The Ethylene Response Factor ERF5 Regulates Anthocyanin Biosynthesis in ‘Zijin’ Mulberry Fruits by Interacting with MYBA and F3H Genes
Abstract
:1. Introduction
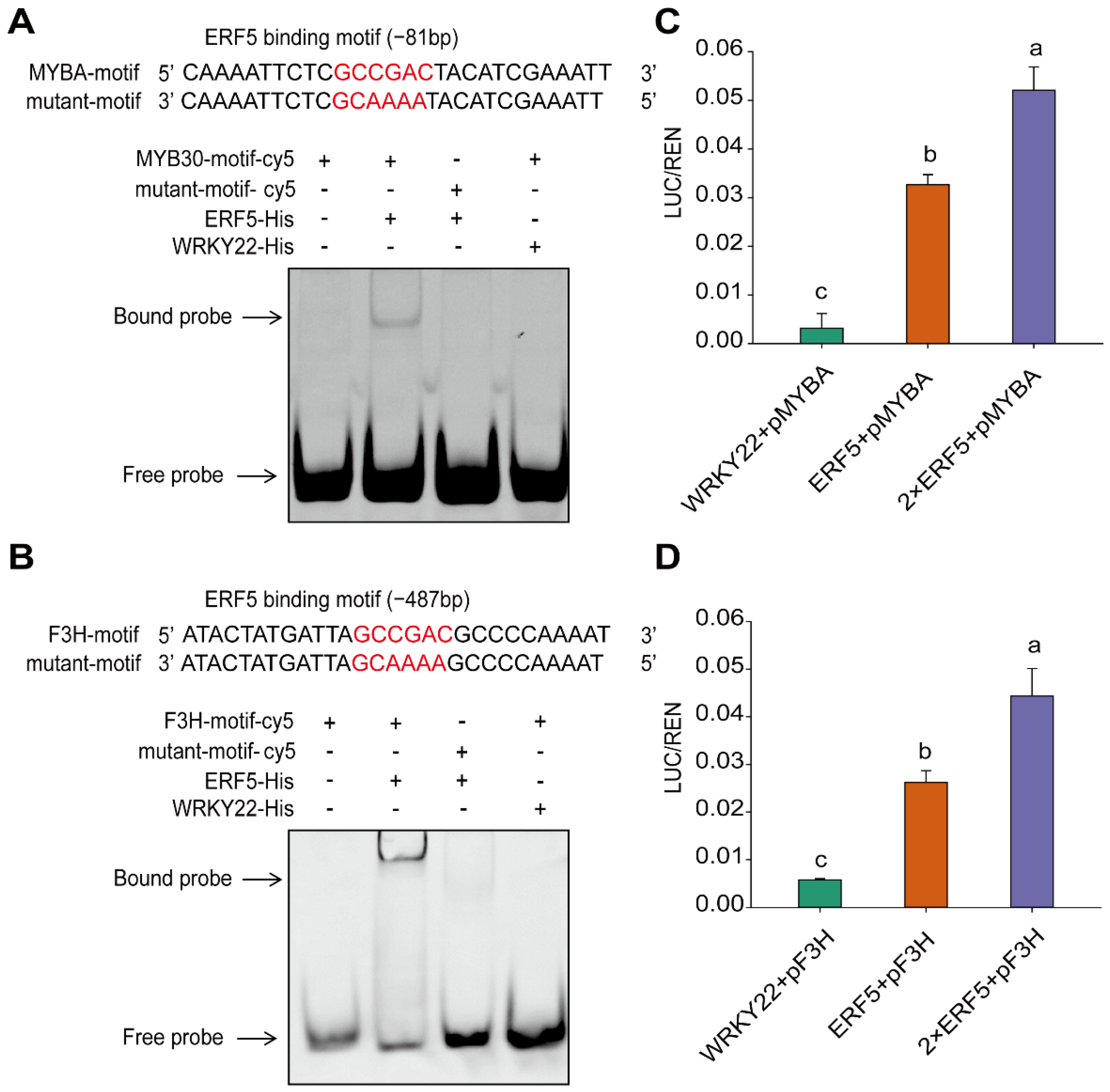
2. Results
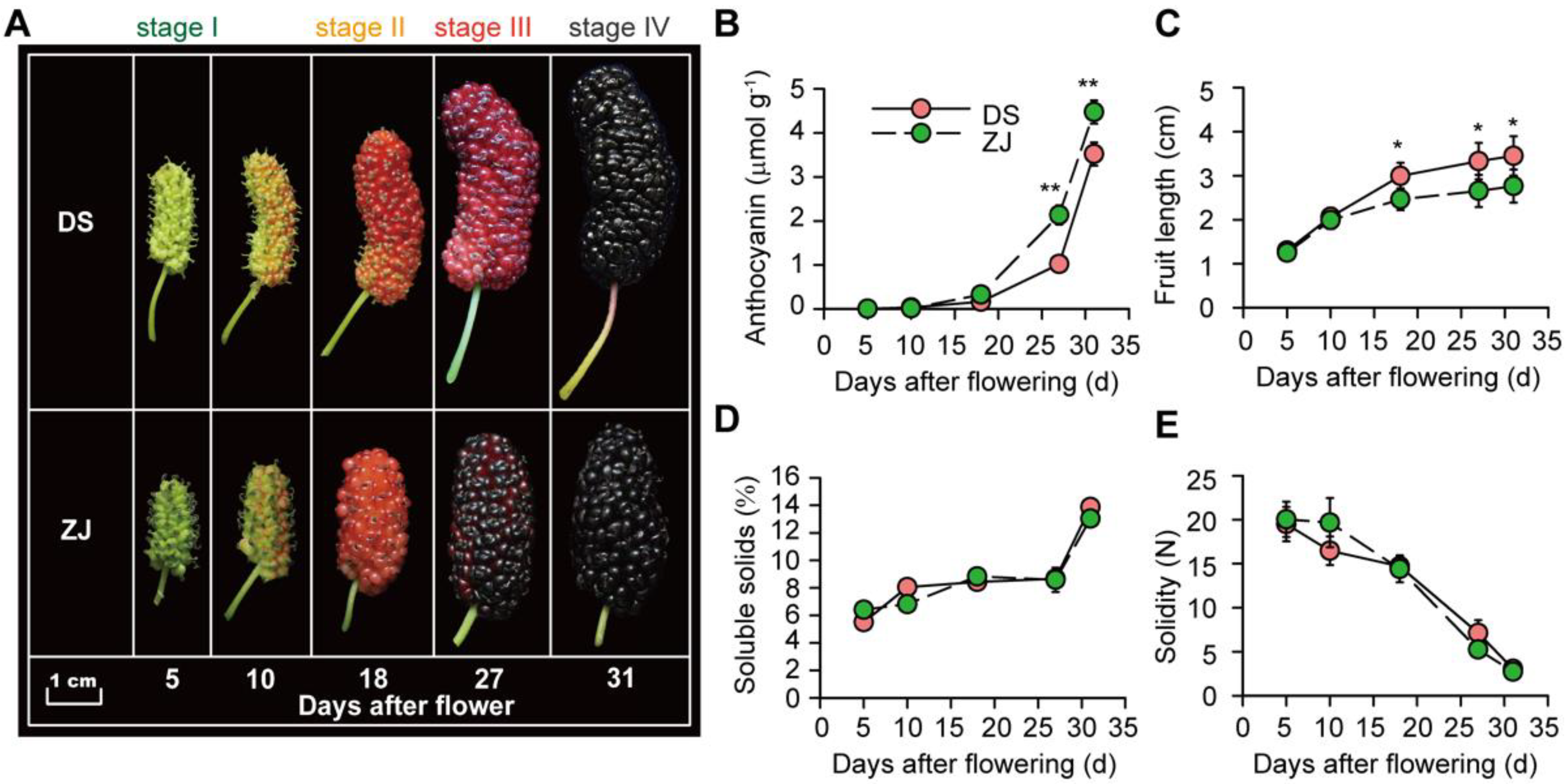
2.1. Morphological and Physiological Traits of Mulberry Fruits
2.2. Quality Control of Mulberry Transcriptome Sequencing
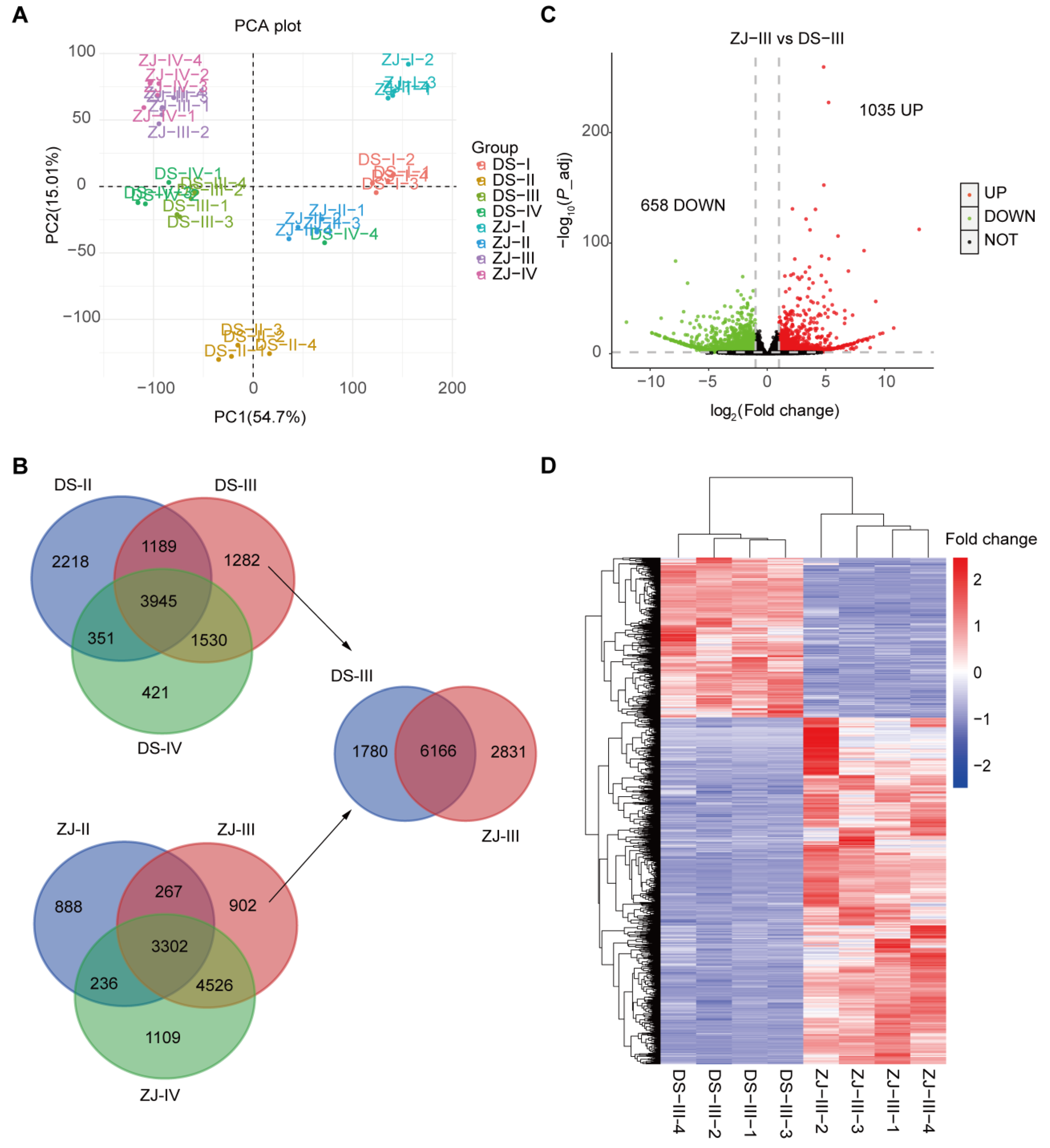
2.3. Principal Component Analysis (PCA) and DEG Profiling in Both Mulberry Genotypes
2.4. Determination of the Differential Molecular Profiling between the Two Mulberry Genotypes at Stage III
2.5. Differentially Abundant Metabolites (DAMs) in DS and ZJ Mulberry Genotypes
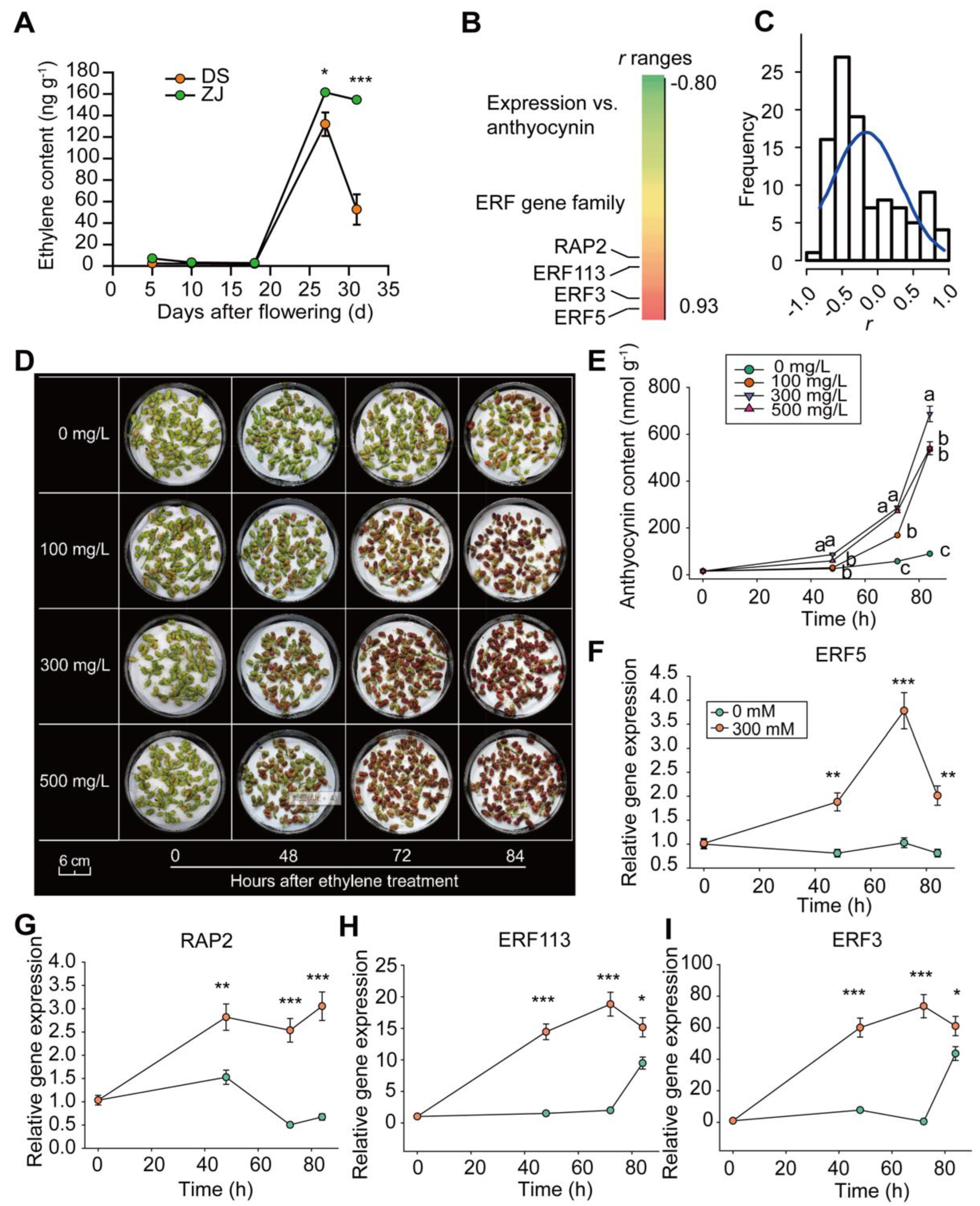
2.6. Ethylene-Induced ERF Gene Family Expression and Anthocyanin Accumulation
2.7. Anthocyanin Over-Accumulates in ZJ Genotype
3. Discussion
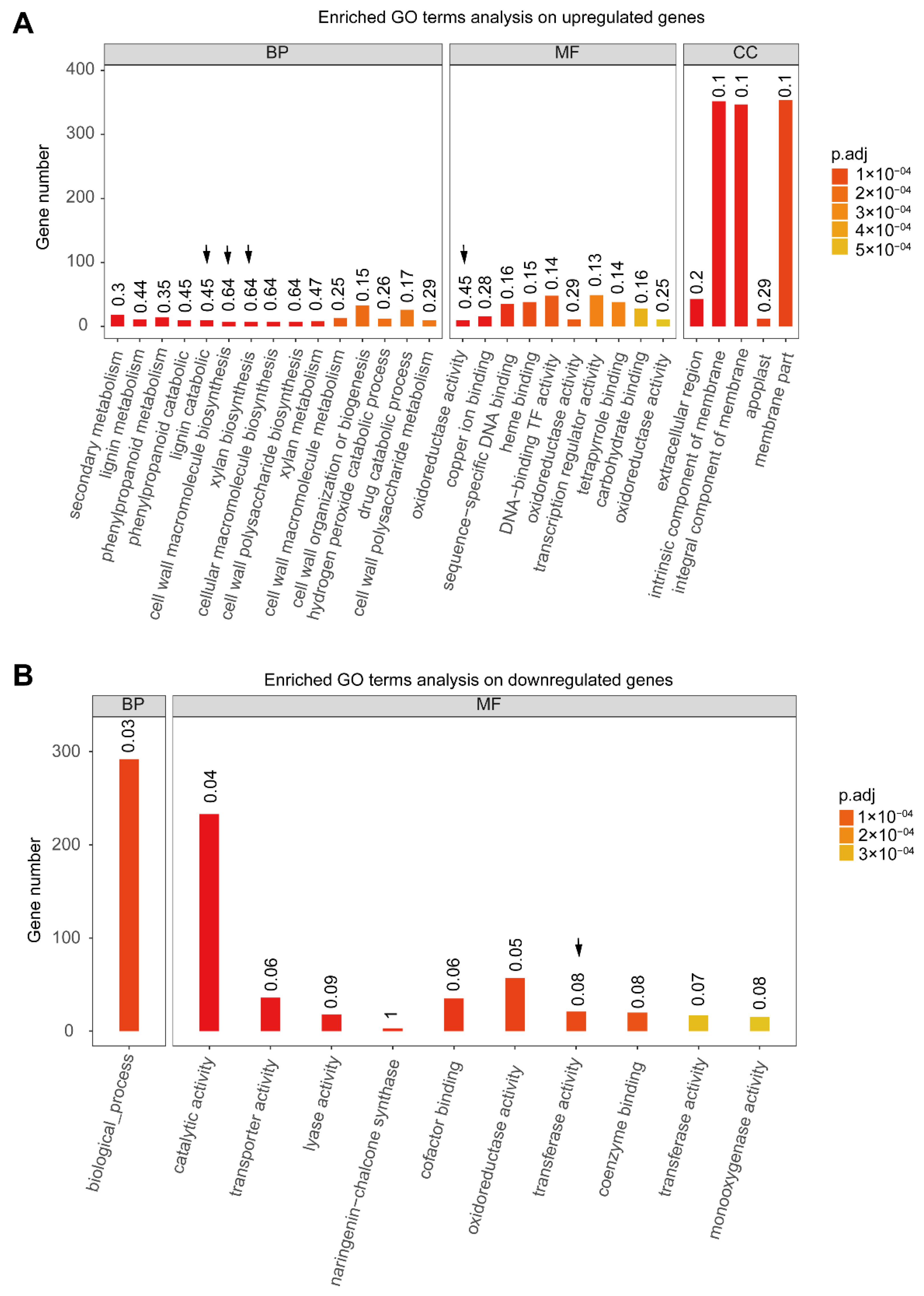
3.1. Flavonoids Biosynthesis Enrichment in Anthocyanin-Abundant Genotypes
3.2. The Metabolome Profiling Concords well with the Anthocyanin Dynamic Changes in Mulberry
3.3. Ethylene Promotes Anthocyanin Accumulation through ERF Gene Upregulation
4. Material and Methods
4.1. Plant Material and Growth Condition
4.2. Physiological Measurements
4.2.1. Determination of Anthocyanin Content
4.2.2. Measurements of Fruit Solidity and Soluble Solid Content (SSC)
4.2.3. Ethylene Determination
