SAGA-Dependent Histone H2Bub1 Deubiquitination Is Essential for Cellular Ubiquitin Balance during Embryonic Development
Abstract
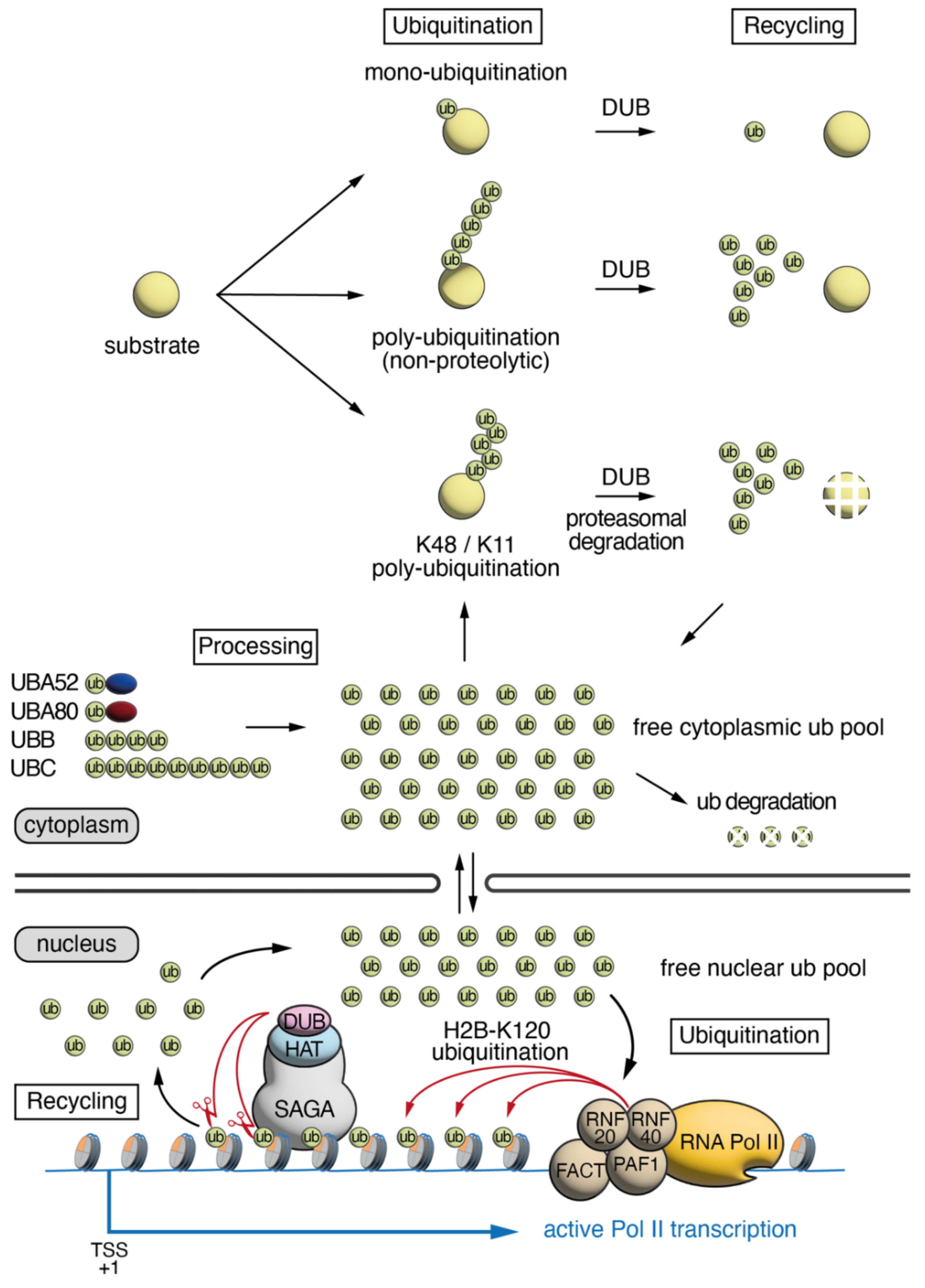
1. Ubiquitin Cellular Dynamics, Abundance and Distribution
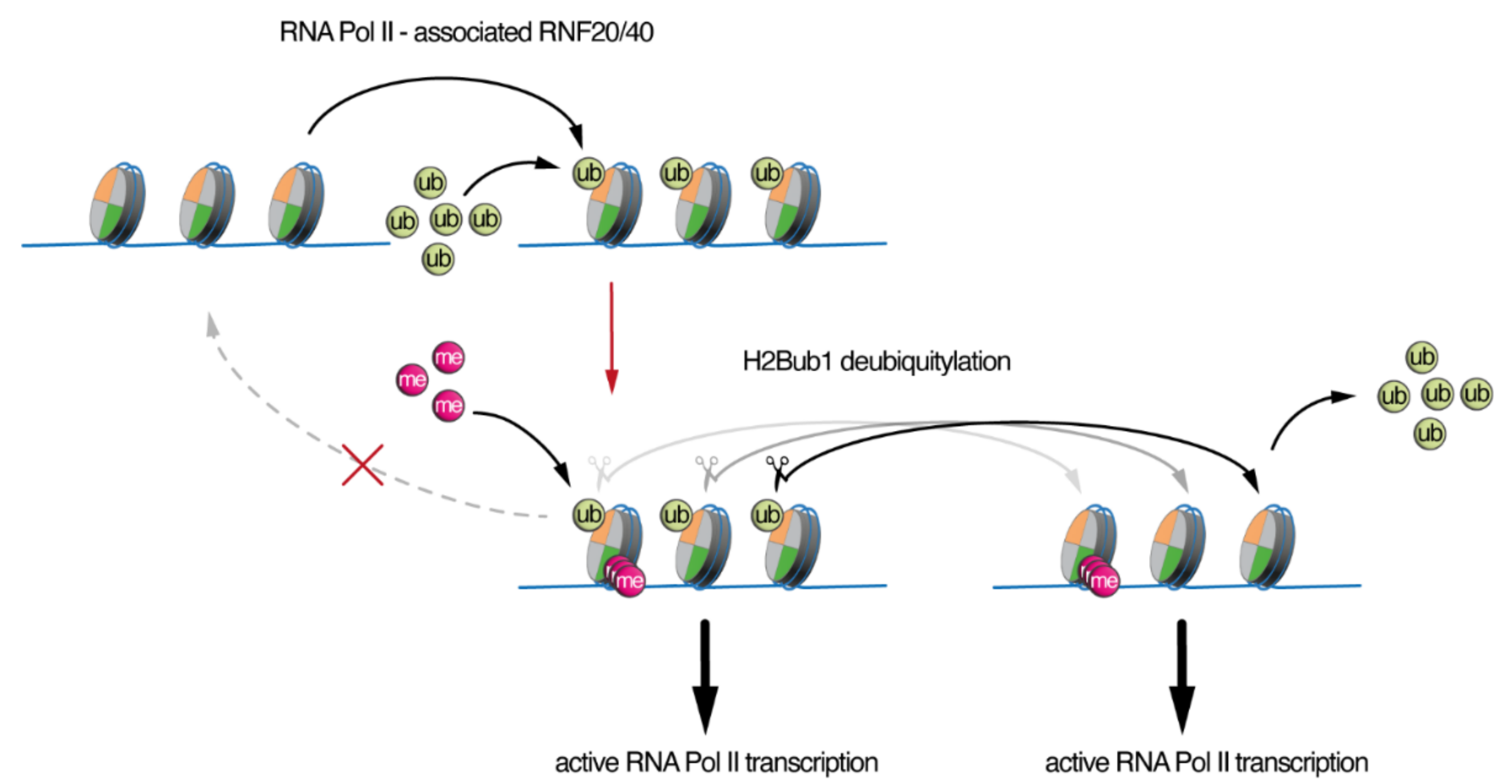
2. Roles of H2Bub1 in Chromatin Compaction and Transcription
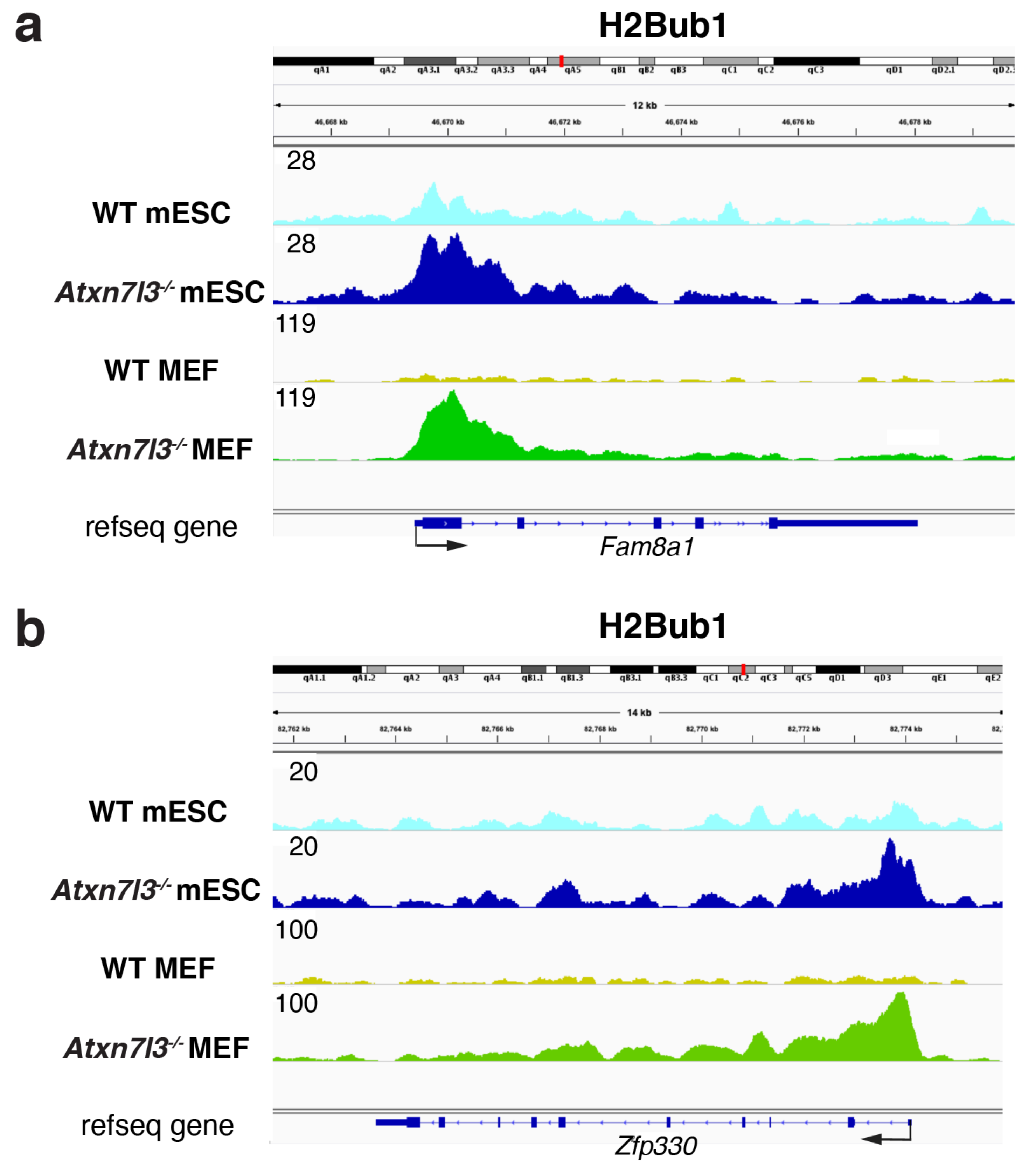
3. SAGA/ATXN7L3-Related DUBs as Free Nuclear Ubiquitin “Generators” during Development
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ryu, K.Y.; Baker, R.T.; Kopito, R.R. Ubiquitin-specific protease 2 as a tool for quantification of total ubiquitin levels in biological specimens. Anal. Biochem. 2006, 353, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Ohtani-Kaneko, R.; Asahara, M.; Takada, K.; Kanda, T.; Iigo, M.; Hara, M.; Yokosawa, H.; Ohkawa, K.; Hirata, K. Nerve growth factor (NGF) induces increase in multi-ubiquitin chains and concomitant decrease in free ubiquitin in nuclei of PC12h. Neurosci. Res. 1996, 26, 349–355. [Google Scholar] [CrossRef]
- Takada, K.; Hibi, N.; Tsukada, Y.; Shibasaki, T.; Ohkawa, K. Ability of ubiquitin radioimmunoassay to discriminate between monoubiquitin and multi-ubiquitin chains. Biochim. Biophys. Acta 1996, 1290, 282–288. [Google Scholar] [CrossRef]
- Osaka, H.; Wang, Y.L.; Takada, K.; Takizawa, S.; Setsuie, R.; Li, H.; Sato, Y.; Nishikawa, K.; Sun, Y.J.; Sakurai, M.; et al. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum. Mol. Genet. 2003, 12, 1945–1958. [Google Scholar] [CrossRef]
- Haas, A.L.; Bright, P.M. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J. Biol. Chem. 1987, 262, 345–351. [Google Scholar] [CrossRef]
- Berndsen, C.E.; Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307. [Google Scholar] [CrossRef]
- Love, K.R.; Catic, A.; Schlieker, C.; Ploegh, H.L. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat. Chem. Biol. 2007, 3, 697–705. [Google Scholar] [CrossRef]
- Kaiser, S.E.; Riley, B.E.; Shaler, T.A.; Trevino, R.S.; Becker, C.H.; Schulman, H.; Kopito, R.R. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 2011, 8, 691–696. [Google Scholar] [CrossRef]
- Goldknopf, I.L.; Taylor, C.W.; Baum, R.M.; Yeoman, L.C.; Olson, M.O.; Prestayko, A.W.; Busch, H. Isolation and characterization of protein A24, a "histone-like" non-histone chromosomal protein. J. Biol. Chem. 1975, 250, 7182–7187. [Google Scholar] [CrossRef]
- Matsui, S.I.; Seon, B.K.; Sandberg, A.A. Disappearance of a structural chromatin protein A24 in mitosis: Implications for molecular basis of chromatin condensation. Proc. Natl. Acad. Sci. USA 1979, 76, 6386–6390. [Google Scholar] [CrossRef]
