Thirty Years of sRNA-Mediated Regulation in Staphylococcus aureus: From Initial Discoveries to In Vivo Biological Implications
Abstract
1. Introduction
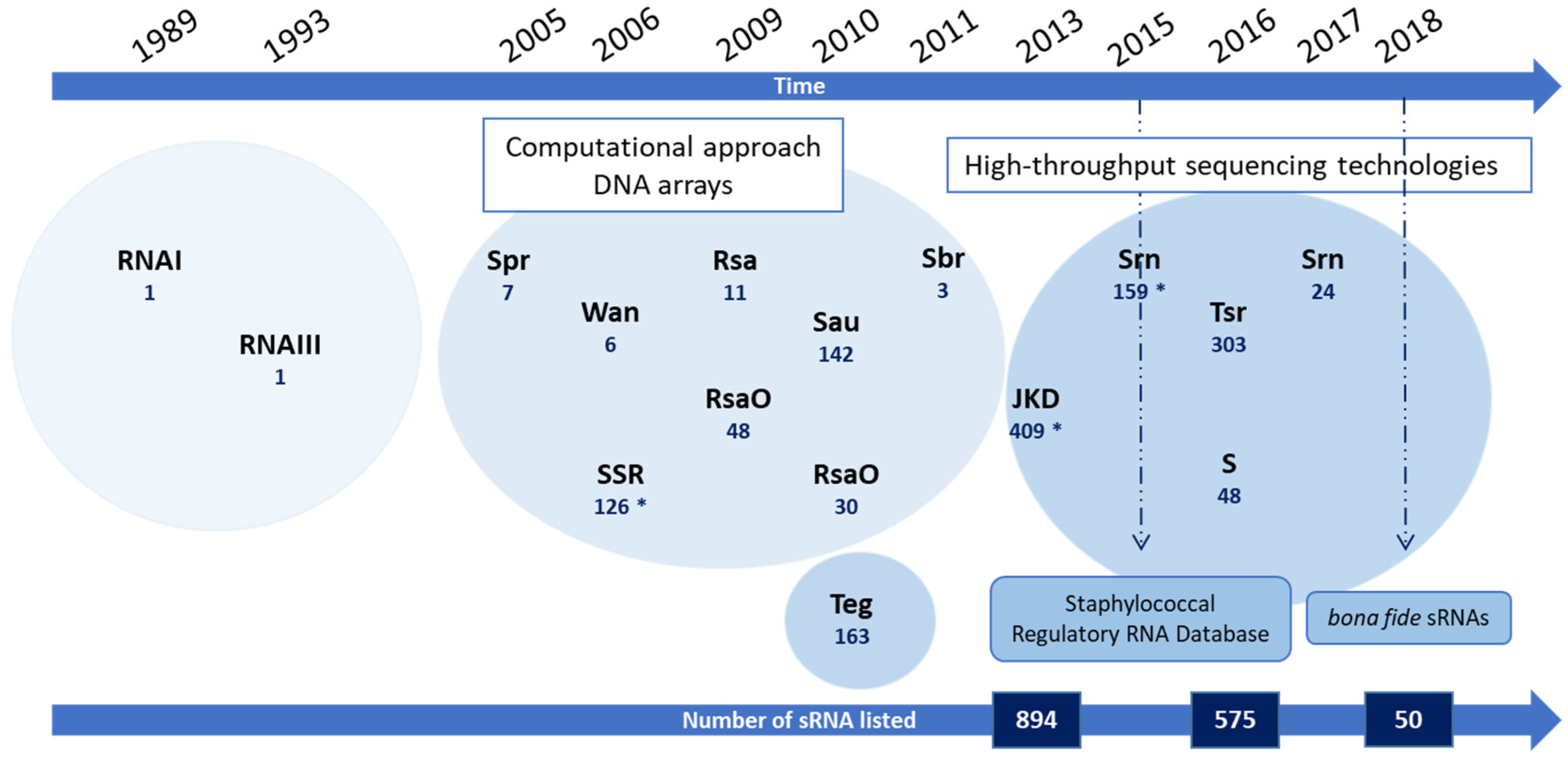
2. From RNAI to Hundreds of sRNA Candidates
| sRNA Family/Name | Signification | Number of sRNA Candidates | Localization | Experimental Approach | Further Validation (Validated) | Strain | Clonal Complex | Reference |
|---|---|---|---|---|---|---|---|---|
| RNAI | 1 | Accessory genome (plasmid) | In silico prediction | Northern blot | NCTC8325 | CC8 | [9] | |
| RNAIII | 1 | Core genome | NB, mutational analyses | NCTC8325 | CC8 | [10,21] | ||
| Spr | Small pathogenicity island RNA | 7 | Accessory genome (pathogenicity islands) | In silico prediction | Northern blot (7) | N315 | CC5 | [15] |
| SSR | Small stable RNAs | 126 | Undefined | DNA arrays (GeneChips) | None | UAMS-1 | CC30 | [16] |
| Wan | Wan | 8 | Undefined | DNA arrays (GeneChips) | Northern blot (8) | N315 | CC5 | [17] |
| Rsa | RNA of S. aureus | 11 | Undefined | In silico prediction | Microrray, Northern blot, RACE (11) | Various strains | [18] | |
| RsaO | RNA of S. aureus Orsay | 48 | Undefined | In silico prediction | Northern blot (7) | N315 | CC5 | [19] |
| RsaO | RNA of S. aureus Orsay | 30 | Undefined | Pyrosequencing | Northern blot (15) | N315 | CC5 | [22] |
| SAU | S. aureus ncRNA | 142 | Undefined | Cloning and sequencing of short cDNAs | Northern blot (18) | A3878 I A3878 III | CC5 | [23] |
| Teg | Transcript from experimental method from Geneva | 163 | Core genome (154 sRNAs) Plasmid (9 sRNAs) | RNA-Seq | RT-qPCR (26) | N315 | CC5 | [12] |
| Sbr | SigB-dependent small RNA | 3 | Core genome | In silico prediction | Northern blot (3) | Various strains | CC8 | [20] |
| JKD sRNA | «JKD6008» S. aureus strains | 409 | Core genome (360 sRNAs) Accessory genome (49 sRNAs) | RNA-Seq | None | JKD6008 JKD6009 | CC8 | [13] |
| Tsr | Tampa small RNA | 39 | Core genome | RNA-Seq | Northern blot (5) | USA300 | CC8 | [24] |
| S | S | 48 | Core genome | Tiling-array | Northern blot (7) | HG001 | CC8 | [25] |
| Srn | Staphylococcal regulatory RNAs | 21 | Core genome (6) Accessory genome (15) | RNA-Seq | Northern blot RT-qPCR (17) | Newman | CC8 | [26] |
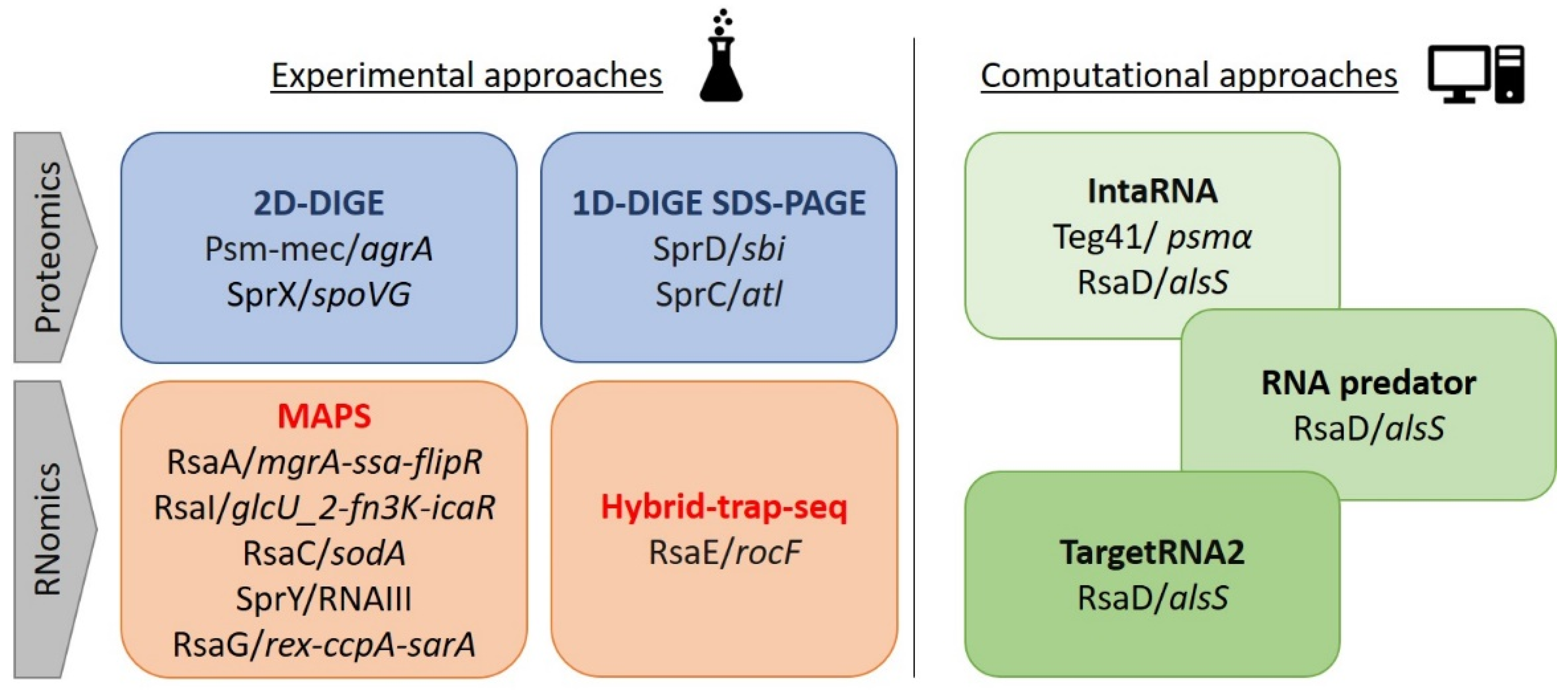
3. On the Quest to Identify sRNA Molecular Targets
3.1. Experimental Approaches
3.2. Computational Tools
4. Insight into the Staphylococcus aureus RNome and Its Functions: Metabolism, Virulence, and Antibiotic Resistance
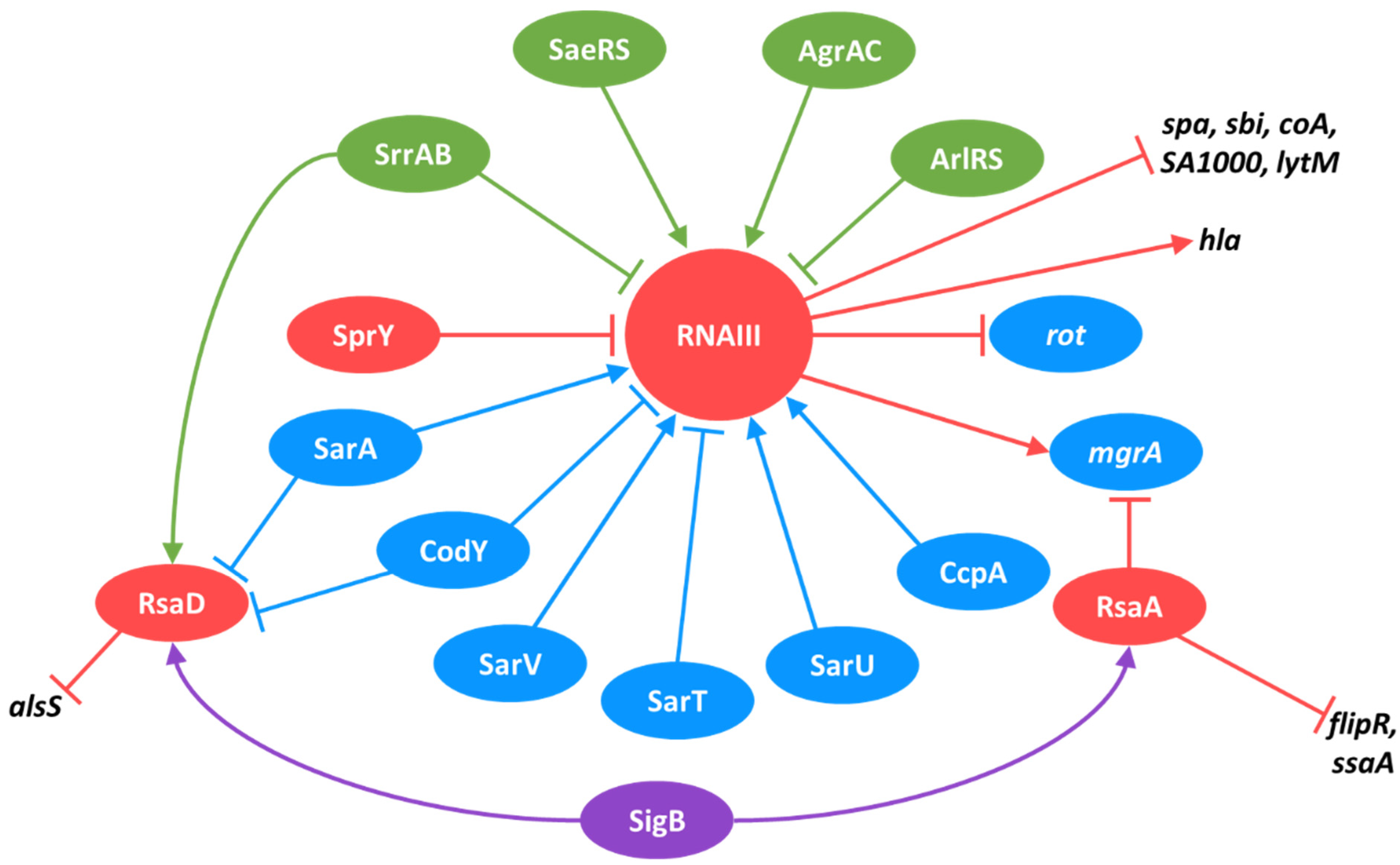
4.1. RNAIII
4.2. Spr sRNAs
4.2.1. SprD
4.2.2. SprC
4.2.3. SprX
4.2.4. Spr sRNAs and Toxin–Antitoxin Systems
4.2.5. Spr sRNAs and the Sponge Mechanism
4.3. Rsa Family
4.3.1. RsaA
4.3.2. RsaC
4.3.3. RsaE
4.3.4. RsaD
4.3.5. RsaI, RsaG, and Nascent Interconnections
4.4. Teg Family
4.5. Other sRNAs
5. In Vivo sRNA Expression in Humans and Animal Models
6. sRNA Interconnections in the Complex Regulatory Network
6.1. The Role of Sigma Factors in Regulatory Network and sRNA Expression
6.2. Two-Component Systems and Their Control over sRNA Expression
6.3. Transcription Factors and Their sRNA Regulons
6.4. sRNAs as Regulators of TCSs, TFs, and Other sRNAs
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, C.M.; Vogel, J. Experimental Approaches for the Discovery and Characterization of Regulatory Small RNA. Curr. Opin. Microbiol. 2009, 12, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.; Srivastava, S. Small RNA-Mediated Regulation in Bacteria: A Growing Palette of Diverse Mechanisms. Gene 2018, 656, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, H.M.; Keogh, R.A.; Wittekind, M.A.; Caillet, A.R.; Wiemels, R.E.; Laner, E.A.; Carroll, R.K. Reading between the Lines: Utilizing RNA-Seq Data for Global Analysis of SRNAs in Staphylococcus aureus. mSphere 2020, 5, e00439-20. [Google Scholar] [CrossRef] [PubMed]
- Carrier, M.-C.; Lalaouna, D.; Massé, E. Broadening the Definition of Bacterial Small RNAs: Characteristics and Mechanisms of Action. Annu. Rev. Microbiol. 2018, 72, 141–161. [Google Scholar] [CrossRef]
- Saberi, F.; Kamali, M.; Najafi, A.; Yazdanparast, A.; Moghaddam, M.M. Natural Antisense RNAs as MRNA Regulatory Elements in Bacteria: A Review on Function and Applications. Cell. Mol. Biol. Lett. 2016, 21, 6. [Google Scholar] [CrossRef]
- Felden, B.; Augagneur, Y. Diversity and Versatility in Small RNA-Mediated Regulation in Bacterial Pathogens. Front. Microbiol. 2021, 12, 719977. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Felden, B.; Vandenesch, F.; Bouloc, P.; Romby, P. The Staphylococcus aureus RNome and Its Commitment to Virulence. PLoS Pathog. 2011, 7, e1002006. [Google Scholar] [CrossRef]
- Novick, R.P.; Iordanescu, S.; Projan, S.J.; Kornblum, J.; Edelman, I. PT181 Plasmid Replication Is Regulated by a Countertranscript-Driven Transcriptional Attenuator. Cell 1989, 59, 395–404. [Google Scholar] [CrossRef]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of Staphylococcal Virulence Factors Is Controlled by a Regulatory RNA Molecule. EMBO J. 1993, 12, 3967–3975. [Google Scholar] [CrossRef]
- Bronesky, D.; Wu, Z.; Marzi, S.; Walter, P.; Geissmann, T.; Moreau, K.; Vandenesch, F.; Caldelari, I.; Romby, P. Staphylococcus aureus RNAIII and Its Regulon Link Quorum Sensing, Stress Responses, Metabolic Adaptation, and Regulation of Virulence Gene Expression. Annu. Rev. Microbiol. 2016, 70, 299–316. [Google Scholar] [CrossRef] [PubMed]
- Beaume, M.; Hernandez, D.; Farinelli, L.; Deluen, C.; Linder, P.; Gaspin, C.; Romby, P.; Schrenzel, J.; Francois, P. Cartography of Methicillin-Resistant S. Aureus Transcripts: Detection, Orientation and Temporal Expression during Growth Phase and Stress Conditions. PLoS ONE 2010, 5, e10725. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Beaume, M.; Harrison, P.F.; Hernandez, D.; Schrenzel, J.; Seemann, T.; Francois, P.; Stinear, T.P. Analysis of the Small RNA Transcriptional Response in Multidrug-Resistant Staphylococcus aureus after Antimicrobial Exposure. Antimicrob. Agents Chemother. 2013, 57, 3864–3874. [Google Scholar] [CrossRef] [PubMed]
- Sassi, M.; Augagneur, Y.; Mauro, T.; Ivain, L.; Chabelskaya, S.; Hallier, M.; Sallou, O.; Felden, B. SRD: A Staphylococcus Regulatory RNA Database. RNA 2015, 21, 1005–1017. [Google Scholar] [CrossRef]
- Pichon, C.; Felden, B. Small RNA Genes Expressed from Staphylococcus aureus Genomic and Pathogenicity Islands with Specific Expression among Pathogenic Strains. Proc. Natl. Acad. Sci. USA 2005, 102, 14249–14254. [Google Scholar] [CrossRef]
- Anderson, K.L.; Roberts, C.; Disz, T.; Vonstein, V.; Hwang, K.; Overbeek, R.; Olson, P.D.; Projan, S.J.; Dunman, P.M. Characterization of the Staphylococcus aureus Heat Shock, Cold Shock, Stringent, and SOS Responses and Their Effects on Log-Phase MRNA Turnover. J. Bacteriol. 2006, 188, 6739–6756. [Google Scholar] [CrossRef]
- Roberts, C.; Anderson, K.L.; Murphy, E.; Projan, S.J.; Mounts, W.; Hurlburt, B.; Smeltzer, M.; Overbeek, R.; Disz, T.; Dunman, P.M. Characterizing the Effect of the Staphylococcus aureus Virulence Factor Regulator, SarA, on Log-Phase MRNA Half-Lives. J. Bacteriol. 2006, 188, 2593–2603. [Google Scholar] [CrossRef]
