Prediction of Time-Series Transcriptomic Gene Expression Based on Long Short-Term Memory with Empirical Mode Decomposition
Abstract
1. Introduction
2. Results and Discussions
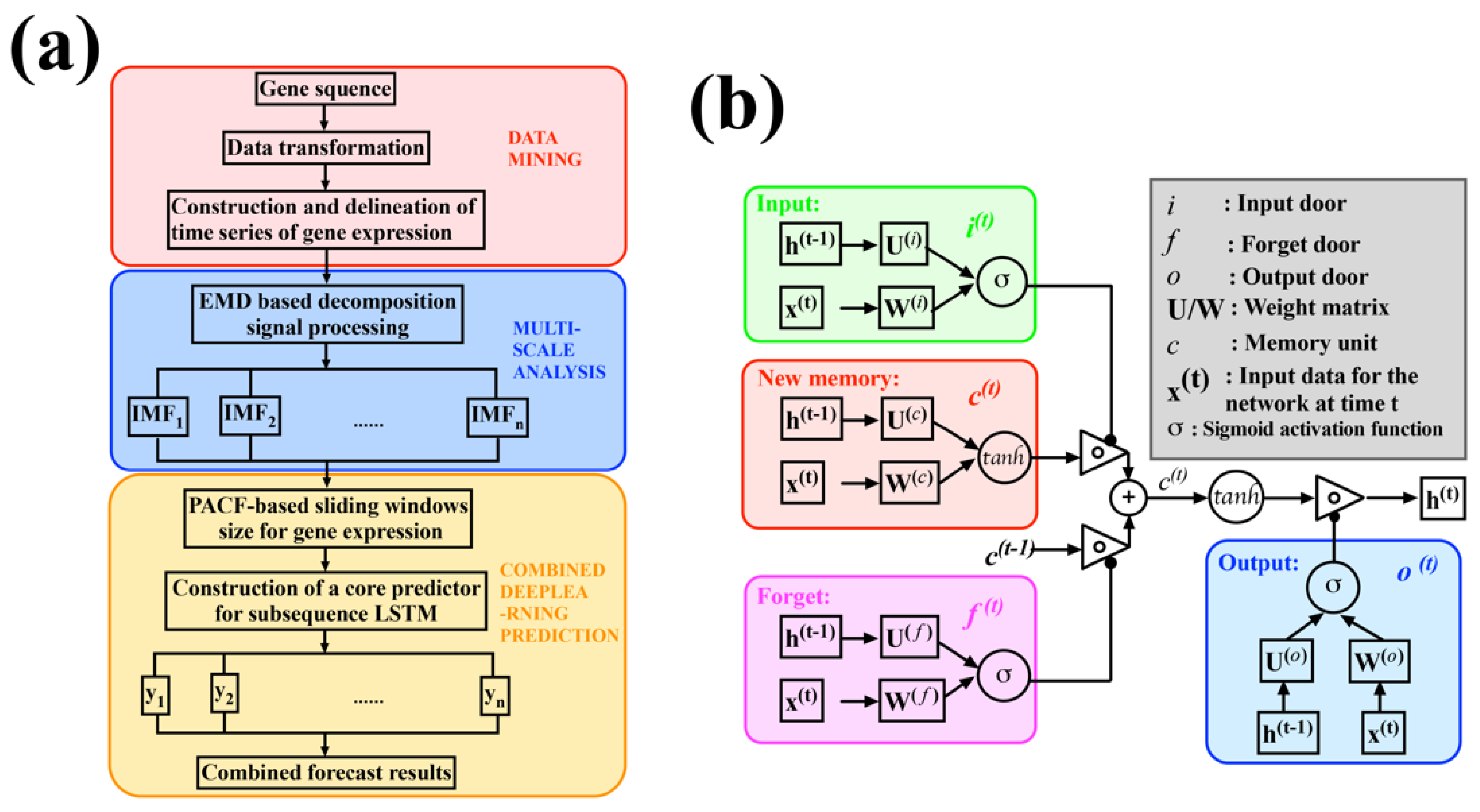
2.1. Multi-Scale Analysis Based on EMD
2.2. Construction of Core Predictor Model Using LSTM
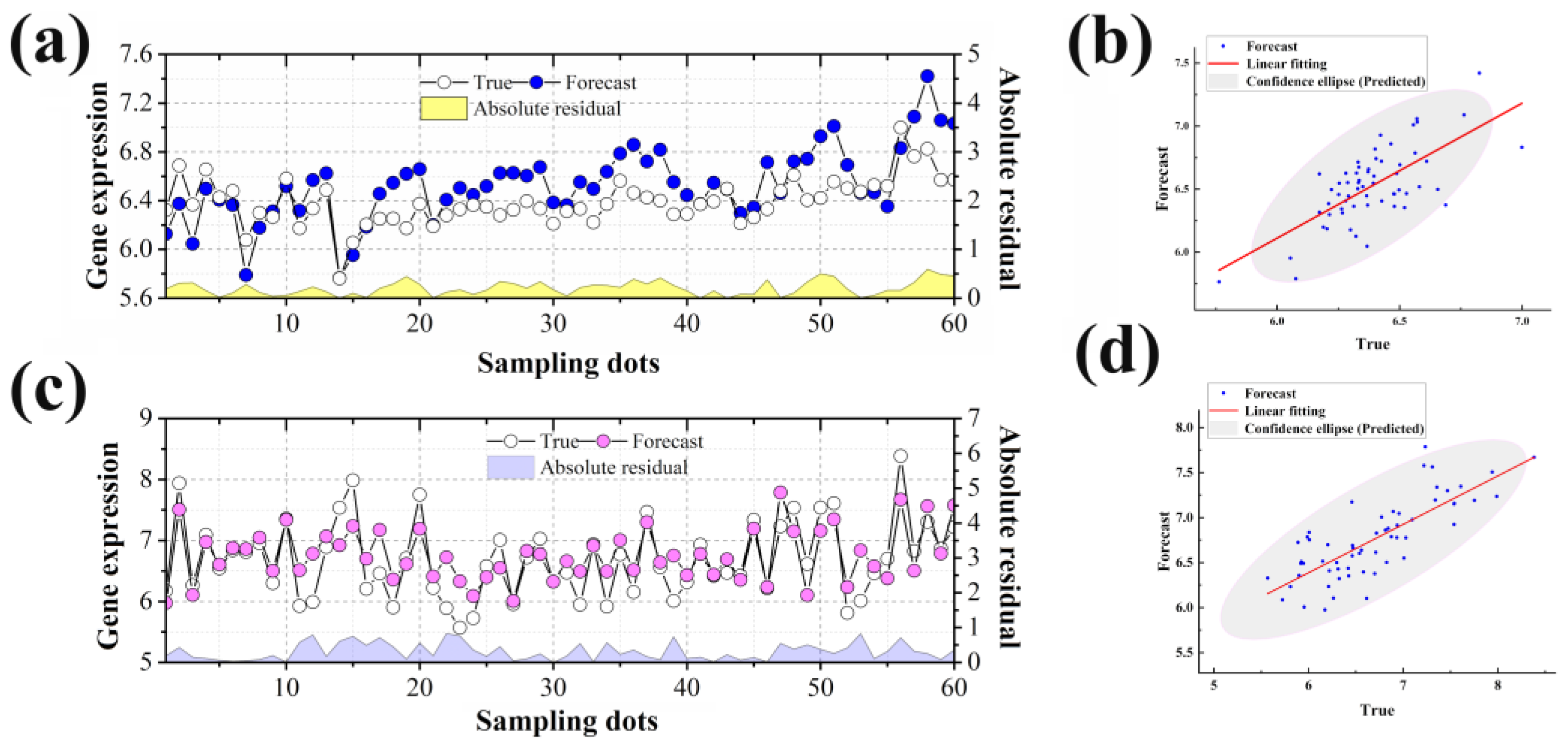
2.3. Target Time Point Prediction Based on LSTM
3. Materials and Methods
3.1. Data Acquisition and Pre-Processing
3.2. EMD-Based Decomposition Signal Processing
3.3. Construction of Core Predictors Using LSTM
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ioannidis, J.P.A. Is Molecular Profiling Ready for Use in Clinical Decision Making? Oncologist 2007, 12, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Gallego, R.I.; Pai, A.A.; Tung, J.; Gilad, Y. RNA-seq: Impact of RNA Degradation On Transcript Quantification. BMC Biol. 2014, 12, 42. [Google Scholar] [CrossRef]
