scholar] [CrossRef]
- Treweek, T.M. Alpha-casein as a molecular chaperone. In Milk Protein; Hurley, W.L., Ed.; InTech: Rijeka, Croatia, 2012; pp. 85–119. [Google Scholar]
- Sønderby, P.; Bukrinski, J.T.; Hebditch, M.; Peters, G.H.; Curtis, R.A.; Harris, P. Self-interaction of human serum albumin: A formulation perspective. ACS Omega 2018, 3, 16105–16117. [Google Scholar] [CrossRef] [PubMed]
- McMillan, W.G.; Mayer, J.E. The statistical thermodynamics of multicomponent systems. J. Chem. Phys. 1945, 13, 276. [Google Scholar] [CrossRef]
- Ohshima, H. The Derjaguin-Landau-Verwey-Overbeek (DLVO) Theory of Colloid Stability. In Electrical Phenomena at Interfaces and Biointerfaces: Fundamentals and Applications in Nano-, Bio-, and Environmental Sciences, 1st ed.; Ohshima, H., Ed.; Wiley Online Library: Hoboken, NJ, USA, 2012; pp. 27–34. [Google Scholar] [CrossRef]
- Hämisch, B.; Büngeler, A.; Kielar Ch Keller, A.; Strube, O.; Huber, K. Self-assembly of fibrinogen in aqueous, thrombin-free solutions of variable ionic strengths. Langmuir 2019, 35, 12113–12122. [Google Scholar] [CrossRef] [PubMed]
- Deszczynski, M.; Harding, S.E.; Winzor, D.J. Negative second virial coefficients as predictors of protein crystal growth: Evidence from sedimentation equilibrium studies that refutes the designation of those light scattering parameters as osmotic virial coefficients. Biophys. Chem. 2006, 120, 106–113. [Google Scholar] [CrossRef]
- Forsman, J.; Jönsson, B.; Woodward, C.E.; Wennerström, H. Attractive surface forces due to liquid density depression. J. Phys. Chem. B 1997, 101, 4253–4259. [Google Scholar] [CrossRef]
- Aymard, P.; Durand, D.; Nicolai, T. The effect of temperature and ionic strength on the dimerisation of β-lactoglobulin. Int. J. Biol. Macromol. 1996, 19, 213–221. [Google Scholar] [CrossRef]
- Arnaudov, L.N.; de Vries, R. Strong impact of ionic strength on the kinetics of fibrilar aggregation of bovine β-lactoglobulin. Biomacromolecules 2006, 7, 3490–3498. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhou, Q.; He, J.; Jiang, Z.; Peng, C.; Tong, R.; Shi, J. Recent advances in the development of protein–protein interactions modulators: Mechanisms and clinical trials. Signal Transduct. Target Ther 2020, 5, 213. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).