- Geissmann, T.; Chevalier, C.; Cros, M.-J.; Boisset, S.; Fechter, P.; Noirot, C.; Schrenzel, J.; François, P.; Vandenesch, F.; Gaspin, C.; et al. A Search for Small Noncoding RNAs in Staphylococcus aureus Reveals a Conserved Sequence Motif for Regulation. Nucleic Acids Res. 2009, 37, 7239–7257. [Google Scholar] [CrossRef]
- Marchais, A.; Naville, M.; Bohn, C.; Bouloc, P.; Gautheret, D. Single-Pass Classification of All Noncoding Sequences in a Bacterial Genome Using Phylogenetic Profiles. Genome Res. 2009, 19, 1084–1092. [Google Scholar] [CrossRef]
- Nielsen, J.S.; Christiansen, M.H.G.; Bonde, M.; Gottschalk, S.; Frees, D.; Thomsen, L.E.; Kallipolitis, B.H. Searching for Small ΣB-Regulated Genes in Staphylococcus aureus. Arch. Microbiol. 2011, 193, 23–34. [Google Scholar] [CrossRef]
- Janzon, L.; Löfdahl, S.; Arvidson, S. Identification and Nucleotide Sequence of the Delta-Lysin Gene, Hld, Adjacent to the Accessory Gene Regulator (Agr) of Staphylococcus aureus. Mol. Gen. Genet. 1989, 219, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Bohn, C.; Rigoulay, C.; Chabelskaya, S.; Sharma, C.M.; Marchais, A.; Skorski, P.; Borezée-Durant, E.; Barbet, R.; Jacquet, E.; Jacq, A.; et al. Experimental Discovery of Small RNAs in Staphylococcus aureus Reveals a Riboregulator of Central Metabolism. Nucleic Acids Res. 2010, 38, 6620–6636. [Google Scholar] [CrossRef] [PubMed]
- Abu-Qatouseh, L.F.; Chinni, S.V.; Seggewiss, J.; Proctor, R.A.; Brosius, J.; Rozhdestvensky, T.S.; Peters, G.; von Eiff, C.; Becker, K. Identification of Differentially Expressed Small Non-Protein-Coding RNAs in Staphylococcus aureus Displaying Both the Normal and the Small-Colony Variant Phenotype. J. Mol. Med. 2010, 88, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Carroll, R.K.; Weiss, A.; Broach, W.H.; Wiemels, R.E.; Mogen, A.B.; Rice, K.C.; Shaw, L.N. Genome-Wide Annotation, Identification, and Global Transcriptomic Analysis of Regulatory or Small RNA Gene Expression in Staphylococcus aureus. mBio 2016, 7, e01990-15. [Google Scholar] [CrossRef] [PubMed]
- Mäder, U.; Nicolas, P.; Depke, M.; Pané-Farré, J.; Debarbouille, M.; van der Kooi-Pol, M.M.; Guérin, C.; Dérozier, S.; Hiron, A.; Jarmer, H.; et al. Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions. PLoS Genet. 2016, 12, e1005962. [Google Scholar] [CrossRef]
- Bronsard, J.; Pascreau, G.; Sassi, M.; Mauro, T.; Augagneur, Y.; Felden, B. SRNA and Cis-Antisense SRNA Identification in Staphylococcus aureus Highlights an Unusual SRNA Gene Cluster with One Encoding a Secreted Peptide. Sci. Rep. 2017, 7, 4565. [Google Scholar] [CrossRef]
- Li, L.; Huang, D.; Cheung, M.K.; Nong, W.; Huang, Q.; Kwan, H.S. BSRD: A Repository for Bacterial Small Regulatory RNA. Nucleic Acids Res. 2013, 41, D233–D238. [Google Scholar] [CrossRef]
- Liu, W.; Rochat, T.; Toffano-Nioche, C.; Le Lam, T.N.; Bouloc, P.; Morvan, C. Assessment of Bona Fide SRNAs in Staphylococcus aureus. Front. Microbiol. 2018, 9, 228. [Google Scholar] [CrossRef]
- Bastock, R.A.; Marino, E.C.; Wiemels, R.E.; Holzschu, D.L.; Keogh, R.A.; Zapf, R.L.; Murphy, E.R.; Carroll, R.K. Staphylococcus aureus Responds to Physiologically Relevant Temperature Changes by Altering Its Global Transcript and Protein Profile. mSphere 2021, 6, e01303-20. [Google Scholar] [CrossRef]
- Tonge, R.; Shaw, J.; Middleton, B.; Rowlinson, R.; Rayner, S.; Young, J.; Pognan, F.; Hawkins, E.; Currie, I.; Davison, M. Validation and Development of Fluorescence Two-Dimensional Differential Gel Electrophoresis Proteomics Technology. Proteomics 2001, 1, 377–396. [Google Scholar] [CrossRef]
- Kaito, C.; Saito, Y.; Ikuo, M.; Omae, Y.; Mao, H.; Nagano, G.; Fujiyuki, T.; Numata, S.; Han, X.; Obata, K.; et al. Mobile Genetic Element SCCmec-Encoded Psm-Mec RNA Suppresses Translation of AgrA and Attenuates MRSA Virulence. PLoS Pathog. 2013, 9, e1003269. [Google Scholar] [CrossRef] [PubMed]
- Eyraud, A.; Tattevin, P.; Chabelskaya, S.; Felden, B. A Small RNA Controls a Protein Regulator Involved in Antibiotic Resistance in Staphylococcus aureus. Nucleic Acids Res. 2014, 42, 4892–4905. [Google Scholar] [CrossRef] [PubMed]
- Chabelskaya, S.; Gaillot, O.; Felden, B. A Staphylococcus aureus Small RNA Is Required for Bacterial Virulence and Regulates the Expression of an Immune-Evasion Molecule. PLoS Pathog. 2010, 6, e1000927. [Google Scholar] [CrossRef]
- Le Pabic, H.; Germain-Amiot, N.; Bordeau, V.; Felden, B. A Bacterial Regulatory RNA Attenuates Virulence, Spread and Human Host Cell Phagocytosis. Nucleic Acids Res. 2015, 43, 9232–9248. [Google Scholar] [CrossRef] [PubMed]
- Desgranges, E.; Caldelari, I.; Marzi, S.; Lalaouna, D. Navigation through the Twists and Turns of RNA Sequencing Technologies: Application to Bacterial Regulatory RNAs. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194506. [Google Scholar] [CrossRef]
- Lalaouna, D.; Massé, E. Identification of SRNA Interacting with a Transcript of Interest Using MS2-Affinity Purification Coupled with RNA Sequencing (MAPS) Technology. Genom. Data 2015, 5, 136–138. [Google Scholar] [CrossRef]
- Bohn, C.; Rigoulay, C.; Bouloc, P. No Detectable Effect of RNA-Binding Protein Hfq Absence in Staphylococcus aureus. BMC Microbiol. 2007, 7, 10. [Google Scholar] [CrossRef]
- Tomasini, A.; Moreau, K.; Chicher, J.; Geissmann, T.; Vandenesch, F.; Romby, P.; Marzi, S.; Caldelari, I. The RNA Targetome of Staphylococcus aureus Non-Coding RNA RsaA: Impact on Cell Surface Properties and Defense Mechanisms. Nucleic Acids Res. 2017, 45, 6746–6760. [Google Scholar] [CrossRef]
- Bronesky, D.; Desgranges, E.; Corvaglia, A.; François, P.; Caballero, C.J.; Prado, L.; Toledo-Arana, A.; Lasa, I.; Moreau, K.; Vandenesch, F.; et al. A Multifaceted Small RNA Modulates Gene Expression upon Glucose Limitation in Staphylococcus aureus. EMBO J. 2019, 38, e99363. [Google Scholar] [CrossRef]
- Lalaouna, D.; Baude, J.; Wu, Z.; Tomasini, A.; Chicher, J.; Marzi, S.; Vandenesch, F.; Romby, P.; Caldelari, I.; Moreau, K. RsaC SRNA Modulates the Oxidative Stress Response of Staphylococcus aureus during Manganese Starvation. Nucleic Acids Res. 2019, 47, 9871–9887. [Google Scholar] [CrossRef]
- Le Huyen, K.B.; Gonzalez, C.D.; Pascreau, G.; Bordeau, V.; Cattoir, V.; Liu, W.; Bouloc, P.; Felden, B.; Chabelskaya, S. A Small Regulatory RNA Alters Staphylococcus aureus Virulence by Titrating RNAIII Activity. Nucleic Acids Res. 2021, 49, 10644–10656. [Google Scholar] [CrossRef] [PubMed]
- Desgranges, E.; Barrientos, L.; Herrgott, L.; Marzi, S.; Toledo-Arana, A.; Moreau, K.; Vandenesch, F.; Romby, P.; Caldelari, I. The 3’UTR-Derived SRNA RsaG Coordinates Redox Homeostasis and Metabolism Adaptation in Response to Glucose-6-Phosphate Uptake in Staphylococcus aureus. Mol. Microbiol. 2022, 117, 193–214. [Google Scholar] [CrossRef] [PubMed]
- Rochat, T.; Bohn, C.; Morvan, C.; Le Lam, T.N.; Razvi, F.; Pain, A.; Toffano-Nioche, C.; Ponien, P.; Jacq, A.; Jacquet, E.; et al. The Conserved Regulatory RNA RsaE Down-Regulates the Arginine Degradation Pathway in Staphylococcus aureus. Nucleic Acids Res. 2018, 46, 8803–8816. [Google Scholar] [CrossRef]
- Melamed, S.; Faigenbaum-Romm, R.; Peer, A.; Reiss, N.; Shechter, O.; Bar, A.; Altuvia, Y.; Argaman, L.; Margalit, H. Mapping the Small RNA Interactome in Bacteria Using RIL-Seq. Nat. Protoc. 2018, 13, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Tjaden, B.; Lory, S. GRIL-Seq Provides a Method for Identifying Direct Targets of Bacterial Small Regulatory RNA by in Vivo Proximity Ligation. Nat. Microbiol. 2016, 2, 16239. [Google Scholar] [CrossRef]