- West, M.H.; Bonner, W.M. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 1980, 8, 4671–4680. [Google Scholar] [CrossRef]
- Robzyk, K.; Recht, J.; Osley, M.A. Rad6-dependent ubiquitination of histone H2B in yeast. Science 2000, 287, 501–504. [Google Scholar] [CrossRef]
- Fuchs, G.; Oren, M. Writing and reading H2B monoubiquitylation. Biochim. Biophys. Acta 2014, 1839, 694–701. [Google Scholar] [CrossRef]
- Fierz, B.; Chatterjee, C.; McGinty, R.K.; Bar-Dagan, M.; Raleigh, D.P.; Muir, T.W. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011, 7, 113–119. [Google Scholar] [CrossRef]
- Chandrasekharan, M.B.; Huang, F.; Sun, Z.W. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. USA 2009, 106, 16686–16691. [Google Scholar] [CrossRef]
- Batta, K.; Zhang, Z.; Yen, K.; Goffman, D.B.; Pugh, B.F. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011, 25, 2254–2265. [Google Scholar] [CrossRef]
- Wright, D.E.; Wang, C.Y.; Kao, C.F. Histone ubiquitylation and chromatin dynamics. Front. Biosci. 2012, 17, 1051–1078. [Google Scholar] [CrossRef]
- Laribee, R.N.; Fuchs, S.M.; Strahl, B.D. H2B ubiquitylation in transcriptional control: A Fact-finding mission. Genes Dev. 2007, 21, 737–743. [Google Scholar] [CrossRef][Green Version]
- Zhu, B.; Zheng, Y.; Pham, A.D.; Mandal, S.S.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell 2005, 20, 601–611. [Google Scholar] [CrossRef]
- Pavri, R.; Zhu, B.; Li, G.; Trojer, P.; Mandal, S.; Shilatifard, A.; Reinberg, D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006, 125, 703–717. [Google Scholar] [CrossRef]
- Fleming, A.B.; Kao, C.F.; Hillyer, C.; Pikaart, M.; Osley, M.A. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 2008, 31, 57–66. [Google Scholar] [CrossRef]
- Bonnet, J.; Wang, C.Y.; Baptista, T.; Vincent, S.D.; Hsiao, W.C.; Stierle, M.; Kao, C.F.; Tora, L.; Devys, D. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Gene Dev. 2014, 28, 1999–2012. [Google Scholar] [CrossRef]
- Shchebet, A.; Karpiuk, O.; Kremmer, E.; Eick, D.; Johnsen, S.A. Phosphorylation by cyclin-dependent kinase-9 controls ubiquitin-conjugating enzyme-2A function. Cell Cycle 2012, 11, 2122–2127. [Google Scholar] [CrossRef]
- Pirngruber, J.; Shchebet, A.; Schreiber, L.; Shema, E.; Minsky, N.; Chapman, R.D.; Eick, D.; Aylon, Y.; Oren, M.; Johnsen, S.A. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3’-end processing. EMBO Rep. 2009, 10, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yu, X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol. Cell 2011, 41, 384–397. [Google Scholar] [CrossRef] [PubMed]
