Biosynthesis and Metabolism of Garlic Odor Compounds in Cultivated Chinese Chives (Allium tuberosum) and Wild Chinese Chives (Allium hookeri)
Abstract
1. Introduction
2. Results
2.1. Germplasm Curation and Nutrition Evaluation
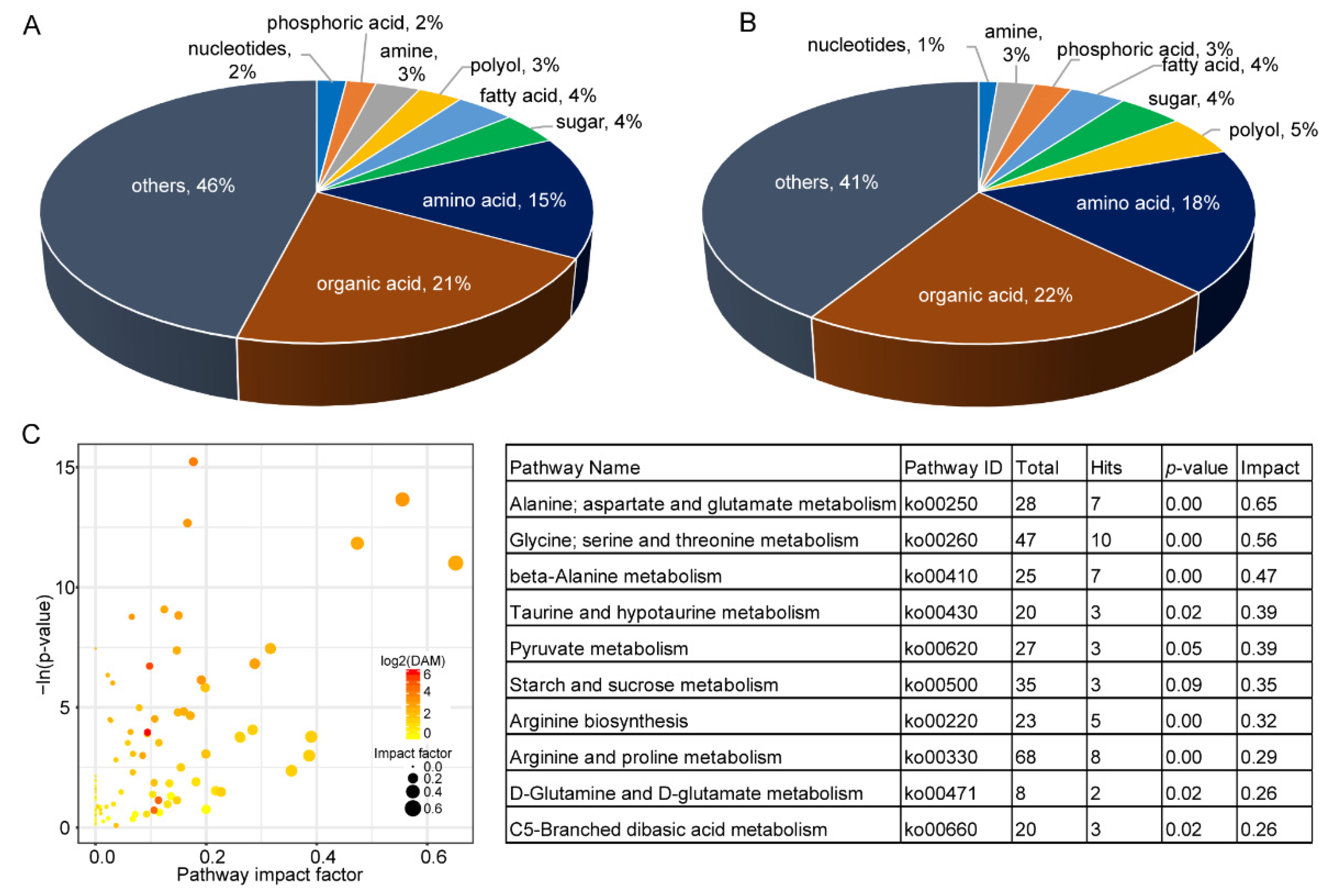
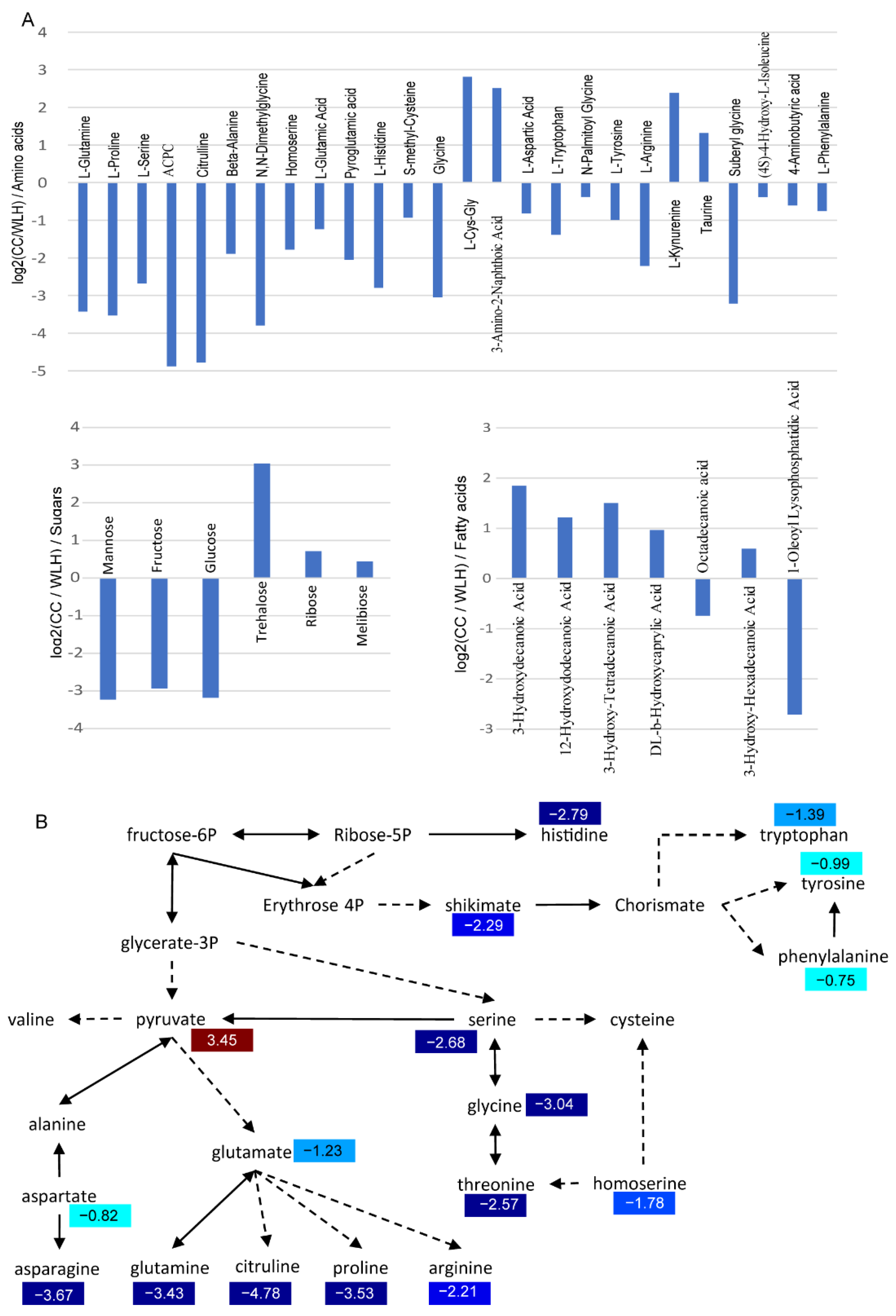
2.2. Metabolite Comparison
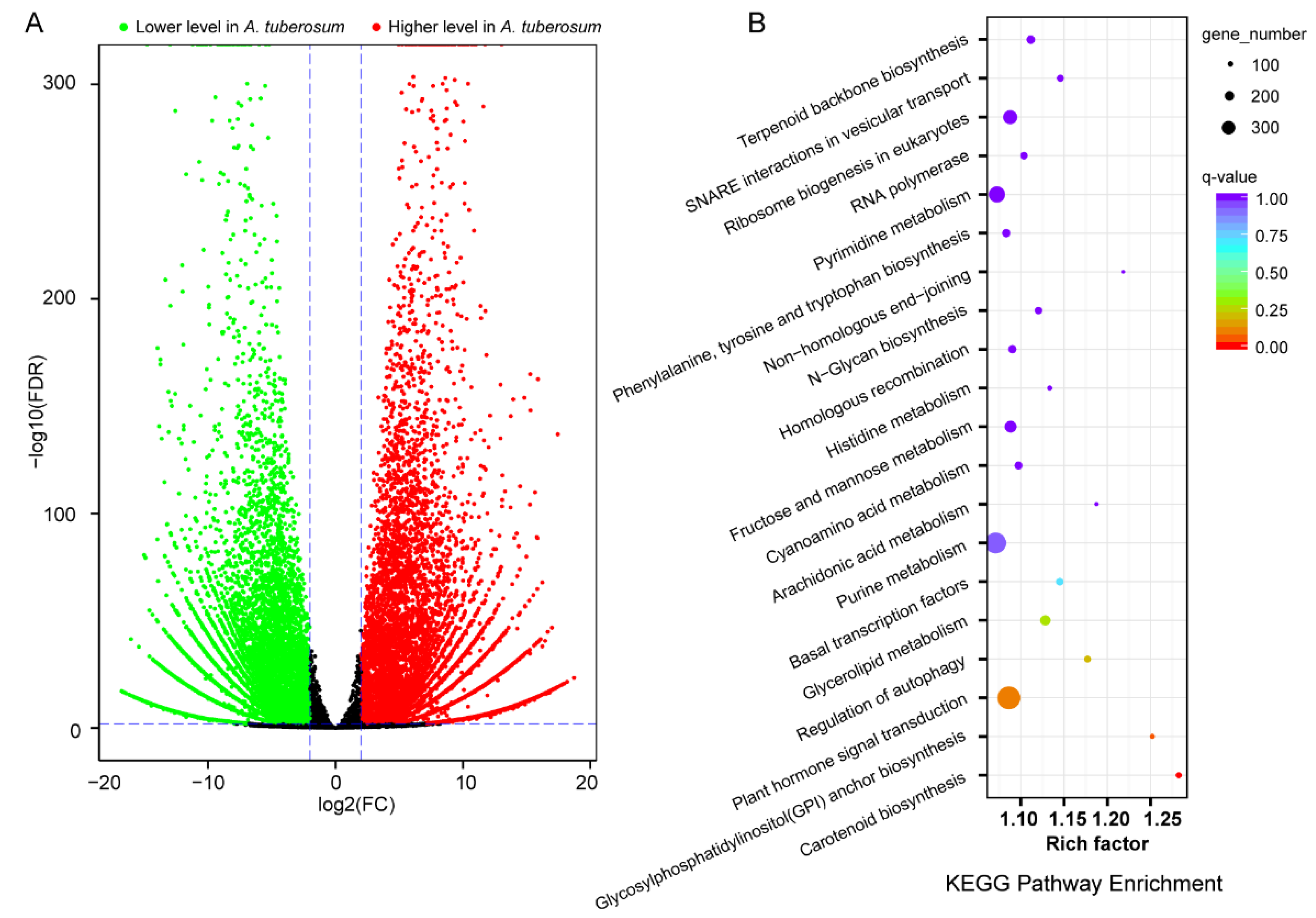
2.3. Leaf Transcriptome Comparisons
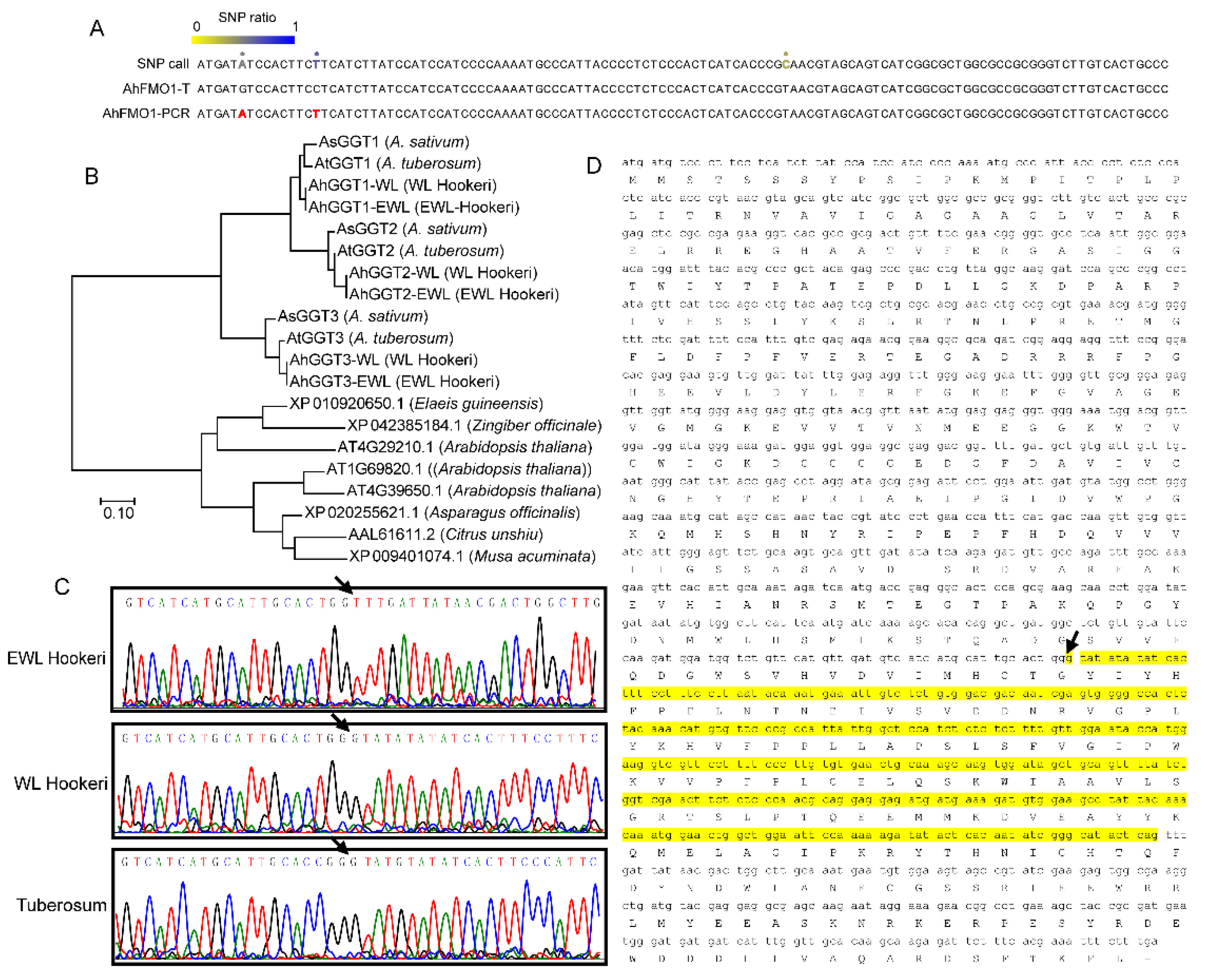
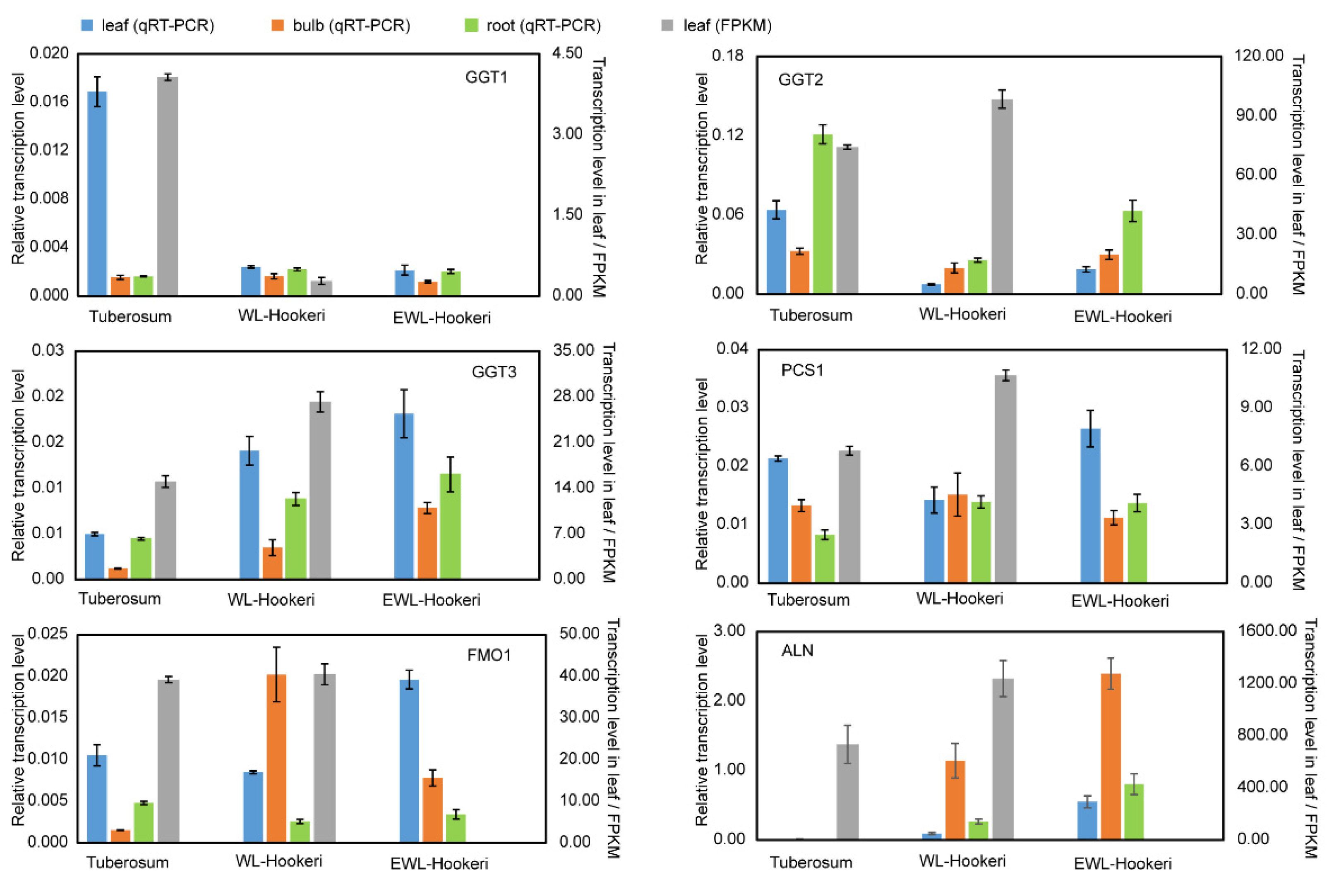
2.4. Gene Cloning of Garlic Odor Compounds
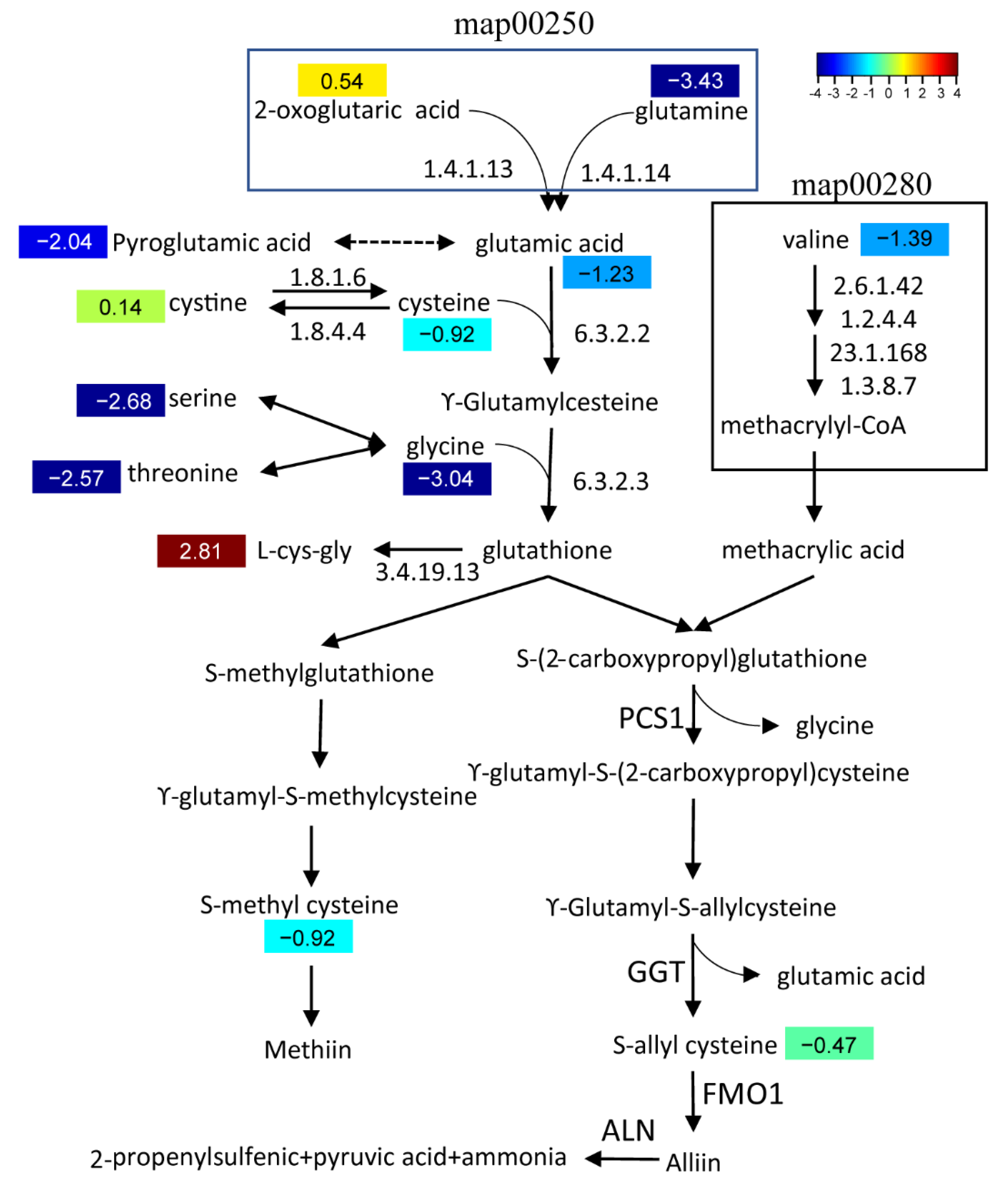
2.5. Integrative Analysis of Garlic Odor Compound Biosynthesis
3. Discussion
4. Conclusions
4.1. Plant Materials
4.2. Physiological Indices
4.3. GC-MS Assay
4.4. LC-MS Assay
4.5. RNA Extraction and RNA-Seq
4.6. Gene Sequence Cloning and qRT-PCR
4.7. Statistical Analysis, Data Processing, and Figure Presentation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oh, M.; Kim, S.-Y.; Park, S.; Kim, K.-N.; Kim, S. Phytochemicals in Chinese chive (Allium tuberosum) induce the skeletal muscle cell proliferation via PI3K/Akt/mTOR and smad pathways in C2C12 Cells. Int. J. Mol. Sci. 2021, 22, 2296. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Mnayer, D.; Tabanelli, G.; Stojanović-Radić, Z.Z.; Sharifi-Rad, M.; Yousaf, Z.; Vallone, L.; Setzer, W.N.; Iriti, M. Plants of the genus Allium as antibacterial agents: From tradition to pharmacy. Cell. Mol. Biol. 2016, 62, 57–68. [Google Scholar] [PubMed]
- Fenwick, G.R.; Hanley, A.B.; Whitaker, J.R. The genus Allium—Part 3. Crit. Rev. Food Sci. Nutr. 1985, 23, 1–73. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, R.M.; Blattner, F.R.; Gurushidze, M. New classification of Allium L. subg. Melanocrommyum (Webb &Berthel) Rouy (Alliaceae) based on molecular and morphological characters. Phyton 2010, 49, 145–220. [Google Scholar]