- Zhang, Y.-F.; Han, K.; Chandler, C.E.; Tjaden, B.; Ernst, R.K.; Lory, S. Probing the SRNA Regulatory Landscape of P. Aeruginosa: Post-Transcriptional Control of Determinants of Pathogenicity and Antibiotic Susceptibility. Mol. Microbiol. 2017, 106, 919–937. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, K.; Xu, S.; Wang, Z.; Fu, H.; Tian, B.; Zheng, X.; Li, W. Detecting RNA-RNA Interactions in E. coli Using a Modified CLASH Method. BMC Genom. 2017, 18, 343. [Google Scholar] [CrossRef][Green Version]
- Wright, P.R.; Georg, J. Workflow for a Computational Analysis of an SRNA Candidate in Bacteria. Methods Mol. Biol. 2018, 1737, 3–30. [Google Scholar] [CrossRef]
- Busch, A.; Richter, A.S.; Backofen, R. IntaRNA: Efficient Prediction of Bacterial SRNA Targets Incorporating Target Site Accessibility and Seed Regions. Bioinformatics 2008, 24, 2849–2856. [Google Scholar] [CrossRef]
- Zapf, R.L.; Wiemels, R.E.; Keogh, R.A.; Holzschu, D.L.; Howell, K.M.; Trzeciak, E.; Caillet, A.R.; King, K.A.; Selhorst, S.A.; Naldrett, M.J.; et al. The Small RNA Teg41 Regulates Expression of the Alpha Phenol-Soluble Modulins and Is Required for Virulence in Staphylococcus aureus. mBio 2019, 10, e02484-18. [Google Scholar] [CrossRef]
- Augagneur, Y.; King, A.N.; Germain-Amiot, N.; Sassi, M.; Fitzgerald, J.W.; Sahukhal, G.S.; Elasri, M.O.; Felden, B.; Brinsmade, S.R. Analysis of the CodY RNome Reveals RsaD as a Stress-Responsive Riboregulator of Overflow Metabolism in Staphylococcus aureus. Mol. Microbiol. 2020, 113, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Eggenhofer, F.; Tafer, H.; Stadler, P.F.; Hofacker, I.L. RNApredator: Fast Accessibility-Based Prediction of SRNA Targets. Nucleic Acids Res. 2011, 39, W149–W154. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.R.; Richter, A.S.; Papenfort, K.; Mann, M.; Vogel, J.; Hess, W.R.; Backofen, R.; Georg, J. Comparative Genomics Boosts Target Prediction for Bacterial Small RNAs. Proc. Natl. Acad. Sci. USA 2013, 110, E3487–E3496. [Google Scholar] [CrossRef] [PubMed]
- Kery, M.B.; Feldman, M.; Livny, J.; Tjaden, B. TargetRNA2: Identifying Targets of Small Regulatory RNAs in Bacteria. Nucleic Acids Res. 2014, 42, W124–W129. [Google Scholar] [CrossRef] [PubMed]
- Georg, J.; Lalaouna, D.; Hou, S.; Lott, S.C.; Caldelari, I.; Marzi, S.; Hess, W.R.; Romby, P. The Power of Cooperation: Experimental and Computational Approaches in the Functional Characterization of Bacterial SRNAs. Mol. Microbiol. 2020, 113, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Luz, B.S.R.D.; Nicolas, A.; Chabelskaya, S.; Rodovalho, V.D.R.; Le Loir, Y.; Azevedo, V.A.D.C.; Felden, B.; Guédon, E. Environmental Plasticity of the RNA Content of Staphylococcus aureus Extracellular Vesicles. Front. Microbiol. 2021, 12, 634226. [Google Scholar] [CrossRef]
- Raina, M.; King, A.; Bianco, C.; Vanderpool, C.K. Dual-Function RNAs. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription Profiling-Based Identification of Staphylococcus aureus Genes Regulated by the Agr and/or SarA Loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef]
- Le, K.Y.; Otto, M. Quorum-Sensing Regulation in Staphylococci-an Overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef]
- Morfeldt, E.; Taylor, D.; von Gabain, A.; Arvidson, S. Activation of Alpha-Toxin Translation in Staphylococcus aureus by the Trans-Encoded Antisense RNA, RNAIII. EMBO J. 1995, 14, 4569–4577. [Google Scholar] [CrossRef]
- Huntzinger, E.; Boisset, S.; Saveanu, C.; Benito, Y.; Geissmann, T.; Namane, A.; Lina, G.; Etienne, J.; Ehresmann, B.; Ehresmann, C.; et al. Staphylococcus aureus RNAIII and the Endoribonuclease III Coordinately Regulate Spa Gene Expression. EMBO J. 2005, 24, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Boisset, S.; Geissmann, T.; Huntzinger, E.; Fechter, P.; Bendridi, N.; Possedko, M.; Chevalier, C.; Helfer, A.C.; Benito, Y.; Jacquier, A.; et al. Staphylococcus aureus RNAIII Coordinately Represses the Synthesis of Virulence Factors and the Transcription Regulator Rot by an Antisense Mechanism. Genes Dev. 2007, 21, 1353–1366. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, C.; Boisset, S.; Romilly, C.; Masquida, B.; Fechter, P.; Geissmann, T.; Vandenesch, F.; Romby, P. Staphylococcus aureus RNAIII Binds to Two Distant Regions of Coa MRNA to Arrest Translation and Promote MRNA Degradation. PLoS Pathog. 2010, 6, e1000809. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mu, C.; Ying, X.; Li, W.; Wu, N.; Dong, J.; Gao, Y.; Shao, N.; Fan, M.; Yang, G. RNAIII Activates Map Expression by Forming an RNA-RNA Complex in Staphylococcus aureus. FEBS Lett. 2011, 585, 899–905. [Google Scholar] [CrossRef]
- Chunhua, M.; Yu, L.; Yaping, G.; Jie, D.; Qiang, L.; Xiaorong, T.; Guang, Y. The Expression of LytM Is Down-Regulated by RNAIII in Staphylococcus aureus. J. Basic Microbiol. 2012, 52, 636–641. [Google Scholar] [CrossRef]
- Chabelskaya, S.; Bordeau, V.; Felden, B. Dual RNA Regulatory Control of a Staphylococcus aureus Virulence Factor. Nucleic Acids Res. 2014, 42, 4847–4858. [Google Scholar] [CrossRef]
- Gupta, R.K.; Luong, T.T.; Lee, C.Y. RNAIII of the Staphylococcus aureus Agr System Activates Global Regulator MgrA by Stabilizing MRNA. Proc. Natl. Acad. Sci. USA 2015, 112, 14036–14041. [Google Scholar] [CrossRef]
- Xue, T.; Zhang, X.; Sun, H.; Sun, B. ArtR, a Novel SRNA of Staphylococcus aureus, Regulates α-Toxin Expression by Targeting the 5’ UTR of SarT MRNA. Med. Microbiol. Immunol. 2014, 203, 1–12. [Google Scholar] [CrossRef]
- Buchad, H.; Nair, M. The Small RNA SprX Regulates the Autolysin Regulator WalR in Staphylococcus aureus. Microbiol. Res. 2021, 250, 126785. [Google Scholar] [CrossRef]
- Gordon, R.J.; Lowy, F.D. Pathogenesis of Methicillin-Resistant Staphylococcus aureus Infection. Clin. Infect. Dis. 2008, 46, S350–S359. [Google Scholar] [CrossRef]
- Cheung, A.L.; Eberhardt, K.J.; Chung, E.; Yeaman, M.R.; Sullam, P.M.; Ramos, M.; Bayer, A.S. Diminished Virulence of a Sar-/Agr- Mutant of Staphylococcus aureus in the Rabbit Model of Endocarditis. J. Clin. Investig. 1994, 94, 1815–1822. [Google Scholar] [CrossRef] [PubMed]
- Mayville, P.; Ji, G.; Beavis, R.; Yang, H.; Goger, M.; Novick, R.P.; Muir, T.W. Structure-Activity Analysis of Synthetic Autoinducing Thiolactone Peptides from Staphylococcus aureus Responsible for Virulence. Proc. Natl. Acad. Sci. USA 1999, 96, 1218–1223. [Google Scholar] [CrossRef] [PubMed]
- Schwan, W.R.; Langhorne, M.H.; Ritchie, H.D.; Stover, C.K. Loss of Hemolysin Expression in Staphylococcus aureus Agr Mutants Correlates with Selective Survival during Mixed Infections in Murine Abscesses and Wounds. FEMS Immunol. Med. Microbiol. 2003, 38, 23–28. [Google Scholar] [CrossRef]
- Wright, J.S.; Jin, R.; Novick, R.P. Transient Interference with Staphylococcal Quorum Sensing Blocks Abscess Formation. Proc. Natl. Acad. Sci. USA 2005, 102, 1691–1696. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, C.P.; Boyle-Vavra, S.; Daum, R.S. Importance of the Global Regulators Agr and SaeRS in the Pathogenesis of CA-MRSA USA300 Infection. PLoS ONE 2010, 5, e15177. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Li, D.; Yan, J.; Liu, Y.; Li, D.; Dong, J.; Gao, Y.; Sun, T.; Yang, G. The Accessory Gene Regulator (Agr) Controls Staphylococcus aureus Virulence in a Murine Intracranial Abscesses Model. Braz. J. Infect. Dis. 2014, 18, 501–506. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and Virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Proctor, R.A.; von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small Colony Variants: A Pathogenic Form of Bacteria That Facilitates Persistent and Recurrent Infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef]