- Henry, K.W.; Wyce, A.; Lo, W.S.; Duggan, L.J.; Emre, N.C.; Kao, C.F.; Pillus, L.; Shilatifard, A.; Osley, M.A.; Berger, S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003, 17, 2648–2663. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.A.; Torok, M.S.; Sun, Z.W.; Schieltz, D.; Allis, C.D.; Yates, J.R., 3rd; Grant, P.A. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 2004, 279, 1867–1871. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lang, G.; Ito, S.; Bonnet, J.; Metzger, E.; Sawatsubashi, S.; Suzuki, E.; Le Guezennec, X.; Stunnenberg, H.G.; Krasnov, A.; et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 2008, 29, 92–101. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Varthi, M.; Sykes, S.M.; Phillips, C.; Warzecha, C.; Zhu, W.; Wyce, A.; Thorne, A.W.; Berger, S.L.; McMahon, S.B. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 2008, 29, 102–111. [Google Scholar] [CrossRef]
- Lang, G.; Bonnet, J.; Umlauf, D.; Karmodiya, K.; Koffler, J.; Stierle, M.; Devys, D.; Tora, L. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol. Cell. Biol. 2011, 31, 3734–3744. [Google Scholar] [CrossRef]
- Herbst, D.A.; Esbin, M.N.; Louder, R.K.; Dugast-Darzacq, C.; Dailey, G.M.; Fang, Q.; Darzacq, X.; Tjian, R.; Nogales, E. Structure of the human SAGA coactivator complex. Nat. Struct. Mol. Biol. 2021, 28, 989–996. [Google Scholar] [CrossRef]
- Atanassov, B.S.; Mohan, R.D.; Lan, X.; Kuang, X.; Lu, Y.; Lin, K.; McIvor, E.; Li, W.; Zhang, Y.; Florens, L.; et al. ATXN7L3 and ENY2 Coordinate Activity of Multiple H2B Deubiquitinases Important for Cellular Proliferation and Tumor Growth. Mol. Cell 2016, 62, 558–571. [Google Scholar] [CrossRef]
- Evangelista, F.M.; Maglott-Roth, A.; Stierle, M.; Brino, L.; Soutoglou, E.; Tora, L. Transcription and mRNA export machineries SAGA and TREX-2 maintain monoubiquitinated H2B balance required for DNA repair. J. Cell Biol. 2018, 217, 3382–3397. [Google Scholar] [CrossRef]
- Wang, F.; El-Saafin, F.; Ye, T.; Stierle, M.; Negroni, L.; Durik, M.; Fischer, V.; Devys, D.; Vincent, S.D.; Tora, L. Histone H2Bub1 deubiquitylation is essential for mouse development, but does not regulate global RNA polymerase II transcription. Cell Death Differ. 2021, 28, 2385–2403. [Google Scholar] [CrossRef]
- Kosinsky, R.L.; Wegwitz, F.; Hellbach, N.; Dobbelstein, M.; Mansouri, A.; Vogel, T.; Begus-Nahrmann, Y.; Johnsen, S.A. Usp22 deficiency impairs intestinal epithelial lineage specification in vivo. Oncotarget 2015, 6, 37906–37918. [Google Scholar] [CrossRef]