- Li, M.J.; Guo, X.L.; Li, J.; Zhou, S.D.; Liu, Q.; He, X.J. Cytotaxonomy of Allium (Amaryllidaceae) subgenera Cyathophora and Amerallium sect. Bromatorrhiza. Phytotaxa 2017, 331, 185–198. [Google Scholar] [CrossRef]
- Xie, D.F.; Tan, J.B.; Yu, Y.; Gui, L.J.; Su, D.M.; Zhou, S.D.; He, X.J. Insights into phylogeny, age and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann. Bot. 2020, 125, 1039–1055. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, Q.; Wang, H. Research progress on the classification of Allium plants. Acta Hortic. Sin. 2021, 48, 1418–1428. [Google Scholar]
- Ayam, V.S. Allium hookeri, Thw. Enum. A lesser known terrestrial perennial herb used as food and its ethnobotanical relevance in Manipur. Afr. J. Food Agric. Nutr. Dev. 2011, 11, 5389–5412. [Google Scholar] [CrossRef]
- Ni, N.C.; He, X.J.; Xu, J.M. Random amplified polymorphic DNA analysis on intraspecific differentiation of Allium hookeri. J. Sichuan Univ. 2002, 39, 944–947. (In Chinese) [Google Scholar]
- Zhang, G.; Wang, M.; Wang, X.; Huang, Z.; Tian, Q.; Wang, C.; Zhang, Z.; Tian, Y.; Tian, X. Breeding on the new Chinese chive varieties of broad leaf and the high quality of the leeks ‘Youkuan No.1’. North. Hortic. 2015, 15, 153–156. (In Chinese) [Google Scholar]
- Singh, K.D.; Chetia, D.; Biplab, D.E. New flavonoid compound from Allium Hookeri thwaites as a gastroprotective agent. Int. J. Pharm. Pharm. Sci. 2018, 10, 24–30. [Google Scholar] [CrossRef][Green Version]
