Antibacterial, Antibiofilm, and Antiviral Farnesol-Containing Nanoparticles Prevent Staphylococcus aureus from Drug Resistance Development
Abstract
:1. Introduction
2. Results and Discussion
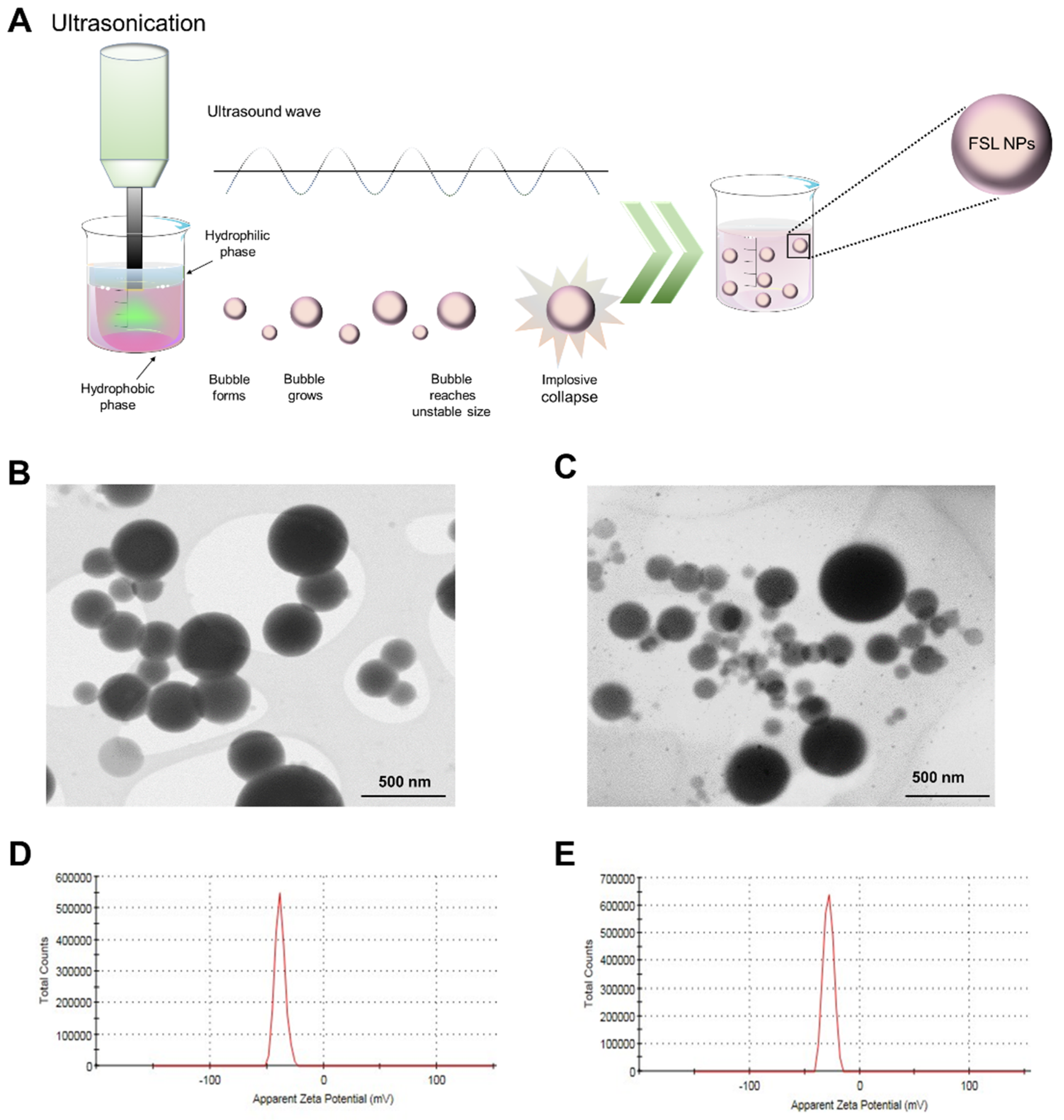
2.1. NP Characterization
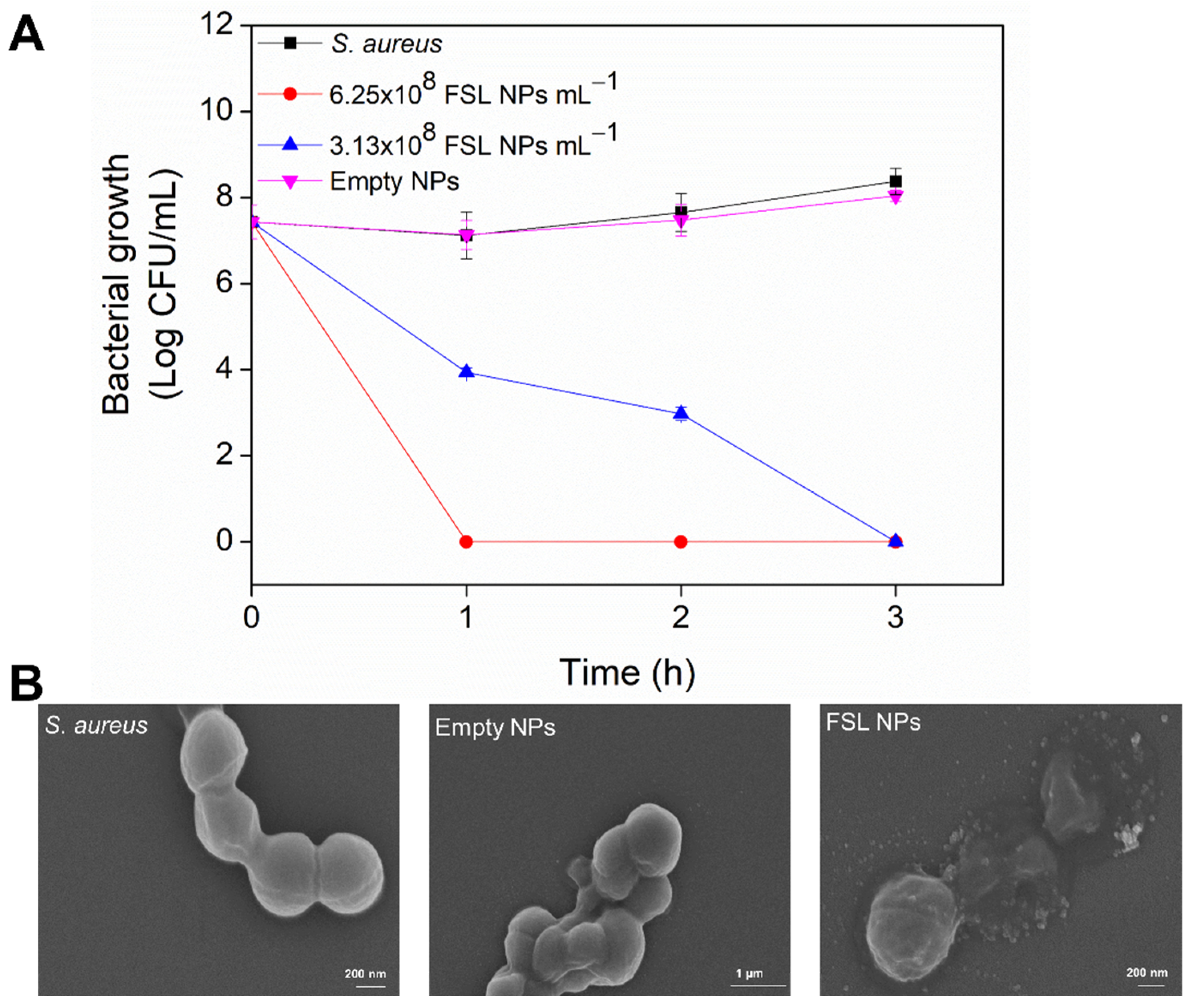
2.2. Antibacterial Efficiency of FSL NPs against Planktonic Bacteria
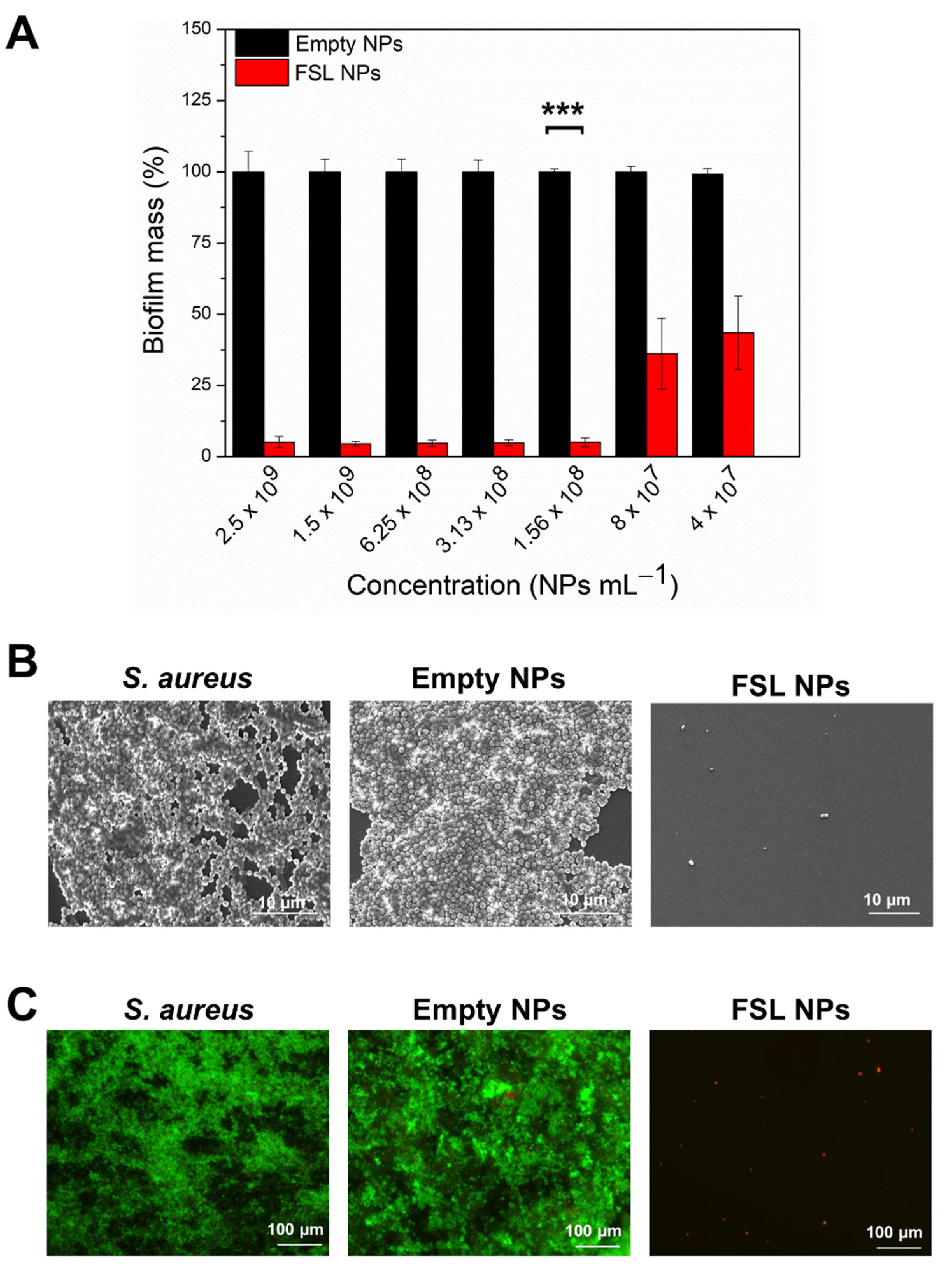