- Fowler, V.G.; Sakoulas, G.; McIntyre, L.M.; Meka, V.G.; Arbeit, R.D.; Cabell, C.H.; Stryjewski, M.E.; Eliopoulos, G.M.; Reller, L.B.; Corey, G.R.; et al. Persistent Bacteremia Due to Methicillin-Resistant Staphylococcus aureus Infection Is Associated with Agr Dysfunction and Low-Level In Vitro Resistance to Thrombin-Induced Platelet Microbicidal Protein. J. Infect. Dis. 2004, 190, 1140–1149. [Google Scholar] [CrossRef]
- Proctor, R.A.; Kriegeskorte, A.; Kahl, B.C.; Becker, K.; Löffler, B.; Peters, G. Staphylococcus aureus Small Colony Variants (SCVs): A Road Map for the Metabolic Pathways Involved in Persistent Infections. Front. Cell. Infect. Microbiol. 2014, 4, 99. [Google Scholar] [CrossRef]
- Chaves-Moreno, D.; Wos-Oxley, M.L.; Jáuregui, R.; Medina, E.; Oxley, A.P.; Pieper, D.H. Exploring the Transcriptome of Staphylococcus aureus in Its Natural Niche. Sci. Rep. 2016, 6, 33174. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Le, K.Y.; Khan, B.A.; Nguyen, T.H.; Hunt, R.L.; Bae, J.S.; Kabat, J.; Zheng, Y.; Cheung, G.Y.C.; Li, M.; et al. Resistance to Leukocytes Ties Benefits of Quorum Sensing Dysfunctionality to Biofilm Infection. Nat. Microbiol. 2019, 4, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.C.; Bae, J.S.; Liu, R.; Hunt, R.L.; Zheng, Y.; Otto, M. Bacterial Virulence Plays a Crucial Role in MRSA Sepsis. PLoS Pathog. 2021, 17, e1009369. [Google Scholar] [CrossRef]
- Nitzan, M.; Fechter, P.; Peer, A.; Altuvia, Y.; Bronesky, D.; Vandenesch, F.; Romby, P.; Biham, O.; Margalit, H. A Defense-Offense Multi-Layered Regulatory Switch in a Pathogenic Bacterium. Nucleic Acids Res. 2015, 43, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.J.; Visai, L.; Kerrigan, S.W.; Speziale, P.; Foster, T.J. The Sbi Protein Is a Multifunctional Immune Evasion Factor of Staphylococcus aureus. Infect. Immun. 2011, 79, 3801–3809. [Google Scholar] [CrossRef] [PubMed]
- Mauro, T.; Rouillon, A.; Felden, B. Insights into the Regulation of Small RNA Expression: SarA Represses the Expression of Two SRNAs in Staphylococcus aureus. Nucleic Acids Res. 2016, 44, 10186–10200. [Google Scholar] [CrossRef]
- Zhao, H.; Hu, F.; Yang, H.; Ding, B.; Xu, X.; He, C.; Cui, Z.; Shu, W.; Liu, Q. Isobaric Tags for Relative and Absolute Quantitation Proteomics Analysis of Gene Regulation by SprC in Staphylococcus aureus. Future Microbiol. 2017, 12, 1181–1199. [Google Scholar] [CrossRef]
- Ménard, G.; Rouillon, A.; Ghukasyan, G.; Emily, M.; Felden, B.; Donnio, P.-Y. Galleria Mellonella Larvae as an Infection Model to Investigate SRNA-Mediated Pathogenesis in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2021, 11, 631710. [Google Scholar] [CrossRef]
- Dejoies, L.; Le Neindre, K.; Reissier, S.; Felden, B.; Cattoir, V. Distinct Expression Profiles of Regulatory RNAs in the Response to Biocides in Staphylococcus aureus and Enterococcus faecium. Sci. Rep. 2021, 11, 6892. [Google Scholar] [CrossRef]
- Ivain, L.; Bordeau, V.; Eyraud, A.; Hallier, M.; Dreano, S.; Tattevin, P.; Felden, B.; Chabelskaya, S. An in Vivo Reporter Assay for SRNA-Directed Gene Control in Gram-Positive Bacteria: Identifying a Novel SRNA Target in Staphylococcus aureus. Nucleic Acids Res. 2017, 45, 4994–5007. [Google Scholar] [CrossRef]
- Chavakis, T.; Wiechmann, K.; Preissner, K.T.; Herrmann, M. Staphylococcus aureus Interactions with the Endothelium: The Role of Bacterial “Secretable Expanded Repertoire Adhesive Molecules” (SERAM) in Disturbing Host Defense Systems. Thromb. Haemost. 2005, 94, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Kathirvel, M.; Buchad, H.; Nair, M. Enhancement of the Pathogenicity of Staphylococcus aureus Strain Newman by a Small Noncoding RNA SprX1. Med. Microbiol. Immunol. 2016, 205, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Delauné, A.; Dubrac, S.; Blanchet, C.; Poupel, O.; Mäder, U.; Hiron, A.; Leduc, A.; Fitting, C.; Nicolas, P.; Cavaillon, J.-M.; et al. The WalKR System Controls Major Staphylococcal Virulence Genes and Is Involved in Triggering the Host Inflammatory Response. Infect. Immun. 2012, 80, 3438–3453. [Google Scholar] [CrossRef] [PubMed]
- Sayed, N.; Jousselin, A.; Felden, B. A Cis-Antisense RNA Acts in Trans in Staphylococcus aureus to Control Translation of a Human Cytolytic Peptide. Nat. Struct. Mol. Biol. 2011, 19, 105–112. [Google Scholar] [CrossRef]
- Pinel-Marie, M.-L.; Brielle, R.; Felden, B. Dual Toxic-Peptide-Coding Staphylococcus aureus RNA under Antisense Regulation Targets Host Cells and Bacterial Rivals Unequally. Cell Rep. 2014, 7, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Germain-Amiot, N.; Augagneur, Y.; Camberlein, E.; Nicolas, I.; Lecureur, V.; Rouillon, A.; Felden, B. A Novel Staphylococcus aureus Cis-Trans Type I Toxin-Antitoxin Module with Dual Effects on Bacteria and Host Cells. Nucleic Acids Res. 2019, 47, 1759–1773. [Google Scholar] [CrossRef]
- Riffaud, C.; Pinel-Marie, M.-L.; Pascreau, G.; Felden, B. Functionality and Cross-Regulation of the Four SprG/SprF Type I Toxin-Antitoxin Systems in Staphylococcus aureus. Nucleic Acids Res. 2019, 47, 1740–1758. [Google Scholar] [CrossRef]
- Riffaud, C.; Pinel-Marie, M.-L.; Felden, B. Cross-Regulations between Bacterial Toxin-Antitoxin Systems: Evidence of an Interconnected Regulatory Network? Trends Microbiol. 2020, 28, 851–866. [Google Scholar] [CrossRef]
- Brielle, R.; Pinel-Marie, M.-L.; Felden, B. Linking Bacterial Type I Toxins with Their Actions. Curr. Opin. Microbiol. 2016, 30, 114–121. [Google Scholar] [CrossRef]
- Pinel-Marie, M.-L.; Brielle, R.; Riffaud, C.; Germain-Amiot, N.; Polacek, N.; Felden, B. RNA Antitoxin SprF1 Binds Ribosomes to Attenuate Translation and Promote Persister Cell Formation in Staphylococcus aureus. Nat. Microbiol. 2021, 6, 209–220. [Google Scholar] [CrossRef]
- Chlebicka, K.; Bonar, E.; Suder, P.; Ostyn, E.; Felden, B.; Wladyka, B.; Pinel-Marie, M.-L. Impacts of the Type I Toxin-Antitoxin System, SprG1/SprF1, on Staphylococcus aureus Gene Expression. Genes 2021, 12, 770. [Google Scholar] [CrossRef] [PubMed]
- Peyrusson, F.; Varet, H.; Nguyen, T.K.; Legendre, R.; Sismeiro, O.; Coppée, J.-Y.; Wolz, C.; Tenson, T.; Van Bambeke, F. Intracellular Staphylococcus aureus Persisters upon Antibiotic Exposure. Nat. Commun. 2020, 11, 2200. [Google Scholar] [CrossRef] [PubMed]
- Desgranges, E.; Marzi, S.; Moreau, K.; Romby, P.; Caldelari, I. Noncoding RNA. Microbiol. Spectr. 2019, 7, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, L.; Mercier, N.; Lalaouna, D.; Caldelari, I. Assembling the Current Pieces: The Puzzle of RNA-Mediated Regulation in Staphylococcus aureus. Front. Microbiol. 2021, 12, 706690. [Google Scholar] [CrossRef]
- Mercier, N.; Prévost, K.; Massé, E.; Romby, P.; Caldelari, I.; Lalaouna, D. MS2-Affinity Purification Coupled with RNA Sequencing in Gram-Positive Bacteria. J. Vis. Exp. 2021, 168, e61731. [Google Scholar] [CrossRef]