- Jang, J.Y.; Lee, M.J.; You, B.R.; Jin, J.S.; Lee, S.H.; Yun, Y.R.; Kim, H.J. Allium hookeri root extract exerts anti-inflammatory effects by nuclear factor-kappaB down-regulation in lipopolysaccharide-induced RAW264.7 cells. BMC Complement. Altern. Med. 2017, 17, 126. [Google Scholar]
- Lee, Y.S.; Lee, S.H.; Gadde, U.D.; Oh, S.T.; Lee, S.J.; Lillehoj, H.S. Allium hookeri supplementation improves intestinal immune response against necrotic enteritis in young broiler chickens. Poult. Sci. 2018, 97, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Rose, P.; Whiteman, M.; Moore, P.K.; Zhu, Y.Z. Bioactive S-alk (en) yl cysteine sulfoxide metabolites in the genus Allium: The chemistry of potential therapeutic agents. Nat. Prod. Rep. 2005, 22, 351–368. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Iwasaki, K.; Mikami, M.; Yagihashi, A. Distribution of eleven flavor precursors, S-Alk(en)yl-L-Cysteine derivatives, in seven Allium vegetables. Food Sci. Technol. Res. 2010, 17, 55–62. [Google Scholar] [CrossRef]
- Van Damme, E.M.J.; Smeets, K.; Torrekens, S.; Vanleuven, F.; Peumans, W.J. Isolation and characterisation of alliinase cDNA clones from garlic (Allium sativum L.) and related species. Eur. J. Biochem. 1992, 209, 751–757. [Google Scholar] [CrossRef]
- Jones, M.G.; Hughes, J.; Tregova, A.; Milne, J.; Tomsett, A.B.; Collin, H.A. Biosynthesis of the flavour precursors of onion and garlic. J. Exp. Bot. 2004, 55, 1903–1918. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Kumagai, H. Characteristics, biosynthesis, decomposition, metabolism and functions of the garlic odour precursor, S-allyl-l-cysteine sulfoxide. Exp. Ther. Med. 2020, 19, 1528–1535. [Google Scholar] [CrossRef]