2.3. Antibiofilm Activity
2.3.1. Inhibition of S. aureus Biofilm Formation
2.3.2. Elimination of the Mature Biofilm with FSL NPs
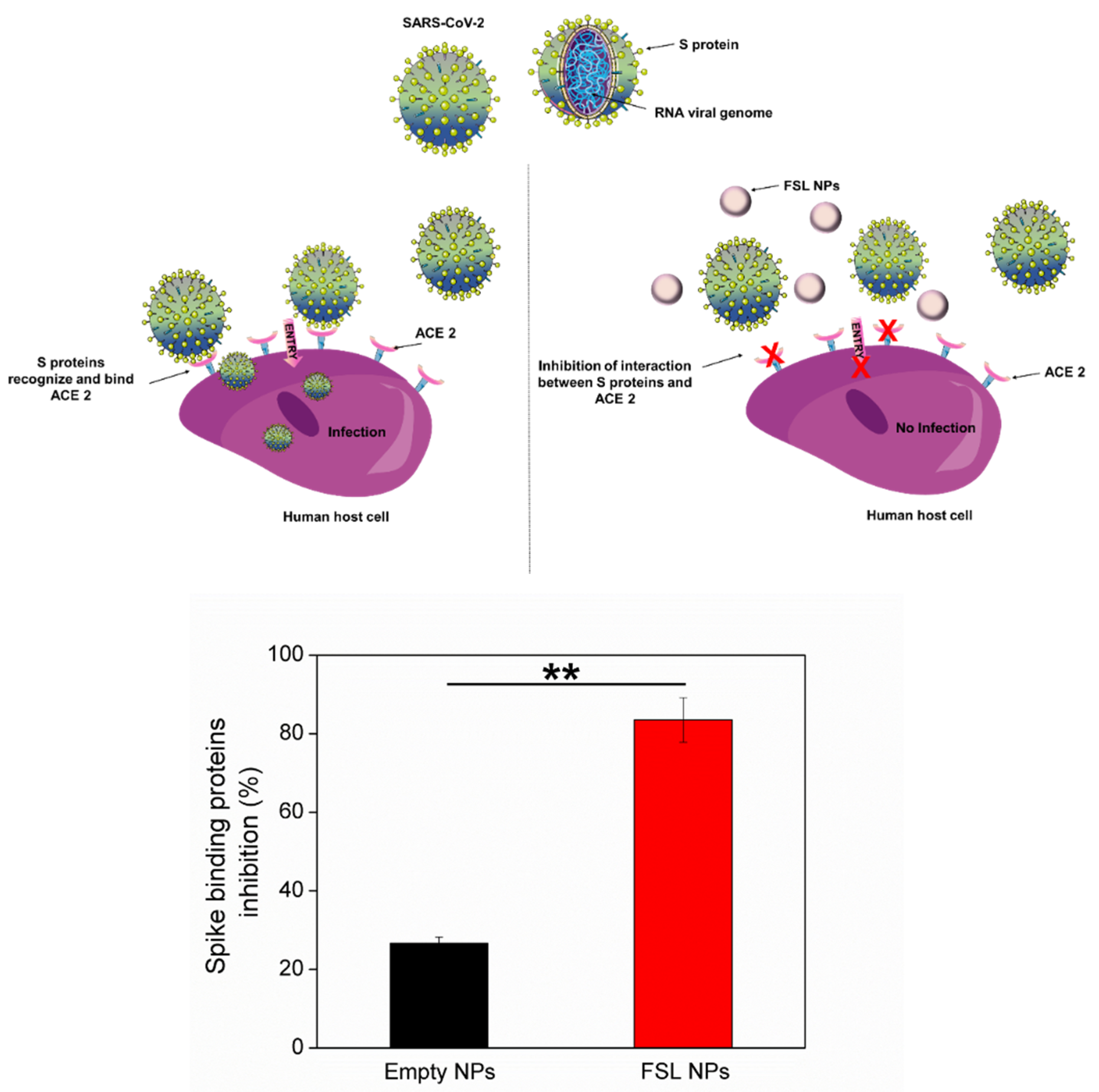
2.4. Antiviral Properties of FSL NPs
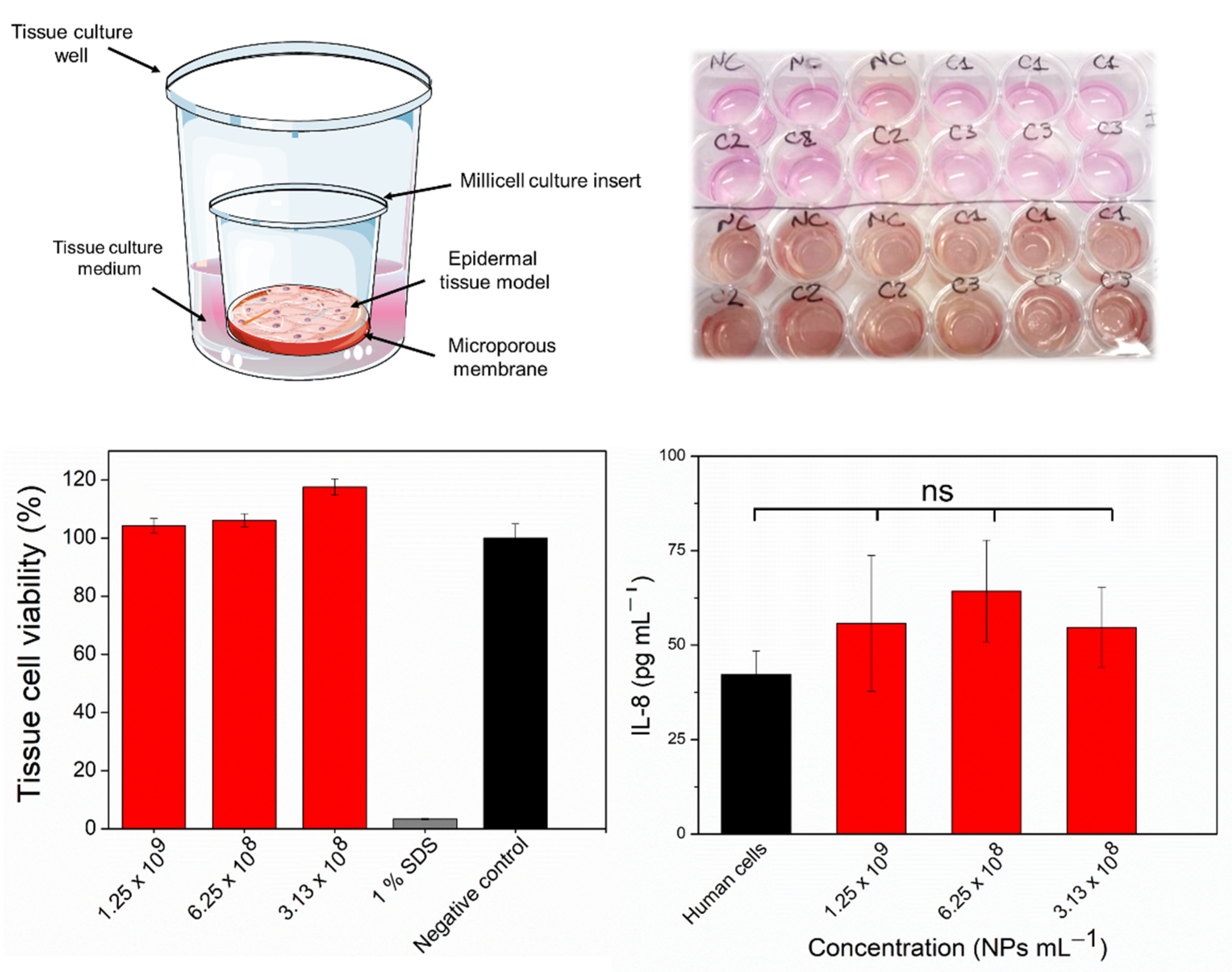
2.5. In Vitro Assessment of Skin Irritation, Tissue Viability, and Immune Response after Exposure to the FSL NPs
3. Materials and Methods
3.1. Materials
3.2. FSL NPs Preparation