4.3. Transcriptome Analysis
4.3.1. RNA Extraction
4.3.2. RNA-Sequencing (RNA-Seq)
4.3.3. De Novo Transcriptome Data Processing and Assembly
4.3.4. Identification of Differentially Expressed Genes (DEGs)
4.3.5. Functional Annotation and Genes Classification
4.4. Quantitative Real-Time (q-RT-PCR) Analysis of Differentially Expressed Unigenes
4.5. LC-MS/MS Metabolism Analysis and Metabolites Identification
4.6. Electrophoresis Mobility Shift Assay (EMSA) Experiments
4.7. Luciferase Assays
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, D.; Dong, Z.; Zhang, Y.; Guo, K.; Guo, P.; Zhao, P.; Xia, Q. Proteomics provides insight into the interaction between mulberry and silkworm. J. Proteome Res. 2017, 16, 2472–2480. [Google Scholar] [CrossRef] [PubMed]
- Harauma, A.; Murayama, T.; Ikeyama, K.; Sano, H.; Arai, H.; Takano, R.; Kita, T.; Hara, S.; Kamei, K.; Yokode, M. Mulberry leaf powder prevents atherosclerosis in apolipoprotein E-deficient mice. Biochem. Biophys. Res. Commun. 2007, 358, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Tang, M.; Wu, J. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar]
- Srivastava, S.; Kapoor, R.; Thathola, A.; Srivastava, R. Mulberry (Moms alba) leaves as human food: A new dimension of sericulture. Int. J. Food Sci. Nutr. 2003, 54, 411–416. [Google Scholar] [CrossRef]