- Yoshimoto, N.; Saito, K. S-Alk(en)ylcysteine sulfoxides in the genus Allium: Proposed biosynthesis, chemical conversion, and bioactivities. J. Exp. Bot. 2019, 70, 4123–4137. [Google Scholar] [CrossRef]
- Jones, M.G.; Collin, H.A.; Tregova, A.; Trueman, L.; Brown, L.; Cosstick, R.; Hughes, J.; Milne, J.; Wilkinson, M.C.; Tomsett, A.B.; et al. The biochemical and physiological genesis of alliin in garlic. Med. Aromat. Plant Sci. Biotechnol. 2007, 1, 21–24. [Google Scholar]
- Yoshimoto, N.; Onuma, M.; Mizuno, S.; Sugino, Y.; Nakabayashi, R.; Imai, S.; Tsuneyoshi, T.; Sumi, S.; Saito, K. Identification of a flavin-containing S-oxygenating monooxygenase involved in alliin biosynthesis in garlic. Plant J. 2015, 83, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.; Tsuge, N.; Tomotake, M.; Nagatome, Y.; Sawada, H.; Nagata, T.; Kumagai, H. Plant biochemistry: An onion enzyme that makes the eyes water. Nature 2002, 419, 685. [Google Scholar] [CrossRef] [PubMed]
- Ohri, D.; Fritsch, R.M.; Hanelt, P. Evolution of genome size in Allium (Alliaceae). Plant Syst. Evol. 1998, 210, 57–86. [Google Scholar] [CrossRef]
- Ricroch, A.; Yockteng, R.; Brown, S.C.; Nadot, S. Evolution of genome size across some cultivated Allium species. Genome 2005, 48, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Finkers, R.; Kaauwen, M.; Ament, K.; Burger-Meijer, K.; Egging, R.; Huits, H.; Kodde, L.; Kroon, L.; Shigyo, M.; Sato, S.; et al. Insights form the first genome assembly of onion (Allium cepa). G3 2021, 11, jkab243. [Google Scholar] [CrossRef]
- Zhou, S.M.; Chen, L.M.; Liu, S.Q.; Wang, X.F.; Sun, X.D. De novo assembly and annotation of the Chinese chive (Allium tuberosum Rottler ex Spr.) transcriptome using the Illumina platform. PLoS ONE 2015, 10, e0133312. [Google Scholar] [CrossRef]