3.3. NPs Characterization
3.4. Antibacterial Activity of FSL NPs
3.4.1. Minimum Inhibitory Concentration
3.4.2. Bacteria Time-Killing Kinetics
3.4.3. Scanning Electron Microscopy of Bacteria
3.4.4. Resistance Development Assay
3.5. Inhibition of S. aureus Attachment and Establishment of Biofilm
3.5.1. Biofilm Inhibition Activity
3.5.2. FSL NP Interaction with Mature Biofilms
3.6. SARS-CoV-2 Spike: ACE2 Inhibitor Screening Assay Kit
3.7. Biocompatibility Assessment
3.7.1. In Vitro Irritation Assay and Tissue Viability after Exposure to the FSL NPs
3.7.2. Immunotoxicity Induction by FSL NPs
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016; p. 84. [Google Scholar] [CrossRef]
- Divakar, S.; Lama, M.; Asad, U.K. Antibiotics versus Biofilm: An Emerging Battleground in Microbial Communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic Resistance of Bacteria in Biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Bi, Y.; Xia, G.; Shi, C.; Wan, J.; Liu, L.; Chen, Y.; Wu, Y.; Zhang, W.; Zhou, M.; He, H.; et al. Therapeutic Strategies against Bacterial Biofilms. Fundam. Res. 2021, 1, 193–212. [Google Scholar] [CrossRef]
- Global Priority List of Antibioticresistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017.