- Bao, T.; Li, Y.; Xie, J.; Jia, Z.; Chen, W. Systematic evaluation of polyphenols composition and antioxidant activity of mulberry cultivars subjected to gastrointestinal digestion and gut microbiota fermentation. J. Funct. Foods 2019, 58, 338–349. [Google Scholar] [CrossRef]
- Yang, B.; Wang, J.S.; Zhao, M.M.; Liu, Y.; Wang, W.; Jiang, Y.M. Identification of polysaccharides from pericarp tissues of litchi (Litchi chinensis Sonn.) fruitin relation to their antioxidant activities. Carbohydr. Res. 2006, 341, 634–638. [Google Scholar] [CrossRef]
- Dugo, P.; Mondello, L.; Errante, G.; Zappia, G.; Dugo, G. Identification of anthocyanins in berries by narrow-bore high-performance liquid chromatography with electrospray ionization detection. J. Agric. Food Chem. 2001, 49, 3987–3992. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Y.; Li, Z.; Liu, M. Role of ethylene response factors (ERFs) in fruit ripening. Food Qual. Saf. 2020, 4, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Li, J.; Essemine, J.; Shang, C.; Zhang, H.; Zhu, X.; Yu, J.; Chen, G.; Qu, M.; Sun, D. Combined proteomics and metabolism analysis unravels prominent roles of antioxidant system in the prevention of alfalfa (Medicago sativa L.) against salt stress. Int. J. Mol. Sci. 2020, 21, 909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, G.F.; Ming, M.L.