- Liu, N.; Tong, J.; Hu, M.; Ji, Y.; Wang, B.; Liang, H.; Liu, M.; Wu, Z. Transcriptome landscapes of multiple tissues highlight the genes involved in the flavor metabolic pathway in Chinese chive (Allium tuberosum). Genomics 2021, 113, 2145–2157. [Google Scholar] [CrossRef]
- Kusano, M.; Kobayashi, M.; Iizuka, Y.; Fukushima, A.; Saito, K. Unbiased profiling of volatile organic compounds in the headspace of Allium plants using an in-tube extraction device. BMC Res. Notes 2016, 9, 133. [Google Scholar] [CrossRef]
- Böttcher, C.; Krähmer, A.; Stürtz, M.; Widder, S.; Schulz, H. Comprehensive metabolite profiling of onion bulbs (Allium cepa) using liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry. Metabolomics 2017, 13, 35. [Google Scholar] [CrossRef]
- Basundari, F.R.A.; Sulistyaningsih, E.; Murti, R.H.; Nuringtyas, T.R. Metabolite profile of two Allium cepa L. aggregatum group cultivars by Nuclear Magnetic Resonance. Biodiversitas J. Biol. Divers. 2021, 22, 3127–3135. [Google Scholar] [CrossRef]
- Soorni, A.; Akrami, A.M.; Abolghasemi, R.; Vahedi, M. Transcriptome and phytochemical analyses provide insights into the organic sulfur pathway in Allium hirtifolium. Sci. Rep. 2021, 11, 768. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.; Hirata, S.; Mukae, T.; Yamada, T.; Sawada, Y.; El-Syaed, M.; Yamada, Y.; Sato, M.; Hirai, M.Y.; Shigyo, M. Comprehensive metabolite profiling in genetic resources of garlic (Allium sativum L.) collected from different geographical regions. Molecules 2021, 26, 1415. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.L.; Koo, I.; Peters, J.M.; Smith, P.B.; Patterson, A.D. Current challenges and recent developments in mass spectrometry-based metabolomics. Annu. Rev. Anal. Chem. 2021, 14, 467–487. [Google Scholar] [CrossRef] [PubMed]