- Sahu, M.K.; George, N.; Rastogi, N.; Bipin, C.; Singh, S.P. Uncommon Pathogens Causing Hospital-Acquired Infections in Postoperative Cardiac Surgical Patients. J. Card. Crit. Care TSS 2020, 3, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Chambers, H.F.; DeLeo, F.R. Waves of Resistance: Staphylococcus aureus in the Antibiotic Era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Kulkarni, A.P.; Nagvekar, V.C.; Veeraraghavan, B.; Warrier, A.R.; Deepak, T.S.; Ahdal, J.; Jain, R. Current Perspectives on Treatment of Gram-Positive Infections in India: What Is the Way Forward? Interdiscip. Perspect. Infect. Dis. 2019, 2019, 8. [Google Scholar] [CrossRef]
- Nazzaro, F.; Fratianni, F.; Coppola, R.; De Feo, V. Essential Oils and Antifungal Activity. Pharmaceuticals 2017, 10, 86. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.Y.; Hwang, S.T.; Sethi, G.; Fan, L.; Arfuso, F.; Ahn, K.S. Potential Anti-Inflammatory and Anti-Cancer Properties of Farnesol. Molecules 2018, 23, 2827. [Google Scholar] [CrossRef] [Green Version]
- Yar, N.; Wittman, E.; Schaut, D.; De Seta, F.; Larsen, B. Effects of Farnesol on Drug-Resistant and Non-Resistant Candida Albicans: Implications for Cosmetic and Pharmaceutical Applications. Adv. Microbiol. 2020, 10, 383–396. [Google Scholar] [CrossRef]
- Inoue, Y.; Togashi, N.; Hamashima, H. Farnesol-Induced Disruption of the Staphylococcus aureus Cytoplasmic Membrane. Biol. Pharm. Bull. 2016, 39, 653–656. [Google Scholar] [CrossRef] [Green Version]
- Koo, H.; Hayacibara, M.F.; Schobel, B.D.; Cury, J.A.; Rosalen, P.L.; Park, Y.K.; Vacca-Smith, A.M.; Bowen, W.H. Inhibition of Streptococcus mutans Biofilm Accumulation and Polysaccharide Production by Apigenin and Tt-Farnesol. J. Antimicrob. Chemother. 2003, 52, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Sims, K.R.; Liu, Y.; Hwang, G.; Jung, H.I.; Koo, H.; Benoit, D.S.W. Enhanced Design and Formulation of Nanoparticles for Anti-Biofilm Drug Delivery. Nanoscale 2019, 11, 219–236. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Busscher, H.J.; Zhao, B.; Li, Y.; Zhang, Z.; Van Der Mei, H.C.; Ren, Y.; Shi, L. Surface-Adaptive, Antimicrobially Loaded, Micellar Nanocarriers with Enhanced Penetration and Killing Efficiency in Staphylococcal Biofilms. ACS Nano 2016, 10, 4779–4789. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, M.M.; Ivanova, K.; Francesko, A.; Rivera, D.; Torrent-Burgués, J.; Gedanken, A.; Mendonza, E.; Tzanov, T. Escherichia Coli and Pseudomonas Aeruginosa Eradication by Nano-Penicillin G. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 2061–2069. [Google Scholar] [CrossRef]
- Fernandes, M.M.; Ivanova, K.; Hoyo, J.; Pérez-Rafael, S.; Francesko, A.; Tzanov, T. Nanotransformation of Vancomycin Overcomes the Intrinsic Resistance of Gram-Negative Bacteria. ACS Appl. Mater. Interfaces 2017, 9, 15022–15030. [Google Scholar] [CrossRef]
- Feger, G.; Angelov, B.; Angelova, A. Prediction of Amphiphilic Cell-Penetrating Peptide Building Blocks from Protein-Derived Amino Acid Sequences for Engineering of Drug Delivery Nanoassemblies. J. Phys. Chem. B 2020, 124, 4069–4078. [Google Scholar] [CrossRef]
- Ivanova, K.; Ivanova, A.; Ramon, E.; Hoyo, J.; Sanchez-Gomez, S.; Tzanov, T. Antibody-Enabled Antimicrobial Nanocapsules for Selective Elimination of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2020, 12, 35918–35927. [Google Scholar] [CrossRef]