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, M.; Chen, G.; Bunce, J.A.; Zhu, X.; Sicher, R.C. Systematic biology analysis on photosynthetic carbon metabolism of maize leaf following sudden heat shock under elevated CO2. Sci. Rep. 2018, 8, 7849. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [Green Version]
- Koyama, T.; Sato, F. The function of ethylene response factor genes in the light-induced anthocyanin production of Arabidopsis thaliana leaves. Plant Biotechnol. 2018, 35, 87–91. [Google Scholar] [CrossRef] [Green Version]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Aramwit, P.; Bang, N.; Srichana, T. The properties and stability of anthocyanins in mulberry fruits. Food Res. Int. 2010, 43, 1093–1097. [Google Scholar] [CrossRef]
- Chao, N.; Wang, R.; Hou, C.; Yu, T.; Miao, K.; Cao, F.; Fang, R.; Liu, L. Functional characterization of two chalcone isomerase (CHI) revealing their responsibility for anthocyanins accumulation in mulberry. Plant Physiol. Biochem. 2021, 161, 65–73. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.; Visser, R.G.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Lynch, J.H.; Guo, L.; Rhodes, D.; Morgan, J.A.; Dudareva, N. Completion of the cytosolic post chorismate phenylalanine biosynthetic pathway in plants. Nat. Commun. 2019, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Zeng, Y.; Wei, L.; Yao, Y.; Dai, J.; Liu, G.; Gui, Z. Comparative transcriptome analysis of mulberry reveals anthocyanin biosynthesis mechanisms in black (Morus atropurpurea Roxb.) and white (Morus alba L.) fruit genotypes. BMC Plant Biol. 2020, 20, 279. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Rogerio, A.; Kanashiro, A.; Fontanari, C.; Da Silva, E.; Lucisano-Valim, Y.; Soares, E.; Faccioli, L. Anti-inflammatory activity of quercetin and isoquercitrin in experimental murine allergic asthma. Inflamm. Res. 2007, 56, 402–408. [Google Scholar] [CrossRef] [PubMed]
- De Masi, L.; Bontempo, P.; Rigano, D.; Stiuso, P.; Carafa, V.; Nebbioso, A.; Piacente, S.; Montoro, P.; Aversano, R.; D’Amelia, V.; et al. Comparative phytochemical characterization, genetic profile, and antiproliferative activity of polyphenol-rich extracts from pigmented tubers of different Solanum Tuberosum varieties. Molecules 2020, 25, 233. [Google Scholar] [CrossRef] [Green Version]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [Green Version]
- Rabino, I.; Mancinelli, A.L. Light, temperature, and anthocyanin production. Plant Physiol. 1986, 81, 922–924. [Google Scholar] [CrossRef] [Green Version]
- Hughes, N.M.; Morley, C.B.; Smith, W.K. Coordination of anthocyanin decline and photosynthetic maturation in juvenile leaves of three deciduous tree species. New Phytol. 2007, 175, 675–685. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, A.; Zhu, P.; Li, J.; Han, L.; Wang, X.; Li, Z. Characterization and expression of genes involved in the ethylene biosynthesis and signal transduction during ripening of mulberry fruit. PLoS ONE 2015, 10, e0122081. [Google Scholar] [CrossRef] [PubMed]