- Soininen, T.H.; Jukarainen, N.; Soininen, P.; Auriola, S.O.K.; Julkunen-Tiitto, R.; Oleszek, W.; Stochmal, A.; Karjalainen, R.O.; Vepsäläinen, J.J. Metabolite profiling of leek (Allium porrum L) cultivars by 1H NMR and HPLC-MS. Phytochem. Anal. 2014, 25, 220–228. [Google Scholar] [CrossRef]
- Trovato, M.; Funck, D.; Forlani, G.; Okumoto, S.; Amir, R. Editorial: Amino acids in plants: Regulation and functions in development and stress defense. Front. Plant Sci. 2021, 12, 772810. [Google Scholar] [CrossRef]
- Jivishov, E. Investigations on Wild Allium Species. Part I: Cysteine Sulfoxides of Flowers. Part 2: Anticancer Activity of Bulb Extracts. Ph.D. Thesis, Philipps University of Marburg, Marburg, Germany, December 2016. [Google Scholar]
- Ueda, Y.; Tsubuku, T.; Miyajima, R. Composition of sulfur-containing components in onion and their flavor characters. Biosci. Biotechnol. Biochem. 1994, 58, 108–110. [Google Scholar] [CrossRef]
- Feinbaum, R.L.; Ausubel, F.M. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol. Cell Biol. 1988, 8, 1985–1992. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinform. Rev. 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. Ital. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Colin, N.D. RSEM: Accurate transcript quantification from RNA Seq data with or without a reference genome. BMC Bioinform. Ital. 2011, 12, 323. [Google Scholar] [CrossRef]
- Liu, M.; Wu, Z.; Jiang, F. Selection and validation of garlic reference genes for quantitative real-time PCR normalization. Plant Cell Tissue Organ Cult. 2015, 122, 435–444. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]