- Resch, A.; Rosenstein, R.; Nerz, C.; Götz, F. Differential Gene Expression Profiling of Staphylococcus aureus Cultivated under Biofilm and Planktonic Conditions. Appl. Environ. Microbiol. 2005, 71, 2663–2676. [Google Scholar] [CrossRef]
- Prat, C.; Bestebroer, J.; de Haas, C.J.C.; van Strijp, J.A.G.; van Kessel, K.P.M. A New Staphylococcal Anti-Inflammatory Protein That Antagonizes the Formyl Peptide Receptor-like 1. J. Immunol. 2006, 177, 8017–8026. [Google Scholar] [CrossRef]
- Panthee, S.; Hamamoto, H.; Paudel, A.; Ohgi, S.; Sekimizu, K. Contribution of RsaC, a Small Non-Coding RNA, towards the Pathogenicity of Staphylococcus aureus in a Mouse Systemic Infection Model. bioRxiv 2021. [Google Scholar] [CrossRef]
- Schoenfelder, S.M.K.; Lange, C.; Prakash, S.A.; Marincola, G.; Lerch, M.F.; Wencker, F.D.R.; Förstner, K.U.; Sharma, C.M.; Ziebuhr, W. The Small Non-Coding RNA RsaE Influences Extracellular Matrix Composition in Staphylococcus Epidermidis Biofilm Communities. PLoS Pathog. 2019, 15, e1007618. [Google Scholar] [CrossRef]
- Lin, M.H.; Shu, J.C.; Lin, L.P.; Chong, K.Y.; Cheng, Y.W.; Du, J.F.; Liu, S.-T. Elucidating the Crucial Role of Poly N-Acetylglucosamine from Staphylococcus aureus in Cellular Adhesion and Pathogenesis. PLoS ONE 2015, 10, e0124216. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Marroquin, S.; Gimza, B.; Tomlinson, B.; Stein, M.; Frey, A.; Keogh, R.A.; Zapf, R.; Todd, D.A.; Cech, N.B.; Carroll, R.K.; et al. MroQ Is a Novel Abi-Domain Protein That Influences Virulence Gene Expression in Staphylococcus aureus via Modulation of Agr Activity. Infect. Immun. 2019, 87, e00002-19. [Google Scholar] [CrossRef] [PubMed]
- Waters, N.R.; Samuels, D.J.; Behera, R.K.; Livny, J.; Rhee, K.Y.; Sadykov, M.R.; Brinsmade, S.R. A Spectrum of CodY Activities Drives Metabolic Reorganization and Virulence Gene Expression in Staphylococcus aureus. Mol. Microbiol. 2016, 101, 495–514. [Google Scholar] [CrossRef] [PubMed]
- Harper, L.; Balasubramanian, D.; Ohneck, E.A.; Sause, W.E.; Chapman, J.; Mejia-Sosa, B.; Lhakhang, T.; Heguy, A.; Tsirigos, A.; Ueberheide, B.; et al. Staphylococcus aureus Responds to the Central Metabolite Pyruvate To Regulate Virulence. mBio 2018, 9, e02272-17. [Google Scholar] [CrossRef]
- Richardson, A.R.; Somerville, G.A.; Sonenshein, A.L. Regulating the Intersection of Metabolism and Pathogenesis in Gram-Positive Bacteria. Microbiol. Spectr. 2015, 3, 3. [Google Scholar] [CrossRef]
- Somerville, G.A.; Saïd-Salim, B.; Wickman, J.M.; Raffel, S.J.; Kreiswirth, B.N.; Musser, J.M. Correlation of Acetate Catabolism and Growth Yield in Staphylococcus aureus: Implications for Host-Pathogen Interactions. Infect. Immun. 2003, 71, 4724–4732. [Google Scholar] [CrossRef]
- Seidl, K.; Stucki, M.; Ruegg, M.; Goerke, C.; Wolz, C.; Harris, L.; Berger-Bächi, B.; Bischoff, M. Staphylococcus aureus CcpA Affects Virulence Determinant Production and Antibiotic Resistance. Antimicrob. Agents Chemother. 2006, 50, 1183–1194. [Google Scholar] [CrossRef]
- Zhu, Y.; Nandakumar, R.; Sadykov, M.R.; Madayiputhiya, N.; Luong, T.T.; Gaupp, R.; Lee, C.Y.; Somerville, G.A. RpiR Homologues May Link Staphylococcus aureus RNAIII Synthesis and Pentose Phosphate Pathway Regulation. J. Bacteriol. 2011, 193, 6187–6196. [Google Scholar] [CrossRef]
- Patel, N.; Nair, M. The Small RNA RsaF Regulates the Expression of Secreted Virulence Factors in Staphylococcus aureus Newman. J. Microbiol. 2021, 59, 920–930. [Google Scholar] [CrossRef]
- Kim, S.; Reyes, D.; Beaume, M.; Francois, P.; Cheung, A. Contribution of Teg49 Small RNA in the 5′ Upstream Transcriptional Region of SarA to Virulence in Staphylococcus aureus. Infect. Immun. 2014, 82, 4369–4379. [Google Scholar] [CrossRef]
- Manna, A.C.; Kim, S.; Cengher, L.; Corvaglia, A.; Leo, S.; Francois, P.; Cheung, A.L. Small RNA Teg49 Is Derived from a SarA Transcript and Regulates Virulence Genes Independent of SarA in Staphylococcus aureus. Infect. Immun. 2018, 86, e00635-17. [Google Scholar] [CrossRef] [PubMed]
- Cengher, L.; Manna, A.C.; Cho, J.; Theprungsirikul, J.; Sessions, K.; Rigby, W.; Cheung, A.L. Regulation of Neutrophil Myeloperoxidase Inhibitor SPIN by the Small RNA Teg49 in Staphylococcus aureus. Mol. Microbiol. 2022, 117, 1447–1463. [Google Scholar] [CrossRef] [PubMed]
- Ploscariu, N.T.; de Jong, N.W.M.; van Kessel, K.P.M.; van Strijp, J.A.G.; Geisbrecht, B.V. Identification and Structural Characterization of a Novel Myeloperoxidase Inhibitor from Staphylococcus delphini. Arch. Biochem. Biophys. 2018, 645, 1–11. [Google Scholar] [CrossRef]
- Guillet, J.; Hallier, M.; Felden, B. Emerging Functions for the Staphylococcus aureus RNome. PLoS Pathog. 2013, 9, e1003767. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.M.; Miller, E.W.; Benson, M.A.; Alonzo, F.; Yoong, P.; Torres, V.J.; Hinrichs, S.H.; Dunman, P.M. Characterization of SSR42, a Novel Virulence Factor Regulatory RNA That Contributes to the Pathogenesis of a Staphylococcus aureus USA300 Representative. J. Bacteriol. 2012, 194, 2924–2938. [Google Scholar] [CrossRef]
- Das, S.; Lindemann, C.; Young, B.C.; Muller, J.; Österreich, B.; Ternette, N.; Winkler, A.-C.; Paprotka, K.; Reinhardt, R.; Förstner, K.U.; et al. Natural Mutations in a Staphylococcus aureus Virulence Regulator Attenuate Cytotoxicity but Permit Bacteremia and Abscess Formation. Proc. Natl. Acad. Sci. USA 2016, 113, E3101–E3110. [Google Scholar] [CrossRef]
- Horn, J.; Klepsch, M.; Manger, M.; Wolz, C.; Rudel, T.; Fraunholz, M. Long Noncoding RNA SSR42 Controls Staphylococcus aureus Alpha-Toxin Transcription in Response to Environmental Stimuli. J. Bacteriol. 2018, 200, e00252-18. [Google Scholar] [CrossRef]
- Qin, L.; McCausland, J.W.; Cheung, G.Y.C.; Otto, M. PSM-Mec-A Virulence Determinant That Connects Transcriptional Regulation, Virulence, and Antibiotic Resistance in Staphylococci. Front. Microbiol. 2016, 7, 1293. [Google Scholar] [CrossRef]
- Kaito, C.; Saito, Y.; Nagano, G.; Ikuo, M.; Omae, Y.; Hanada, Y.; Han, X.; Kuwahara-Arai, K.; Hishinuma, T.; Baba, T.; et al. Transcription and Translation Products of the Cytolysin Gene Psm-Mec on the Mobile Genetic Element SCCmec Regulate Staphylococcus aureus Virulence. PLoS Pathog. 2011, 7, e1001267. [Google Scholar] [CrossRef]
- Gillet, Y.; Issartel, B.; Vanhems, P.; Fournet, J.-C.; Lina, G.; Bes, M.; Vandenesch, F.; Piémont, Y.; Brousse, N.; Floret, D.; et al. Association between Staphylococcus aureus Strains Carrying Gene for Panton-Valentine Leukocidin and Highly Lethal Necrotising Pneumonia in Young Immunocompetent Patients. Lancet 2002, 359, 753–759. [Google Scholar] [CrossRef]
- Chatterjee, S.S.; Chen, L.; Joo, H.-S.; Cheung, G.Y.C.; Kreiswirth, B.N.; Otto, M. Distribution and Regulation of the Mobile Genetic Element-Encoded Phenol-Soluble Modulin PSM-Mec in Methicillin-Resistant Staphylococcus aureus. PLoS ONE 2011, 6, e28781. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.L.; Bayer, A.S.; Zhang, G.; Gresham, H.; Xiong, Y.-Q. Regulation of Virulence Determinants in Vitro and in Vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2004, 40, 1–9. [Google Scholar] [CrossRef]
- Abdelnour, A.; Arvidson, S.; Bremell, T.; Rydén, C.; Tarkowski, A. The Accessory Gene Regulator (Agr) Controls Staphylococcus aureus Virulence in a Murine Arthritis Model. Infect. Immun. 1993, 61, 3879–3885. [Google Scholar] [CrossRef] [PubMed]