- Ivanova, A.; Ivanova, K.; Hoyo, J.; Heinze, T.; Sanchez-Gomez, S.; Tzanov, T. Layer-By-Layer Decorated Nanoparticles with Tunable Antibacterial and Antibiofilm Properties against Both Gram-Positive and Gram-Negative Bacteria. ACS Appl. Mater. Interfaces 2018, 10, 3314–3323. [Google Scholar] [CrossRef]
- Maillard, J.; Bloom, S.F.; Courvalin, P.; Essack, S.Y.; Gandra, S.; Gerba, C.P.; Ba, J.R.R.; Scott, E.A. Reducing Antibiotic Prescribing and Addressing the Global Problem of Antibiotic Resistance by Targeted Hygiene in the Home and Everyday Life Settings: A Position Paper. Am. J. Infect. Control. 2020, 48, 1090–1099. [Google Scholar] [CrossRef]
- Hesham, H.A.; Salman, A.I.G. Nanoparticles Comprising Esters of Poly (Methyl Vinyl Ether-Co-Maleic Anhydride) and Uses Thereof. WIPO Patent No. WO2012140252, 18 October 2012. [Google Scholar]
- Gupta, V.; Trivedi, P. In Vitro and in Vivo Characterization of Pharmaceutical Topical Nanocarriers Containing Anticancer Drugs for Skin Cancer Treatment. In Lipid Nanocarriers for Drug Targeting; Grumezescu, A.M., Ed.; William Andrew Publishing: Norwich, NY, USA, 2018; pp. 563–627. ISBN 9780128136874. [Google Scholar]
- Albert, C.; Huang, N.; Tsapis, N.; Geiger, S.; Rosilio, V.; Mekhloufi, G.; Chapron, D.; Robin, B.; Beladjine, M.; Nicolas, V.; et al. Bare and Sterically Stabilized PLGA Nanoparticles for the Stabilization of Pickering Emulsions. Langmuir 2018, 34, 13935–13945. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabra-Rizk, M.; Meiller, T.F.; James, C.E.; Shirtliff, M.E. Effect of Farnesol on Staphylococcus aureus Biofilm Formation and Antimicrobial Susceptibility Effect of Farnesol on Staphylococcus aureus Biofilm Formation and Antimicrobial Susceptibility. Antimicrob. Agents Chemother. 2006, 50, 1463–1469. [Google Scholar] [CrossRef] [Green Version]
- Stautz, J.; Hellmich, Y.; Fuss, M.F.; Silberberg, J.M.; Devlin, J.R.; Stockbridge, R.B.; Hänelt, I. Molecular Mechanisms for Bacterial Potassium Homeostasis. J. Mol. Biol. 2021, 433, 166968. [Google Scholar] [CrossRef]
- Chouhan, S.; Sharma, K.; Guleria, S. Antimicrobial Activity of Some Essential Oils—Present Status and Future Perspectives. Medicines 2017, 4, 58. [Google Scholar] [CrossRef] [Green Version]
- Masters, E.A.; Muthukrishnan, G.; Ho, L.; Gill, A.L.; de Mesy Bentley, K.L.; Galloway, C.A.; McGrath, J.L.; Awad, H.A.; Gill, S.R.; Schwarz, E.M. Staphylococcus aureus Cell Wall Biosynthesis Modulates Bone Invasion and Osteomyelitis Pathogenesis. Front. Microbiol. 2021, 12, 723498. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [Green Version]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial Biofilm Formation on Implantable Devices and Approaches to Its Treatment and Prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef] [Green Version]
- Unnanuntana, A.; Bonsignore, L.; Shirtliff, M.E.; Greenfield, E.M. The Effects of Farnesol on Staphylococcus aureus Biofilms and Osteoblasts: An in Vitro Study. J. Bone Jt. Surg. 2009, 91, 2683–2692. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, M.; Azevedo, N.F.; Ivask, A. Propidium Iodide Staining Underestimates Viability of Adherent Bacterial Cells. Sci. Rep. 2019, 9, 6483. [Google Scholar] [CrossRef] [Green Version]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K.; Ren, Q. Critical Aspects of Using Bacterial Cell Viability Assays with the Fluorophores SYTO9 and Propidium Iodide. BMC Microbiol. 2015, 15, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanova, A.; Ivanova, K.; Tzanov, T. Inhibition of Quorum-Sensing: A New Paradigm in Controlling Bacterial Virulence and Biofilm Formation. In Biotechnological Applications of Quorum Sensing Inhibitors; Kalia, V.C., Ed.; Springer: Singapore, 2018; pp. 3–21. ISBN 978-981-10-9026-4. [Google Scholar]