| Tuberosum | WL Hookeri | EWL Hookeri | Funckiaefolium | |
|---|---|---|---|---|
| Dry matter (mg/gFW) | 67.09 ± 2.09 c | 95.01 ± 4.44 b | 96.75 ± 3.74 b | 105.52 ± 3.86 a |
| Soluble sugar (mg/gFW) | 10.87 ± 0.95 c | 15.49 ± 0.29 a | 14.79 ± 0.36 b | 10.49 ± 0.75 c |
| Vitamin C (mg/gFW) | 0.23 ± 0.02 ab | 0.21 ± 0.01 b | 0.23 ± 0.01 ab | 0.24 ± 0.02 a |
| Soluble protein (mg/gFW) | 2.37 ± 0.10 c | 2.97 ± 0.05 a | 2.94 ± 0.03 a | 2.88 ± 0.03 b |
| GO ID | GO Term | Annotated | DEG N | Expected N | p-KS |
|---|---|---|---|---|---|
| GO:0015979 | photosynthesis | 240 | 205 | 185.14 | 4.3 × 10−12 |
| GO:0009658 | chloroplast organization | 81 | 76 | 62.49 | 1.1 × 10−8 |
| GO:0018298 | protein-chromophore linkage | 62 | 52 | 47.83 | 9.9 × 10−8 |
| GO:0009765 | photosynthesis, light harvesting | 59 | 50 | 45.51 | 1.5 × 10−7 |
| GO:0055114 | oxidation-reduction process | 1986 | 1489 | 1532.04 | 1.5 × 10−5 |
| GO:0016117 | carotenoid biosynthetic process | 26 | 26 | 20.06 | 1.6 × 10−5 |
| GO:0009793 | embryo development ending in seed dormancy | 153 | 135 | 118.03 | 2.1 × 10−5 |
| GO:0000413 | protein peptidyl-prolyl isomerization | 91 | 82 | 70.2 | 2.5 × 10−5 |
| GO:0006418 | tRNA aminoacylation for protein translation | 107 | 88 | 82.54 | 3.8 × 10−5 |
| GO:0010027 | thylakoid membrane organization | 22 | 21 | 16.97 | 5.4 × 10−5 |
| GO:0046835 | carbohydrate phosphorylation | 48 | 42 | 37.03 | 1.4 × 10−4 |
| GO:0010207 | photosystem II assembly | 19 | 19 | 14.66 | 2.0 × 10−4 |
| GO:0006400 | tRNA modification | 81 | 74 | 62.49 | 4.2 × 10−4 |
| GO:0006000 | fructose metabolic process | 6 | 6 | 4.63 | 5.2 × 10−4 |