- Fouda, M.A.; Elzefzafy, N.A.; Taha, I.I.; Mohemmed, O.M.; Wahab, A.A.A.; Farahat, I.G. Effect of Seasonal Variation in Ambient Temperature On RNA Quality of Breast Cancer Tissue in a Remote Biobank Setting. Exp. Mol. Pathol. 2020, 112, 104334. [Google Scholar] [CrossRef] [PubMed]
- Opitz, L.; Salinas-Riester, G.; Grade, M.; Jung, K.; Jo, P.; Emons, G.; Ghadimi, B.M.; Beißbarth, T.; Gaedcke, J. Impact of RNA Degradation On Gene Expression Profiling. BMC Med. Genom. 2010, 3, 36. [Google Scholar] [CrossRef]
- Shen, Y.; Li, R.; Tian, F.; Chen, Z.; Lu, N.; Bai, Y.; Ge, Q.; Lu, Z. Impact of RNA Integrity and Blood Sample Storage Conditions On the Gene Expression Analysis. Oncol. Targets Ther. 2018, 11, 3573–3581. [Google Scholar] [CrossRef]
- Jin, C.; Cukier, R.I. Machine Learning Can be Used to Distinguish Protein Families and Generate New Proteins Belonging to those Families. J. Chem. Phys. 2019, 151, 175102. [Google Scholar] [CrossRef]
- El-Attar, N.E.; Moustafa, B.M.; Awad, W.A. Deep Learning Model to Detect Diabetes Mellitus Based on DNA Sequence. Intell. Autom. Soft Comput. 2022, 31, 325–338. [Google Scholar] [CrossRef]