- Gillaspy, A.F.; Hickmon, S.G.; Skinner, R.A.; Thomas, J.R.; Nelson, C.L.; Smeltzer, M.S. Role of the Accessory Gene Regulator (Agr) in Pathogenesis of Staphylococcal Osteomyelitis. Infect. Immun. 1995, 63, 3373–3380. [Google Scholar] [CrossRef]
- Pragman, A.A.; Schlievert, P.M. Virulence Regulation in Staphylococcus aureus: The Need for in Vivo Analysis of Virulence Factor Regulation. FEMS Immunol. Med. Microbiol. 2004, 42, 147–154. [Google Scholar] [CrossRef]
- Goerke, C.; Fluckiger, U.; Steinhuber, A.; Bisanzio, V.; Ulrich, M.; Bischoff, M.; Patti, J.M.; Wolz, C. Role of Staphylococcus aureus Global Regulators Sae and SigmaB in Virulence Gene Expression during Device-Related Infection. Infect. Immun. 2005, 73, 3415–3421. [Google Scholar] [CrossRef]
- Yarwood, J.M.; McCormick, J.K.; Paustian, M.L.; Kapur, V.; Schlievert, P.M. Repression of the Staphylococcus aureus Accessory Gene Regulator in Serum and In Vivo. J. Bacteriol. 2002, 184, 1095–1101. [Google Scholar] [CrossRef]
- Song, J.; Lays, C.; Vandenesch, F.; Benito, Y.; Bes, M.; Chu, Y.; Lina, G.; Romby, P.; Geissmann, T.; Boisset, S. The Expression of Small Regulatory RNAs in Clinical Samples Reflects the Different Life Styles of Staphylococcus aureus in Colonization vs. Infection. PLoS ONE 2012, 7, e37294. [Google Scholar] [CrossRef]
- Bordeau, V.; Cady, A.; Revest, M.; Rostan, O.; Sassi, M.; Tattevin, P.; Donnio, P.-Y.; Felden, B. Staphylococcus aureus Regulatory RNAs as Potential Biomarkers for Bloodstream Infections. Emerg. Infect. Dis. 2016, 22, 1570–1578. [Google Scholar] [CrossRef]
- Burian, M.; Wolz, C.; Goerke, C. Regulatory Adaptation of Staphylococcus aureus during Nasal Colonization of Humans. PLoS ONE 2010, 5, e10040. [Google Scholar] [CrossRef]
- Burian, M.; Rautenberg, M.; Kohler, T.; Fritz, M.; Krismer, B.; Unger, C.; Hoffmann, W.H.; Peschel, A.; Wolz, C.; Goerke, C. Temporal Expression of Adhesion Factors and Activity of Global Regulators during Establishment of Staphylococcus aureus Nasal Colonization. J. Infect. Dis. 2010, 201, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Date, S.V.; Modrusan, Z.; Lawrence, M.; Morisaki, J.H.; Toy, K.; Shah, I.M.; Kim, J.; Park, S.; Xu, M.; Basuino, L.; et al. Global Gene Expression of Methicillin-Resistant Staphylococcus aureus USA300 during Human and Mouse Infection. J. Infect. Dis. 2014, 209, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Szafranska, A.K.; Oxley, A.P.A.; Chaves-Moreno, D.; Horst, S.A.; Roßlenbroich, S.; Peters, G.; Goldmann, O.; Rohde, M.; Sinha, B.; Pieper, D.H.; et al. High-Resolution Transcriptomic Analysis of the Adaptive Response of Staphylococcus aureus during Acute and Chronic Phases of Osteomyelitis. mBio 2014, 5, e01775-14. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Schilcher, K.; Burcham, L.R.; Kwiecinski, J.M.; Johnson, P.M.; Head, S.R.; Heinrichs, D.E.; Horswill, A.R.; Doran, K.S. Identification of Key Determinants of Staphylococcus aureus Vaginal Colonization. mBio 2019, 10, e02321-19. [Google Scholar] [CrossRef]
- Ibberson, C.B.; Whiteley, M. The Staphylococcus aureus Transcriptome during Cystic Fibrosis Lung Infection. mBio 2019, 10, e02774-19. [Google Scholar] [CrossRef]
- Herbert, S.; Ziebandt, A.-K.; Ohlsen, K.; Schäfer, T.; Hecker, M.; Albrecht, D.; Novick, R.; Götz, F. Repair of Global Regulators in Staphylococcus aureus 8325 and Comparative Analysis with Other Clinical Isolates. Infect. Immun. 2010, 78, 2877. [Google Scholar] [CrossRef]
- Mrochen, D.M.; Fernandes de Oliveira, L.M.; Raafat, D.; Holtfreter, S. Staphylococcus aureus Host Tropism and Its Implications for Murine Infection Models. Int. J. Mol. Sci. 2020, 21, 7061. [Google Scholar] [CrossRef]
- Bubeck Wardenburg, J.; Palazzolo-Ballance, A.M.; Otto, M.; Schneewind, O.; DeLeo, F.R. Panton-Valentine Leukocidin Is Not a Virulence Determinant in Murine Models of Community-Associated Methicillin-Resistant Staphylococcus aureus Disease. J. Infect. Dis. 2008, 198, 1166–1170. [Google Scholar] [CrossRef]
- Diep, B.A.; Chan, L.; Tattevin, P.; Kajikawa, O.; Martin, T.R.; Basuino, L.; Mai, T.T.; Marbach, H.; Braughton, K.R.; Whitney, A.R.; et al. Polymorphonuclear Leukocytes Mediate Staphylococcus aureus Panton-Valentine Leukocidin-Induced Lung Inflammation and Injury. Proc. Natl. Acad. Sci. USA 2010, 107, 5587–5592. [Google Scholar] [CrossRef]
- Burian, M.; Plange, J.; Schmitt, L.; Kaschke, A.; Marquardt, Y.; Huth, L.; Baron, J.M.; Hornef, M.W.; Wolz, C.; Yazdi, A.S. Adaptation of Staphylococcus aureus to the Human Skin Environment Identified Using an Ex Vivo Tissue Model. Front. Microbiol. 2021, 12, 728989. [Google Scholar] [CrossRef]
- Mannala, G.K.; Rupp, M.; Alagboso, F.; Kerschbaum, M.; Pfeifer, C.; Sommer, U.; Kampschulte, M.; Domann, E.; Alt, V. Galleria Mellonella as an Alternative In Vivo Model to Study Bacterial Biofilms on Stainless Steel and Titanium Implants. Altex 2021, 38, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Hermansen, G.M.M.; Sazinas, P.; Kofod, D.; Millard, A.; Andersen, P.S.; Jelsbak, L. Transcriptomic Profiling of Interacting Nasal Staphylococci Species Reveals Global Changes in Gene and Non-Coding RNA Expression. FEMS Microbiol. Lett. 2018, 365, fny004. [Google Scholar] [CrossRef] [PubMed]
- Pynnonen, M.; Stephenson, R.E.; Schwartz, K.; Hernandez, M.; Boles, B.R. Hemoglobin Promotes Staphylococcus aureus Nasal Colonization. PLoS Pathog. 2011, 7, e1002104. [Google Scholar] [CrossRef] [PubMed]
- Kahl, B.C.; Becker, K.; Löffler, B. Clinical Significance and Pathogenesis of Staphylococcal Small Colony Variants in Persistent Infections. Clin. Microbiol. Rev. 2016, 29, 401–427. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent Bacterial Infections and Persister Cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Loss, G.; Simões, P.M.; Valour, F.; Cortês, M.F.; Gonzaga, L.; Bergot, M.; Trouillet-Assant, S.; Josse, J.; Diot, A.; Ricci, E.; et al. Staphylococcus aureus Small Colony Variants (SCVs): News From a Chronic Prosthetic Joint Infection. Front. Cell. Infect. Microbiol. 2019, 9, 363. [Google Scholar] [CrossRef]
- Romilly, C.; Lays, C.; Tomasini, A.; Caldelari, I.; Benito, Y.; Hammann, P.; Geissmann, T.; Boisset, S.; Romby, P.; Vandenesch, F. A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in Staphylococcus aureus. PLoS Pathog. 2014, 10, e1003979. [Google Scholar] [CrossRef]
- Wagner, E.G.H.; Romby, P. Small RNAs in Bacteria and Archaea: Who They Are, What They Do, and How They Do It. Adv. Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef]