- Xie, W.; Nagarajan, S.; Baumgart, S.J.; Kosinsky, R.L.; Najafova, Z.; Kari, V.; Hennion, M.; Indenbirken, D.; Bonn, S.; Grundhoff, A.; et al. RNF40 regulates gene expression in an epigenetic context-dependent manner. Genome Biol. 2017, 18, 32. [Google Scholar] [CrossRef]
- Baptista, T.; Devys, D. Saccharomyces cerevisiae Metabolic Labeling with 4-thiouracil and the Quantification of Newly Synthesized mRNA As a Proxy for RNA Polymerase II Activity. J. Vis. Exp. 2018, 140, e57982. [Google Scholar] [CrossRef]
- Briggs, S.D.; Xiao, T.; Sun, Z.W.; Caldwell, J.A.; Shabanowitz, J.; Hunt, D.F.; Allis, C.D.; Strahl, B.D. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature 2002, 418, 498. [Google Scholar] [CrossRef]
- Dover, J.; Schneider, J.; Tawiah-Boateng, M.A.; Wood, A.; Dean, K.; Johnston, M.; Shilatifard, A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002, 277, 28368–28371. [Google Scholar] [CrossRef]
- Ng, H.H.; Xu, R.M.; Zhang, Y.; Struhl, K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002, 277, 34655–34657. [Google Scholar] [CrossRef]
- Sun, Z.W.; Allis, C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002, 418, 104–108. [Google Scholar] [CrossRef]
- Lee, J.S.; Shukla, A.; Schneider, J.; Swanson, S.K.; Washburn, M.P.; Florens, L.; Bhaumik, S.R.; Shilatifard, A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 2007, 131, 1084–1096. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.A.; McGinty, R.K.; Nguyen, U.T.; Muir, T.W.; Allis, C.D.; Roeder, R.G. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol. Cell 2013, 49, 1121–1133. [Google Scholar] [CrossRef]
- Xie, W.; Miehe, M.; Laufer, S.; Johnsen, S.A. The H2B ubiquitin-protein ligase RNF40 is required for somatic cell reprogramming. Cell Death Dis. 2020, 11, 287. [Google Scholar] [CrossRef]
- Vethantham, V.; Yang, Y.; Bowman, C.; Asp, P.; Lee, J.H.; Skalnik, D.G.; Dynlacht, B.D. Dynamic loss of H2B ubiquitylation without corresponding changes in H3K4 trimethylation during myogenic differentiation. Mol. Cell Biol. 2012, 32, 1044–1055. [Google Scholar] [CrossRef]
- Ryu, K.Y.; Maehr, R.; Gilchrist, C.A.; Long, M.A.; Bouley, D.M.; Mueller, B.; Ploegh, H.L.; Kopito, R.R. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J. 2007, 26, 2693–2706. [Google Scholar] [CrossRef]
- Ryu, K.Y.; Sinnar, S.A.; Reinholdt, L.G.; Vaccari, S.; Hall, S.; Garcia, M.A.; Zaitseva, T.S.; Bouley, D.M.; Boekelheide, K.; Handel, M.A.; et al. The mouse polyubiquitin gene Ubb is essential for meiotic progression. Mol. Cell Biol 2008, 28, 1136–1146. [Google Scholar] [CrossRef]