- Gomes, F.; Teixeira, P.; Cerca, N.; Azeredo, J.; Oliveira, R. Effect of Farnesol on Structure and Composition of Staphylococcus epidermidis Biofilm Matrix. Curr. Microbiol. 2011, 63, 354–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef] [Green Version]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and Distributions of SARS-CoV-2 Spike Proteins on Intact Virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef]
- Horev, B.; Klein, M.I.; Hwang, G.; Li, Y.; Kim, D.; Koo, H.; Benoit, D.S.W. PH-Activated Nanoparticles for Controlled Topical Delivery of Farnesol to Disrupt Oral Biofilm Virulence. ACS Nano 2015, 9, 2390–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanova, A.; Ivanova, K.; Tied, A.; Heinze, T.; Tzanov, T. Layer-By-Layer Coating of Aminocellulose and Quorum Quenching Acylase on Silver Nanoparticles Synergistically Eradicate Bacteria and Their Biofilms. Adv. Funct. Mater. 2020, 30, 2001284. [Google Scholar] [CrossRef] [Green Version]
- Ching, C.; Zaman, M.H. Development and Selection of Low-Level Multi-Drug Resistance over an Extended Range of Sub-Inhibitory Ciprofloxacin Concentrations in Escherichia Coli. Sci. Rep. 2020, 10, 8754. [Google Scholar] [CrossRef]
- Bengalli, R.; Colantuoni, A.; Perelshtein, I.; Gedanken, A.; Collini, M.; Mantecca, P.; Fiandra, L. In Vitro Skin Toxicity of CuO and ZnO Nanoparticles: Application in the Safety Assessment of Antimicrobial Coated Textiles. NanoImpact 2021, 21, 100282. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanova, A.; Ivanova, K.; Fiandra, L.; Mantecca, P.; Catelani, T.; Natan, M.; Banin, E.; Jacobi, G.; Tzanov, T. Antibacterial, Antibiofilm, and Antiviral Farnesol-Containing Nanoparticles Prevent Staphylococcus aureus from Drug Resistance Development. Int. J. Mol. Sci. 2022, 23, 7527. https://doi.org/10.3390/ijms23147527
Ivanova A, Ivanova K, Fiandra L, Mantecca P, Catelani T, Natan M, Banin E, Jacobi G, Tzanov T. Antibacterial, Antibiofilm, and Antiviral Farnesol-Containing Nanoparticles Prevent Staphylococcus aureus from Drug Resistance Development. International Journal of Molecular Sciences. 2022; 23(14):7527. https://doi.org/10.3390/ijms23147527
Chicago/Turabian StyleIvanova, Aleksandra, Kristina Ivanova, Luisa Fiandra, Paride Mantecca, Tiziano Catelani, Michal Natan, Ehud Banin, Gila Jacobi, and Tzanko Tzanov. 2022. "Antibacterial, Antibiofilm, and Antiviral Farnesol-Containing Nanoparticles Prevent Staphylococcus aureus from Drug Resistance Development" International Journal of Molecular Sciences 23, no. 14: 7527. https://doi.org/10.3390/ijms23147527
APA StyleIvanova, A., Ivanova, K., Fiandra, L., Mantecca, P., Catelani, T., Natan, M., Banin, E., Jacobi, G., & Tzanov, T. (2022). Antibacterial, Antibiofilm, and Antiviral Farnesol-Containing Nanoparticles Prevent Staphylococcus aureus from Drug Resistance Development. International Journal of Molecular Sciences, 23(14), 7527. https://doi.org/10.3390/ijms23147527