- Liang, J.; Huang, X.; Li, W.; Hu, Y. Identification and External Validation of the Hub Genes Associated with Cardiorenal Syndrome through Time-Series and Network Analyses. Aging 2022, 14, 1351–1373. [Google Scholar] [CrossRef]
- Zhou, J.; Theesfeld, C.L.; Yao, K.; Chen, K.M.; Wong, A.K.; Troyanskaya, O.G. Deep Learning Sequence-Based Ab Initio Prediction of Variant Effects On Expression and Disease Risk. Nat. Genet. 2018, 50, 1171–1179. [Google Scholar] [CrossRef]
- Karim, F.; Majumdar, S.; Darabi, H.; Chen, S. LSTM Fully Convolutional Networks for Time Series Classification. IEEE Access. 2018, 6, 1662–1669. [Google Scholar] [CrossRef]
- Lakizadeh, A.; Jalili, S.; Marashi, S. PCD-GED: Protein Complex Detection Considering PPI Dynamics Based On Time Series Gene Expression Data. J. Theor. Biol. 2015, 378, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Wise, A.; Bar-Joseph, Z. SMARTS: Reconstructing Disease Response Networks From Multiple Individuals Using Time Series Gene Expression Data. Bioinformatics 2015, 31, 1250–1257. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qian, B.; Xiao, Y.; Zheng, Z.; Zhou, M.; Zhuang, W.; Li, S.; Ma, Q. Dynamic Multi-Scale Convolutional Neural Network for Time Series Classification. IEEE Access. 2020, 8, 109732–109746. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, J.; Li, Q.; Liu, T. BiLSTM-5mC: A Bidirectional Long Short-Term Memory-Based Approach for Predicting 5-Methylcytosine Sites in Genome-Wide DNA Promoters. Molecules 2021, 26, 7414. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zheng, X.Y. Walsh Transform and Empirical Mode Decomposition Applied to Reconstruction of Velocity and Displacement from Seismic Acceleration Measurement. Appl. Sci. 2020, 10, 3509. [Google Scholar] [CrossRef]
- Yang, C.; Ling, B.W.; Kuang, W.; Gu, J. Decimations of Intrinsic Mode Functions Via Semi-Infinite Programming Based Optimal Adaptive Nonuniform Filter Bank Design Approach. Signal Process. 