- Ryu, K.Y.; Garza, J.C.; Lu, X.Y.; Barsh, G.S.; Kopito, R.R. Hypothalamic neurodegeneration and adult-onset obesity in mice lacking the Ubb polyubiquitin gene. Proc. Natl. Acad. Sci USA 2008, 105, 4016–4021. [Google Scholar] [CrossRef]
- Ryu, K.Y.; Fujiki, N.; Kazantzis, M.; Garza, J.C.; Bouley, D.M.; Stahl, A.; Lu, X.Y.; Nishino, S.; Kopito, R.R. Loss of polyubiquitin gene Ubb leads to metabolic and sleep abnormalities in mice. Neuropathol. Appl. Neurobiol. 2010, 36, 285–299. [Google Scholar] [CrossRef]
- Sinnar, S.A.; Small, C.L.; Evanoff, R.M.; Reinholdt, L.G.; Griswold, M.D.; Kopito, R.R.; Ryu, K.Y. Altered testicular gene expression patterns in mice lacking the polyubiquitin gene Ubb. Mol. Reprod. Dev. 2011, 78, 415–425. [Google Scholar] [CrossRef][Green Version]
- Ryu, H.W.; Park, C.W.; Ryu, K.Y. Disruption of polyubiquitin gene Ubb causes dysregulation of neural stem cell differentiation with premature gliogenesis. Sci. Rep. 2014, 4, 7026. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.W.; Park, C.W.; Ryu, K.Y. Restoration of cellular ubiquitin reverses impairments in neuronal development caused by disruption of the polyubiquitin gene Ubb. Biochem. Biophys. Res. Commun. 2014, 453, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.; Park, C.W.; Ryu, K.Y.; Chung, H. Disruption of the polyubiquitin gene Ubb causes retinal degeneration in mice. Biochem. Biophys. Res. Commun. 2019, 513, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Park, C.W.; Jung, B.K.; Ryu, K.Y. Disruption of the polyubiquitin gene Ubb reduces the self-renewal capacity of neural stem cells. Biochem. Biophys. Res. Commun. 2020, 527, 372–378. [Google Scholar] [CrossRef]
- Koutelou, E.; Wang, L.; Schibler, A.C.; Chao, H.P.; Kuang, X.; Lin, K.; Lu, Y.; Shen, J.; Jeter, C.R.; Salinger, A.; et al. USP22 controls multiple signaling pathways that are essential for vasculature formation in the mouse placenta. Development 2019, 146, dev174037. [Google Scholar] [CrossRef]
- Park, H.; Yoon, M.S.; Ryu, K.Y. Disruption of polyubiquitin gene Ubc leads to defective proliferation of hepatocytes and bipotent fetal liver epithelial progenitor cells. Biochem. Biophys. Res. Commun. 2013, 435, 434–440. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Pfeiffer, H.K.; Thorne, A.W.; McMahon, S.B. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle 2008, 7, 1522–1524. [Google Scholar] [CrossRef]