- Dastghey, S.; Otto, M. Staphylococcal Adaptation to Diverse Physiologic Niches: An Overview of Transcriptomic and Phenotypic Changes in Different Biological Environments. Future Microbiol. 2015, 10, 1981–1995. [Google Scholar] [CrossRef]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Bronner, S.; Monteil, H.; Prévost, G. Regulation of Virulence Determinants in Staphylococcus aureus: Complexity and Applications. FEMS Microbiol. Rev. 2004, 28, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Österberg, S.; del Peso-Santos, T.; Shingler, V. Regulation of Alternative Sigma Factor Use. Annu. Rev. Microbiol. 2011, 65, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Deora, R.; Misra, T.K. Characterization of the Primary Sigma Factor of Staphylococcus aureus. J. Biol. Chem. 1996, 271, 21828–21834. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morikawa, K.; Inose, Y.; Okamura, H.; Maruyama, A.; Hayashi, H.; Takeyasu, K.; Ohta, T. A New Staphylococcal Sigma Factor in the Conserved Gene Cassette: Functional Significance and Implication for the Evolutionary Processes. Genes Cells 2003, 8, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Shaw, L.N.; Lindholm, C.; Prajsnar, T.K.; Miller, H.K.; Brown, M.C.; Golonka, E.; Stewart, G.C.; Tarkowski, A.; Potempa, J. Identification and Characterization of Sigma, a Novel Component of the Staphylococcus aureus Stress and Virulence Responses. PLoS ONE 2008, 3, e3844. [Google Scholar] [CrossRef]
- Senn, M.M.; Giachino, P.; Homerova, D.; Steinhuber, A.; Strassner, J.; Kormanec, J.; Flückiger, U.; Berger-Bächi, B.; Bischoff, M. Molecular Analysis and Organization of the SigmaB Operon in Staphylococcus aureus. J. Bacteriol. 2005, 187, 8006–8019. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Goerke, C.; Wolz, C.; Seidl, K.; Homerova, D.; Schulthess, B.; Kormanec, J.; Berger-Bächi, B.; Bischoff, M. SigmaB and the SigmaB-Dependent ArlRS and YabJ-SpoVG Loci Affect Capsule Formation in Staphylococcus aureus. Infect. Immun. 2007, 75, 4562–4571. [Google Scholar] [CrossRef]
- Depke, M.; Burian, M.; Schäfer, T.; Bröker, B.M.; Ohlsen, K.; Völker, U. The Alternative Sigma Factor B Modulates Virulence Gene Expression in a Murine Staphylococcus aureus Infection Model but Does Not Influence Kidney Gene Expression Pattern of the Host. Int. J. Med. Microbiol. 2012, 302, 33–39. [Google Scholar] [CrossRef]
- Giachino, P.; Engelmann, S.; Bischoff, M. Sigma(B) Activity Depends on RsbU in Staphylococcus aureus. J. Bacteriol. 2001, 183, 1843–1852. [Google Scholar] [CrossRef]
- Chan, P.F.; Foster, S.J.; Ingham, E.; Clements, M.O. The Staphylococcus aureus Alternative Sigma Factor SigmaB Controls the Environmental Stress Response but Not Starvation Survival or Pathogenicity in a Mouse Abscess Model. J. Bacteriol. 1998, 180, 6082–6089. [Google Scholar] [CrossRef]
- Ranganathan, N.; Johnson, R.; Edwards, A.M. The General Stress Response of Staphylococcus aureus Promotes Tolerance of Antibiotics and Survival in Whole Human Blood. Microbiology 2020, 166, 1088–1094. [Google Scholar] [CrossRef] [PubMed]
- Lauderdale, K.J.; Boles, B.R.; Cheung, A.L.; Horswill, A.R. Interconnections between Sigma B, Agr, and Proteolytic Activity in Staphylococcus aureus Biofilm Maturation. Infect. Immun. 2009, 77, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Fugère, A.; Pépin Gaudreau, K.; Brouillette, E.; Frost, E.H.; Cantin, A.M.; Malouin, F. SigB Is a Dominant Regulator of Virulence in Staphylococcus aureus Small-Colony Variants. PLoS ONE 2013, 8, e65018. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Löffler, B.; Proctor, R.A. Persistence of Staphylococcus aureus: Multiple Metabolic Pathways Impact the Expression of Virulence Factors in Small-Colony Variants (SCVs). Front. Microbiol. 2020, 11, 1028. [Google Scholar] [CrossRef] [PubMed]
- Gertz, S.; Engelmann, S.; Schmid, R.; Ziebandt, A.K.; Tischer, K.; Scharf, C.; Hacker, J.; Hecker, M. Characterization of the Sigma(B) Regulon in Staphylococcus aureus. J. Bacteriol. 2000, 182, 6983–6991. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, M.; Dunman, P.; Kormanec, J.; Macapagal, D.; Murphy, E.; Mounts, W.; Berger-Bächi, B.; Projan, S. Microarray-Based Analysis of the Staphylococcus aureus SigmaB Regulon. J. Bacteriol. 2004, 186, 4085–4099. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.S.; Olsen, A.S.; Bonde, M.; Valentin-Hansen, P.; Kallipolitis, B.H. Identification of a Sigma B-Dependent Small Noncoding RNA in Listeria Monocytogenes. J. Bacteriol. 2008, 190, 6264–6270. [Google Scholar] [CrossRef]
- Villanueva, M.; García, B.; Valle, J.; Rapún, B.; Ruiz de Los Mozos, I.; Solano, C.; Martí, M.; Penadés, J.R.; Toledo-Arana, A.; Lasa, I. Sensory Deprivation in Staphylococcus aureus. Nat. Commun. 2018, 9, 523. [Google Scholar] [CrossRef]
- Dubrac, S.; Msadek, T. Identification of Genes Controlled by the Essential YycG/YycF Two-Component System of Staphylococcus aureus. J. Bacteriol. 2004, 186, 1175–1181. [Google Scholar] [CrossRef]
- Bleul, L.; Francois, P.; Wolz, C. Two-Component Systems of S. Aureus: Signaling and Sensing Mechanisms. Genes 2021, 13, 34. [Google Scholar] [CrossRef]
- Qiu, Y.; Xu, D.; Xia, X.; Zhang, K.; Aadil, R.M.; Batool, Z.; Wang, J. Five Major Two Components Systems of Staphylococcus aureus for Adaptation in Diverse Hostile Environment. Microb. Pathog. 2021, 159, 105119. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P. Autoinduction and Signal Transduction in the Regulation of Staphylococcal Virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar] [CrossRef] [PubMed]
- Reyes, D.; Andrey, D.O.; Monod, A.; Kelley, W.L.; Zhang, G.; Cheung, A.L. Coordinated Regulation by AgrA, SarA, and SarR to Control Agr Expression in Staphylococcus aureus. J. Bacteriol. 2011, 193, 6020–6031. [Google Scholar] [CrossRef] [PubMed]
- Fournier, B.; Klier, A.; Rapoport, G. The Two-Component System ArlS-ArlR Is a Regulator of Virulence Gene Expression in Staphylococcus aureus. Mol. Microbiol. 2001, 41, 247–261. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, J.W.; Moon, B.Y.; Lee, J.; Fortin, Y.J.; Austin, F.W.; Yang, S.-J.; Seo, K.S. Characterization of a Novel Two-Component Regulatory System, HptRS, the Regulator for the Hexose Phosphate Transport System in Staphylococcus aureus. Infect. Immun. 2015, 83, 1620–1628. [Google Scholar] [CrossRef]
- Cheung, A.L.; Ying, P. Regulation of Alpha- and Beta-Hemolysins by the Sar Locus of Staphylococcus aureus. J. Bacteriol. 1994, 176, 580–585. [Google Scholar] [CrossRef]
- Chien, Y.; Cheung, A.L. Molecular Interactions between Two Global Regulators, Sar and Agr, in Staphylococcus aureus. J. Biol. Chem. 1998, 273, 2645–2652. [Google Scholar] [CrossRef]
- Oriol, C.; Cengher, L.; Manna, A.C.; Mauro, T.; Pinel-Marie, M.-L.; Felden, B.; Cheung, A.; Rouillon, A. Expanding the Staphylococcus aureus SarA Regulon to Small RNAs. mSystems 2021, 6, e0071321. [Google Scholar] [CrossRef]
- Majerczyk, C.D.; Dunman, P.M.; Luong, T.T.; Lee, C.Y.; Sadykov, M.R.; Somerville, G.A.; Bodi, K.; Sonenshein, A.L. Direct Targets of CodY in Staphylococcus aureus. J. Bacteriol. 2010, 192, 2861–2877. [Google Scholar] [CrossRef]
- Stenz, L.; Francois, P.; Whiteson, K.; Wolz, C.; Linder, P.; Schrenzel, J. The CodY Pleiotropic Repressor Controls Virulence in Gram-Positive Pathogens. FEMS Immunol. Med. Microbiol. 2011, 62, 123–139. [Google Scholar] [CrossRef]
- Schmidt, K.A.; Manna, A.C.; Gill, S.; Cheung, A.L. SarT, a Repressor of Alpha-Hemolysin in Staphylococcus aureus. Infect. Immun. 2001, 69, 4749–4758. [Google Scholar] [CrossRef] [PubMed]
- Ingavale, S.S.; Van Wamel, W.; Cheung, A.L. Characterization of RAT, an Autolysis Regulator in Staphylococcus aureus. Mol. Microbiol. 2003, 48, 1451–1466. [Google Scholar] [CrossRef] [PubMed]
- Manna, A.C.; Cheung, A.L. SarU, a SarA Homolog, Is Repressed by SarT and Regulates Virulence Genes in Staphylococcus aureus. Infect. Immun. 2003, 71, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Manna, A.C.; Ingavale, S.S.; Maloney, M.; van Wamel, W.; Cheung, A.L. Identification of SarV (SA2062), a New Transcriptional Regulator, Is Repressed by SarA and MgrA (SA0641) and Involved in the Regulation of Autolysis in Staphylococcus aureus. J. Bacteriol. 2004, 186, 5267–5280. [Google Scholar] [CrossRef]