2019, 159, 53–71. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, S. BP Neural Network Modeling with Sensitivity Analysis On Monotonicity Based Spearman Coefficient. Chemometr. Intell. Lab. 2020, 200, 103977. [Google Scholar] [CrossRef]
- Waindim, M.; Agostini, L.; Larchêveque, L.; Adler, M.; Gaitonde, D.V. Dynamics of separation bubble dilation and collapse in shock wave/turbulent boundary layer interactions. Shock Waves 2020, 30, 63–75. [Google Scholar] [CrossRef]
- Jia, E.; Zhou, Y.; Shi, H.; Pan, M.; Zhao, X.; Ge, Q. Effects of Brain Tissue Section Processing and Storage Time On Gene Expression. Anal. Chim. Acta. 2021, 1142, 38–47. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.E.; Shen, Z.; Long, S.R.; Wu, M.C.; Shih, H.H.; Zheng, Q.; Yen, N.-C.; Tung, C.C.; Liu, H.H. The empirical mode decomposition and the Hilbert spectrum for nonlinear and non-stationary time series analysis. Proc. R. Soc. Lond. A 1998, 454, 903–995. [Google Scholar] [CrossRef]
- Xie, W.; Luo, J.; Pan, C.; Liu, Y. SG-LSTM-FRAME: A Computational Frame Using Sequence and Geometrical Information Via LSTM to Predict miRNA–Gene Associations. Brief Bioinform. 2021, 22, 2032–2042. [Google Scholar] [CrossRef] [PubMed]

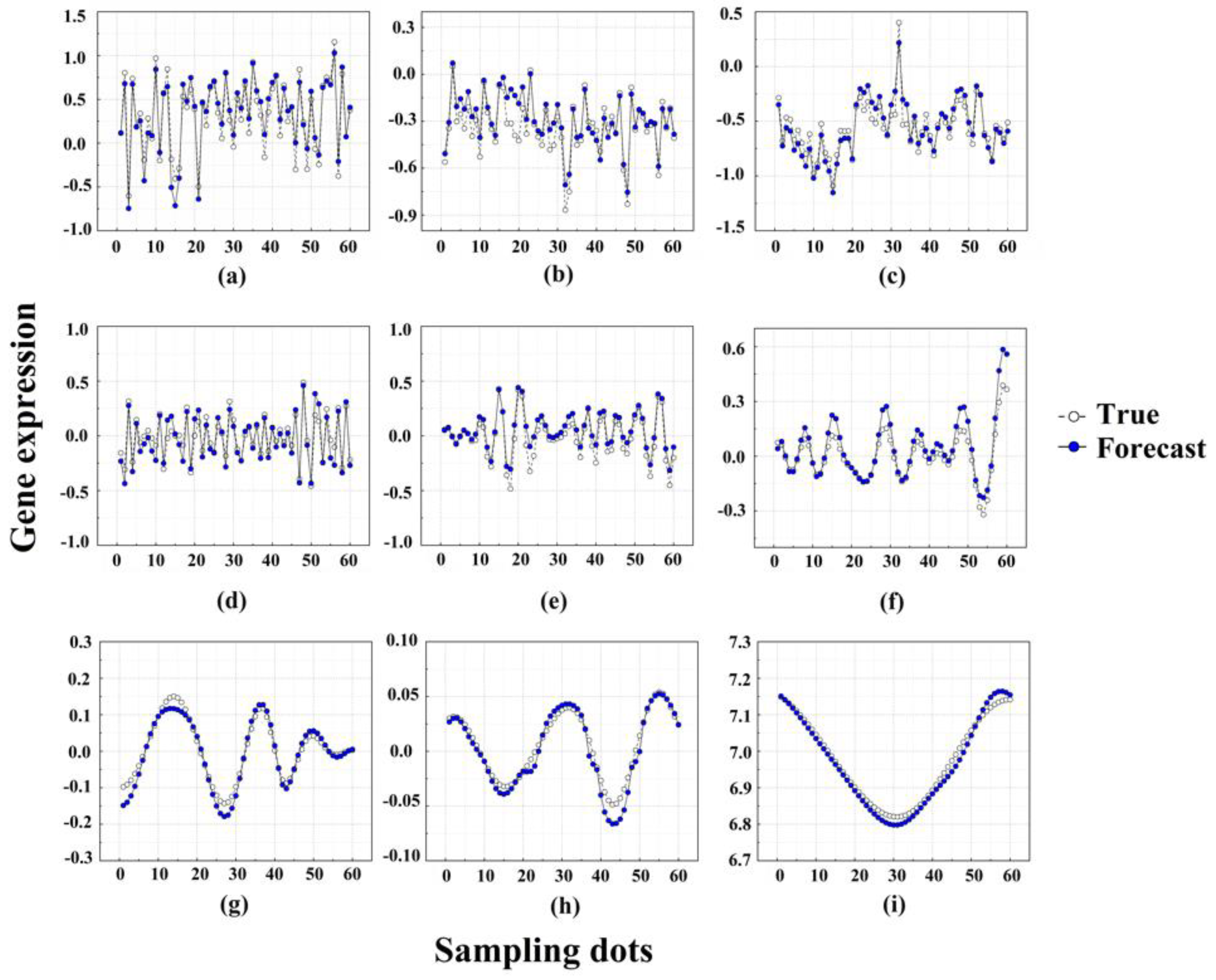
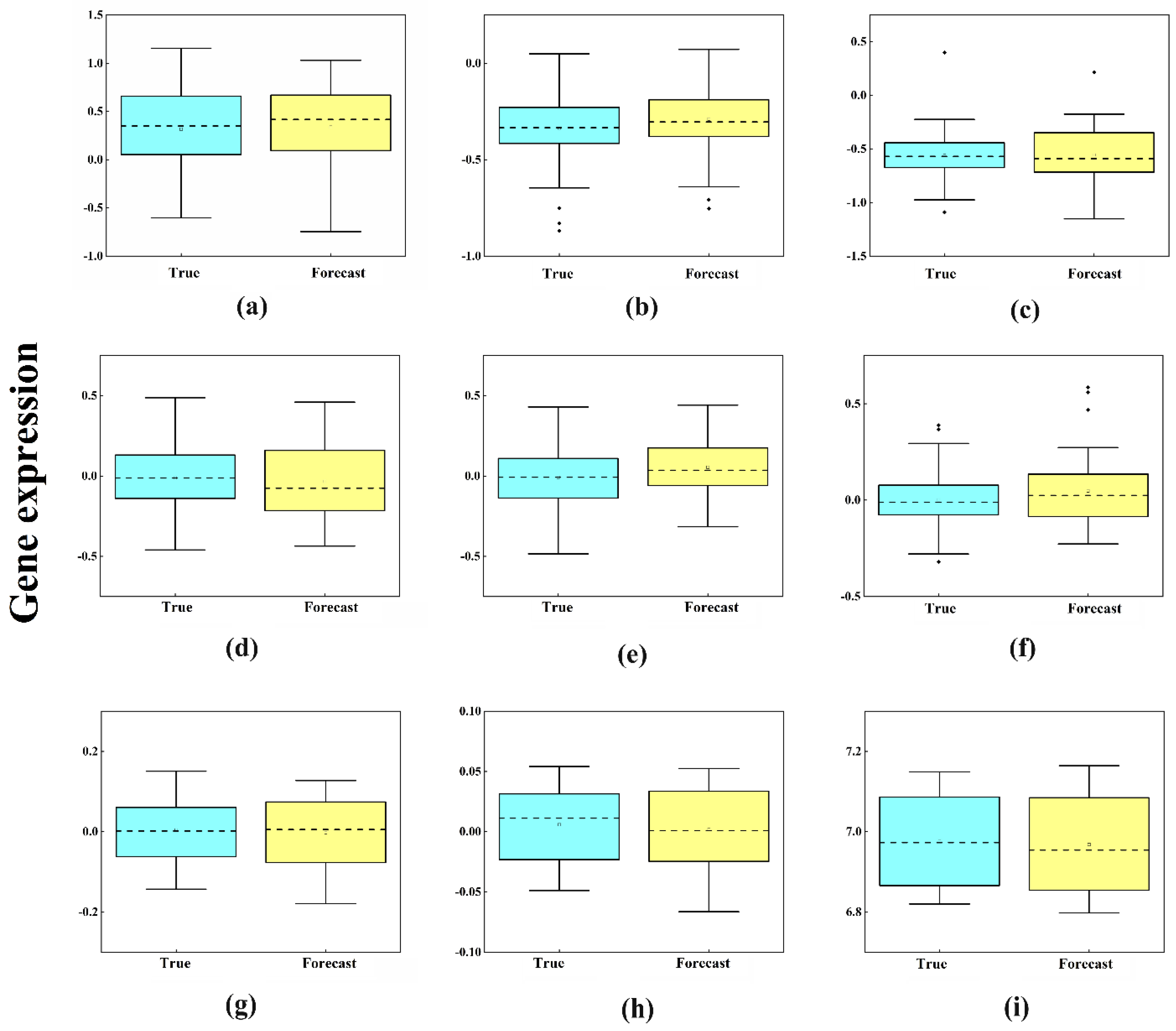
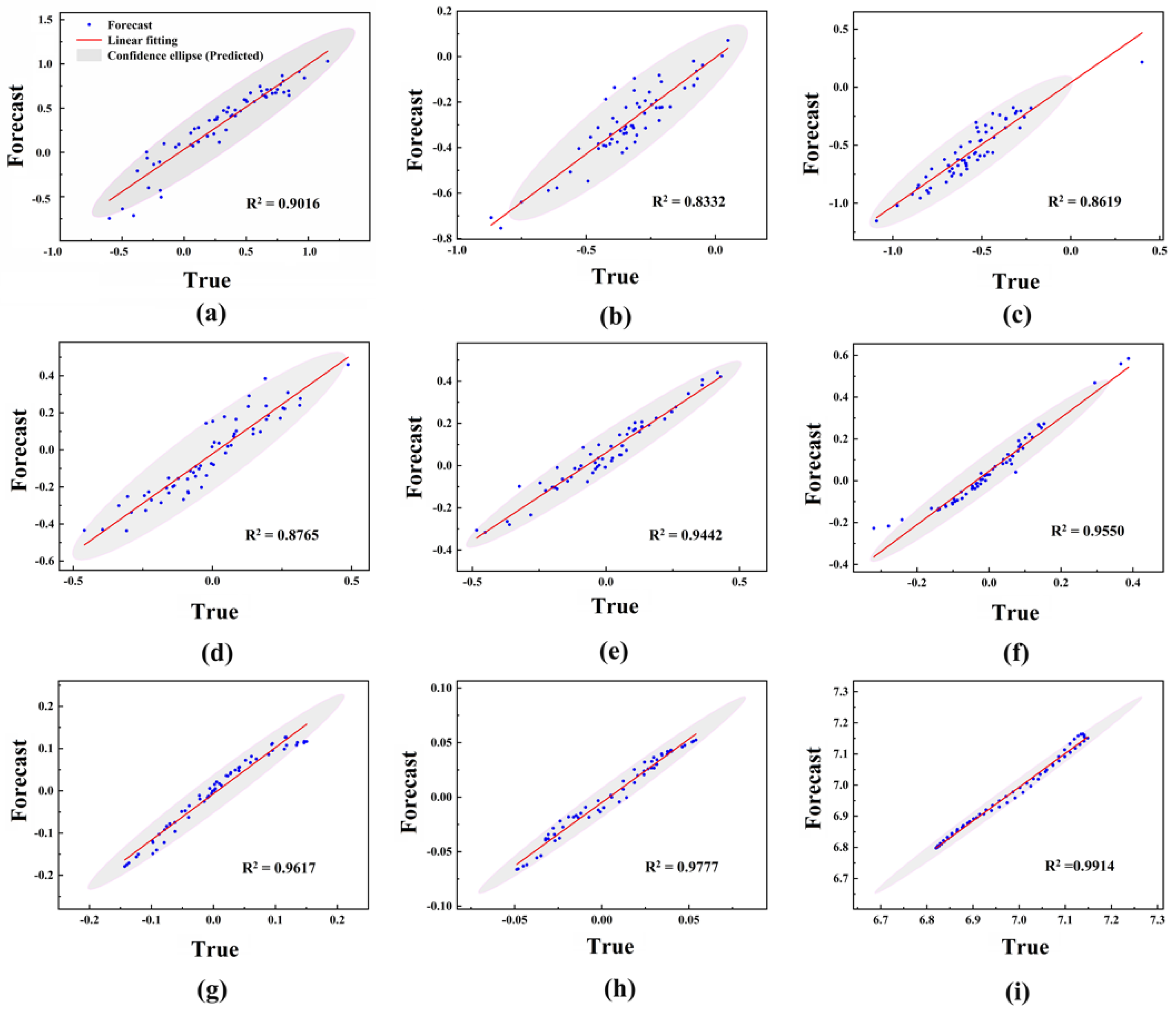
| IMF1 | IMF2 | IMF3 | IMF4 | IMF5 | IMF6 | IMF7 | IMF8 | IMF9 | |
|---|---|---|---|---|---|---|---|---|---|
| RMSE | 0.1332 | 0.0871 | 0.0959 | 0.0806 | 0.0817 | 0.0688 | 0.0202 | 0.0081 | 0.0172 |
| MAE | 0.1059 | 0.0661 | 0.0812 | 0.0647 | 0.0639 | 0.0492 | 0.0160 | 0.0060 | 0.0151 |
| R2 | 0.9016 | 0.8332 | 0.8619 | 0.8765 | 0.9442 | 0.9550 | 0.9617 | 0.9777 | 0.9914 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Jia, E.; Shi, H.; Liu, Z.; Sheng, Y.; Pan, M.; Tu, J.; Ge, Q.; Lu, Z. Prediction of Time-Series Transcriptomic Gene Expression Based on Long Short-Term Memory with Empirical Mode Decomposition. Int. J. Mol. Sci. 2022, 23, 7532. https://doi.org/10.3390/ijms23147532
Zhou Y, Jia E, Shi H, Liu Z, Sheng Y, Pan M, Tu J, Ge Q, Lu Z. Prediction of Time-Series Transcriptomic Gene Expression Based on Long Short-Term Memory with Empirical Mode Decomposition. International Journal of Molecular Sciences. 2022; 23(14):7532. https://doi.org/10.3390/ijms23147532
Chicago/Turabian StyleZhou, Ying, Erteng Jia, Huajuan Shi, Zhiyu Liu, Yuqi Sheng, Min Pan, Jing Tu, Qinyu Ge, and Zuhong Lu. 2022. "Prediction of Time-Series Transcriptomic Gene Expression Based on Long Short-Term Memory with Empirical Mode Decomposition" International Journal of Molecular Sciences 23, no. 14: 7532. https://doi.org/10.3390/ijms23147532
APA StyleZhou, Y., Jia, E., Shi, H., Liu, Z., Sheng, Y., Pan, M., Tu, J., Ge, Q., & Lu, Z. (2022). Prediction of Time-Series Transcriptomic Gene Expression Based on Long Short-Term Memory with Empirical Mode Decomposition. International Journal of Molecular Sciences, 23(14), 7532. https://doi.org/10.3390/ijms23147532