- Atanassov, B.S.; Evrard, Y.A.; Multani, A.S.; Zhang, Z.; Tora, L.; Devys, D.; Chang, S.; Dent, S.Y. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol. Cell 2009, 35, 352–364. [Google Scholar] [CrossRef]
- Gennaro, V.J.; Stanek, T.J.; Peck, A.R.; Sun, Y.; Wang, F.; Qie, S.; Knudsen, K.E.; Rui, H.; Butt, T.; Diehl, J.A.; et al. Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc. Natl. Acad. Sci. USA 2018, 115, E9298–E9307. [Google Scholar] [CrossRef]
- Atanassov, B.S.; Dent, S.Y. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 2011, 12, 924–930. [Google Scholar] [CrossRef]
- Armour, S.M.; Bennett, E.J.; Braun, C.R.; Zhang, X.Y.; McMahon, S.B.; Gygi, S.P.; Harper, J.W.; Sinclair, D.A. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol. Cell Biol. 2013, 33, 1487–1502. [Google Scholar] [CrossRef]
- Lin, Z.; Yang, H.; Kong, Q.; Li, J.; Lee, S.M.; Gao, B.; Dong, H.; Wei, J.; Song, J.; Zhang, D.D.; et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 2012, 46, 484–494. [Google Scholar] [CrossRef]
- Kobayashi, T.; Iwamoto, Y.; Takashima, K.; Isomura, A.; Kosodo, Y.; Kawakami, K.; Nishioka, T.; Kaibuchi, K.; Kageyama, R. Deubiquitinating enzymes regulate Hes1 stability and neuronal differentiation. FEBS J. 2015, 282, 2411–2423. [Google Scholar] [CrossRef]
- Lambies, G.; Miceli, M.; Martinez-Guillamon, C.; Olivera-Salguero, R.; Pena, R.; Frias, C.P.; Calderon, I.; Atanassov, B.S.; Dent, S.Y.R.; Arribas, J.; et al. TGFbeta-Activated USP27X Deubiquitinase Regulates Cell Migration and Chemoresistance via Stabilization of Snail1. Cancer Res. 2019, 79, 33–46. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, P.; Hu, X.; Kim, J.; Yao, F.; Xiao, Z.; Zeng, L.; Chang, L.; Sun, Y.; Ma, L. USP51 promotes deubiquitination and stabilization of ZEB1. Am. J. Cancer Res. 2017, 7, 2020–2031. [Google Scholar]
- McCann, J.J.; Vasilevskaya, I.A.; Poudel Neupane, N.; Shafi, A.A.; McNair, C.; Dylgjeri, E.; Mandigo, A.C.; Schiewer, M.J.; Schrecengost, R.S.; Gallagher, P.; et al. USP22 Functions as an Oncogenic Driver in Prostate Cancer by Regulating Cell Proliferation and DNA Repair. Cancer Res. 2020, 80, 430–443. [Google Scholar] [CrossRef]
- Prokakis, E.; Dyas, A.; Grun, R.; Fritzsche, S.; Bedi, U.; Kazerouni, Z.B.; Kosinsky, R.L.; Johnsen, S.A.; Wegwitz, F. USP22 promotes HER2-driven mammary carcinoma aggressiveness by suppressing the unfolded protein response. Oncogene 2021, 40, 4004–4018. [Google Scholar] [CrossRef]
- Roedig, J.; Kowald, L.; Juretschke, T.; Karlowitz, R.; Ahangarian Abhari, B.; Roedig, H.; Fulda, S.; Beli, P.; van Wijk, S.J. USP22 controls necroptosis by regulating receptor-interacting protein kinase 3 ubiquitination. EMBO Rep. 2021, 22, e50163. [Google Scholar] [CrossRef]