- Loayza, F.E.; Masarirambi, M.T.; Brecht, J.K.; Sargent, S.A.; Sims, C.A. Physiological response of mature green tomatoes to treatment with ethylene at high temperature. Horttechnology 2020, 30, 773–780. [Google Scholar] [CrossRef]
- Li, J.; Essemine, J.; Bunce, J.A.; Shang, C.; Zhang, H.; Sun, D.; Qu, M. Roles of heat shock protein and reprogramming of photosynthetic carbon metabolism in thermotolerance under elevated CO2 in maize. Environ. Exp. Bot. 2019, 168, 103869. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 2011, 29, 644. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010, 1. [Google Scholar] [CrossRef] [Green Version]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Perveen, S.; Qu, M.; Chen, F.; Essemine, J.; Khan, N.; Lyu, M.-J.A.; Chang, T.; Song, Q.; Chen, G.-Y.; Zhu, X.-G. Overexpression of maize transcription factor mEmBP-1 increases photosynthesis, biomass, and yield in rice. J. Exp. Bot. 2020, 71, 4944–4957. [Google Scholar] [CrossRef]
- Moyle, R.L.; Carvalhais, L.C.; Pretorius, L.-S.; Nowak, E.; Subramaniam, G.; Dalton-Morgan, J.; Schenk, P.M. An optimized transient dual luciferase assay for quantifying microRNA directed repression of targeted sequences. Front. Plant Sci. 2017, 8, 1631. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, R.; Han, G.; Zhu, Z.; Essemine, J.; Dong, Z.; Li, Y.; Deng, W.; Qu, M.; Zhang, C.; Yu, C. The Ethylene Response Factor ERF5 Regulates Anthocyanin Biosynthesis in ‘Zijin’ Mulberry Fruits by Interacting with MYBA and F3H Genes. Int. J. Mol. Sci. 2022, 23, 7615. https://doi.org/10.3390/ijms23147615
Mo R, Han G, Zhu Z, Essemine J, Dong Z, Li Y, Deng W, Qu M, Zhang C, Yu C. The Ethylene Response Factor ERF5 Regulates Anthocyanin Biosynthesis in ‘Zijin’ Mulberry Fruits by Interacting with MYBA and F3H Genes. International Journal of Molecular Sciences. 2022; 23(14):7615. https://doi.org/10.3390/ijms23147615
Chicago/Turabian StyleMo, Rongli, Guangming Han, Zhixian Zhu, Jemaa Essemine, Zhaoxia Dong, Yong Li, Wen Deng, Mingnan Qu, Cheng Zhang, and Cui Yu. 2022. "The Ethylene Response Factor ERF5 Regulates Anthocyanin Biosynthesis in ‘Zijin’ Mulberry Fruits by Interacting with MYBA and F3H Genes" International Journal of Molecular Sciences 23, no. 14: 7615. https://doi.org/10.3390/ijms23147615
APA StyleMo, R., Han, G., Zhu, Z., Essemine, J., Dong, Z., Li, Y., Deng, W., Qu, M., Zhang, C., & Yu, C. (2022). The Ethylene Response Factor ERF5 Regulates Anthocyanin Biosynthesis in ‘Zijin’ Mulberry Fruits by Interacting with MYBA and F3H Genes. International Journal of Molecular Sciences, 23(14), 7615. https://doi.org/10.3390/ijms23147615