- Horsburgh, M.J.; Wharton, S.J.; Cox, A.G.; Ingham, E.; Peacock, S.; Foster, S.J. MntR Modulates Expression of the PerR Regulon and Superoxide Resistance in Staphylococcus aureus through Control of Manganese Uptake. Mol. Microbiol. 2002, 44, 1269–1286. [Google Scholar] [CrossRef]
- Balleza, E.; López-Bojorquez, L.N.; Martínez-Antonio, A.; Resendis-Antonio, O.; Lozada-Chávez, I.; Balderas-Martínez, Y.I.; Encarnación, S.; Collado-Vides, J. Regulation by Transcription Factors in Bacteria: Beyond Description. FEMS Microbiol. Rev. 2009, 33, 133–151. [Google Scholar] [CrossRef]
- Ibarra, J.A.; Pérez-Rueda, E.; Carroll, R.K.; Shaw, L.N. Global Analysis of Transcriptional Regulators in Staphylococcus aureus. BMC Genom. 2013, 14, 126. [Google Scholar] [CrossRef]
- Roux, A.; Todd, D.A.; Velázquez, J.V.; Cech, N.B.; Sonenshein, A.L. CodY-Mediated Regulation of the Staphylococcus aureus Agr System Integrates Nutritional and Population Density Signals. J. Bacteriol. 2014, 196, 1184–1196. [Google Scholar] [CrossRef]
- Mlynek, K.D.; Sause, W.E.; Moormeier, D.E.; Sadykov, M.R.; Hill, K.R.; Torres, V.J.; Bayles, K.W.; Brinsmade, S.R. Nutritional Regulation of the Sae Two-Component System by CodY in Staphylococcus aureus. J. Bacteriol. 2018, 200, e00012-18. [Google Scholar] [CrossRef]
- Durand, S.; Braun, F.; Lioliou, E.; Romilly, C.; Helfer, A.-C.; Kuhn, L.; Quittot, N.; Nicolas, P.; Romby, P.; Condon, C. A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in Bacillus Subtilis. PLoS Genet. 2015, 11, e1004957. [Google Scholar] [CrossRef]
- Correia Santos, S.; Bischler, T.; Westermann, A.J.; Vogel, J. MAPS Integrates Regulation of Actin-Targeting Effector SteC into the Virulence Control Network of Salmonella Small RNA PinT. Cell Rep. 2021, 34, 108722. [Google Scholar] [CrossRef] [PubMed]
- Roncarati, D.; Scarlato, V.; Vannini, A. Targeting of Regulators as a Promising Approach in the Search for Novel Antimicrobial Agents. Microorganisms 2022, 10, 185. [Google Scholar] [CrossRef] [PubMed]


| Name | Consensual Name | Length (nt) | ORF | Direct mRNA Targets | Mechanisms of Action | Function |
|---|---|---|---|---|---|---|
| RNAIII | Srn_3910 | 514 | Yes (Hld) | spa, sbi, coa, sa1000, lytM, rot mgrA, hla | Translation inhibition (lytM, sbi), translation inhibition and mRNA cleavage (rot, spa, coa, SA1000), translation activation (hla), mRNA stabilization (mgrA) | Provirulent |
| Psm-mec | N/A | 143–157 | Yes (Psm-mec) | agrA | Translation inhibition | Antivirulent |
| SprC | Srn_3610 | 154 | No | atl | Translation inhibition | Antivirulent/Provirulent |
| SprD | Srn_3800 | 145 | No | sbi | Translation inhibition | Provirulent |
| SprF1 | Srn_3830 | 138 | No | sprG1, ribosomes | Translation attenuation | Persistence |
| SprX | Srn_3820 | 150 | No | spoVG, walR, ecb, clfB, hld | Translation inhibition (spoVG, ecb) | Provirulent |
| SprY | Srn_9630 | 128 | No | RNAIII | Seric blocking of mRNA binding sites | Antivirulent |
| RsaA | Srn_1510 | 143 | No | mgrA, flip-r, ssaA | Translation inhibition | Antivirulent |
| RsaC | Srn_1590 | Strain-dependent | No | sodA, sarA | Translation inhibition (sodA) | Provirulent, metabolism |
| RsaD | Srn_1640 | 177 | No | alsS | Translation inhibition | Metabolism |
| RsaE | Srn_2130 | 459 | No | rocF | Translation inhibition | Metabolism |
| RsaF | N/A | 105 | No | hysA, splD | Unknown | Undefined |
| RsaG | Srn_0510 | 194 | No | rex, | Metabolism | |
| RsaI | Srn_4390 | 111 | No | glcU_2, fn3K, icaR, rsaG | Translation inhibition (glcU_2, fn3K, IcaR) | Metabolism |
| ArtR | Srn_4050 | 346 | No | sarT | Translation inhibition and mRNA degradation | Undefined |
| SSR42 | Srn_4470 | 1232 | Yes (unknown peptide) | sae | Unknown (mRNA stabilization?) | Provirulent |
| Teg49 | Srn_1550 | 196 | Yes (unknown peptide) | sarA spn | mRNA stabilization | Undefined |
| Teg41 | Srn_1080 | 205 | Yes (unknown peptide) | psmα | Unknown (mRNA stabilization or translation initiation?) | Provirulent |
| Murine Osteomyelitis | Human Serum | Human Cystic Fribrosis | Murine Vaginal Colonization | Murine Liver | |
|---|---|---|---|---|---|
| Conditions | In vivo | Ex vivo | In vivo | In vivo | In vivo |
| sRNA expression | >15 differentially expressed sRNAs | 42 upregulated 41 downregulated | 122 upregulated | 60 upregulated | 17 upregulated 17 downregulated |
| Comparator | BHI medium | TSB medium | Chemically defined medium, synthetic fibrosis media | Laboratory media | TSB medium |
| Kinetics | Yes Acute infection (7 days), chronic infection (28 days) | No | No | Yes 5 h, 24 h, 72 h | Yes 6 h, 24 h, 48 h |
| References | [143] | [24] | [3] | [3] | [108] |
| Regulator | Name | Functions | sRNA Targets | References |
|---|---|---|---|---|
| TCSs | SaeRS | Regulation of virulence factors | RNAIII (+) | [160,182] |
| SrrAB | Oxydative stress | RsaE (+), RsaD (+), RNAIII (−) | [11,160] | |
| AgrAC | Regulation of virulence factors, activation of quorum sensing | RNAIII (+), ArtR (−) | [68,183] | |
| ArlRS | Autolysis regulation | RNAIII (−) | [184] | |
| HptRS | Hexose phosphate transport | RsaG (+) | [39,185] | |
| TFs | SarA | Global regulator of virulence determinant | Many sRNAs, including RsaD (−), sprG2 (−), Spr2AS (−), SprC (−), Srn_9340 (−), RNAIII (+) | [86,132,186,187,188] |
| CodY | Adaptive response to starvation, regulation of virulence factors | Many sRNAs, including RsaD (−), RNAIII (−) | [51,189,190] | |
| CcpA | Adaptive response to carbon source, modulation of virulence factors | RsaI (−), RNAIII (+) | [39,117] | |
| SarT | Repressor of alpha hemolysin synthesis | RNAIII (−) | [132,191] | |
| MgrA | Global regulator of virulence factors | RNAIII (+) | [192] | |
| SarU | Positive regulator of agr | RNAIII (+) | [193] | |
| SarV | Autolysis regulator | RNAIII (+) | [194] | |
| MntR | Control of manganese uptake | RsaC (−) | [40,195] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menard, G.; Silard, C.; Suriray, M.; Rouillon, A.; Augagneur, Y. Thirty Years of sRNA-Mediated Regulation in Staphylococcus aureus: From Initial Discoveries to In Vivo Biological Implications. Int. J. Mol. Sci. 2022, 23, 7346. https://doi.org/10.3390/ijms23137346
Menard G, Silard C, Suriray M, Rouillon A, Augagneur Y. Thirty Years of sRNA-Mediated Regulation in Staphylococcus aureus: From Initial Discoveries to In Vivo Biological Implications. International Journal of Molecular Sciences. 2022; 23(13):7346. https://doi.org/10.3390/ijms23137346
Chicago/Turabian StyleMenard, Guillaume, Chloé Silard, Marie Suriray, Astrid Rouillon, and Yoann Augagneur. 2022. "Thirty Years of sRNA-Mediated Regulation in Staphylococcus aureus: From Initial Discoveries to In Vivo Biological Implications" International Journal of Molecular Sciences 23, no. 13: 7346. https://doi.org/10.3390/ijms23137346
APA StyleMenard, G., Silard, C., Suriray, M., Rouillon, A., & Augagneur, Y. (2022). Thirty Years of sRNA-Mediated Regulation in Staphylococcus aureus: From Initial Discoveries to In Vivo Biological Implications. International Journal of Molecular Sciences, 23(13), 7346. https://doi.org/10.3390/ijms23137346