- Stanek, T.J.; Gennaro, V.J.; Tracewell, M.A.; Di Marcantonio, D.; Pauley, K.L.; Butt, S.; McNair, C.; Wang, F.; Kossenkov, A.V.; Knudsen, K.E.; et al. The SAGA complex regulates early steps in transcription via its deubiquitylase module subunit USP22. EMBO J. 2021, 40, e102509. [Google Scholar] [CrossRef]
- Ben Yehuda, A.; Risheq, M.; Novoplansky, O.; Bersuker, K.; Kopito, R.R.; Goldberg, M.; Brandeis, M. Ubiquitin Accumulation on Disease Associated Protein Aggregates Is Correlated with Nuclear Ubiquitin Depletion, Histone De-Ubiquitination and Impaired DNA Damage Response. PLoS ONE 2017, 12, e0169054. [Google Scholar] [CrossRef]
- Mimnaugh, E.G.; Chen, H.Y.; Davie, J.R.; Celis, J.E.; Neckers, L. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: Effects on replication, transcription, translation, and the cellular stress response. Biochemistry 1997, 36, 14418–14429. [Google Scholar] [CrossRef]
- Dantuma, N.P.; Groothuis, T.A.; Salomons, F.A.; Neefjes, J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol. 2006, 173, 19–26. [Google Scholar] [CrossRef]
- Prenzel, T.; Begus-Nahrmann, Y.; Kramer, F.; Hennion, M.; Hsu, C.; Gorsler, T.; Hintermair, C.; Eick, D.; Kremmer, E.; Simons, M.; et al. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res. 2011, 71, 5739–5753. [Google Scholar] [CrossRef]
- Basar, M.A.; Beck, D.B.; Werner, A. Deubiquitylases in developmental ubiquitin signaling and congenital diseases. Cell Death Differ. 2021, 28, 538–556. [Google Scholar] [CrossRef]
- McDonell, L.M.; Mirzaa, G.M.; Alcantara, D.; Schwartzentruber, J.; Carter, M.T.; Lee, L.J.; Clericuzio, C.L.; Graham, J.M., Jr.; Morris-Rosendahl, D.J.; Polster, T.; et al. Mutations in STAMBP, encoding a deubiquitinating enzyme, cause microcephaly-capillary malformation syndrome. Nat. Genet. 2013, 45, 556–562. [Google Scholar] [CrossRef]
- Hao, Y.H.; Fountain, M.D., Jr.; Fon Tacer, K.; Xia, F.; Bi, W.; Kang, S.H.; Patel, A.; Rosenfeld, J.A.; Le Caignec, C.; Isidor, B.; et al. USP7 Acts as a Molecular Rheostat to Promote WASH-Dependent Endosomal Protein Recycling and Is Mutated in a Human Neurodevelopmental Disorder. Mol. Cell 2015, 59, 956–969. [Google Scholar] [CrossRef]
- Homan, C.C.; Kumar, R.; Nguyen, L.S.; Haan, E.; Raymond, F.L.; Abidi, F.; Raynaud, M.; Schwartz, C.E.; Wood, S.A.; Gecz, J.; et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am. J. Hum. Genet. 2014, 94, 470–478. [Google Scholar] [CrossRef]
- Reijnders, M.R.; Zachariadis, V.; Latour, B.; Jolly, L.; Mancini, G.M.; Pfundt, R.; Wu, K.M.; van Ravenswaaij-Arts, C.M.; Veenstra-Knol, H.E.; Anderlid, B.M.; et al. De Novo Loss-of-Function Mutations in USP9X Cause a Female-Specific Recognizable Syndrome with Developmental Delay and Congenital Malformations. Am. J. Hum. Genet. 2016, 98, 373–381. [Google Scholar] [CrossRef]
- Santiago-Sim, T.; Burrage, L.C.; Ebstein, F.; Tokita, M.J.; Miller, M.; Bi, W.; Braxton, A.A.; Rosenfeld, J.A.; Shahrour, M.; Lehmann, A.; et al. Biallelic Variants in OTUD6B Cause an Intellectual Disability Syndrome Associated with Seizures and Dysmorphic Features. Am. J. Hum. Genet. 2017, 100, 676–688. [Google Scholar] [CrossRef]
- Ben-Shachar, S.; Lanpher, B.; German, J.R.; Qasaymeh, M.; Potocki, L.; Nagamani, S.C.; Franco, L.M.; Malphrus, A.; Bottenfield, G.W.; Spence, J.E.; et al. Microdeletion 15q13.3: A locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J. Med. Genet. 2009, 46, 382–388. [Google Scholar] [CrossRef]
- Hu, H.; Haas, S.A.; Chelly, J.; Van Esch, H.; Raynaud, M.; de Brouwer, A.P.; Weinert, S.; Froyen, G.; Frints, S.G.; Laumonnier, F.; et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol. Psychiatry 2016, 21, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; Wang, X. From Discovery to Bedside: Targeting the Ubiquitin System. Cell Chem. Biol. 2019, 26, 156–177. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Saafin, F.; Devys, D.; Johnsen, S.A.; Vincent, S.D.; Tora, L. SAGA-Dependent Histone H2Bub1 Deubiquitination Is Essential for Cellular Ubiquitin Balance during Embryonic Development. Int. J. Mol. Sci. 2022, 23, 7459. https://doi.org/10.3390/ijms23137459
El-Saafin F, Devys D, Johnsen SA, Vincent SD, Tora L. SAGA-Dependent Histone H2Bub1 Deubiquitination Is Essential for Cellular Ubiquitin Balance during Embryonic Development. International Journal of Molecular Sciences. 2022; 23(13):7459. https://doi.org/10.3390/ijms23137459
Chicago/Turabian StyleEl-Saafin, Farrah, Didier Devys, Steven A. Johnsen, Stéphane D. Vincent, and László Tora. 2022. "SAGA-Dependent Histone H2Bub1 Deubiquitination Is Essential for Cellular Ubiquitin Balance during Embryonic Development" International Journal of Molecular Sciences 23, no. 13: 7459. https://doi.org/10.3390/ijms23137459
APA StyleEl-Saafin, F., Devys, D., Johnsen, S. A., Vincent, S. D., & Tora, L. (2022). SAGA-Dependent Histone H2Bub1 Deubiquitination Is Essential for Cellular Ubiquitin Balance during Embryonic Development. International Journal of Molecular Sciences, 23(13), 7459. https://doi.org/10.3390/ijms23137459