| GO:0006003 | fructose 2,6-bisphosphate metabolic process | 6 | 6 | 4.63 | 5.2 × 10−4 |
| GO:0048366 | leaf development | 88 | 79 | 67.89 | 6.0 × 10−4 |
| GO:1901566 | organonitrogen compound biosynthetic process | 877 | 689 | 676.54 | 6.6 × 10−4 |
| GO:0001731 | formation of translation preinitiation complex | 41 | 32 | 31.63 | 7.2 × 10−4 |
| GO:0009814 | defense response, incompatible interaction | 38 | 35 | 29.31 | 7.7 × 10−4 |
| GO:0009657 | plastid organization | 111 | 105 | 85.63 | 8.5 × 10−4 |
| Length of AA 1 | Identity with Transcript | Identity with NCBI Database 2 | |
|---|---|---|---|
| AhGGT1 | 593 | 99.1% | 90.35%; BAQ21911.1 |
| AhGGT1E | 593 | 99.3% | 89.92%; BAQ21911.1 |
| AtGGT1 | 629 | 99.8% | 92.24%; BAQ21911.1 |
| AhGGT2 | 622 | 99.5% | 87.24%; BAQ21912.1 |
| AhGGT2E | 622 | 100% | 88.27%; BAQ21912.1 |
| AtGGT2 | 622 | 99.5% | 89.23%; BAQ21912.1 |
| AhGGT3 | 626 | 98.9% | 86.92%; BAQ21913.1 |
| AhGGT3E | 626 | 98.9% | 87.20%; BAQ21913.1 |
| AtGGT3 | 584 | 99.3% | 86.11%; BAQ21913.1 |
| AhPCS1 | 505 | 98.8% | 89.35%; AAO13809.1 |
| AhPCS1E | 505 | 97.7% | 89.16%; AAO13809.1 |
| AtPCS1 | 505 | 98.8% | 92.49%; AAO13809.1 |
| AhFMO1 | 458 | 99.4% | 87.00%; 6WPU_A |
| AhFMO1E | 317 | 92.4% | 88.73%; 6WPU_A |
| AtFMO1 | 462 | 97.5% | 91.88%; 6WPU_A |
| AhALN | 471 | 91.9% | 91.15%; AYN25508.1 |
| AhALNE | 471 | 91.7% | 90.88%; AYN25510.1 |
| AtALN | 471 | 100% | 100%; BAA20358.1 |
| Group | Rank | Gene | SD | CV |
|---|---|---|---|---|
| Leaf | 1 | EF-1α | 0.37 | 2.07 |
| 2 | 18SrRNA | 0.57 | 5.44 | |
| 3 | Actin | 1.27 | 5.92 | |
| Bulb | 1 | EF-1α | 0.58 | 3.35 |
| 2 | 18SrRNA | 1.12 | 11.02 | |
| 3 | Actin | 1.32 | 6.16 | |
| Root | 1 | 18SrRNA | 0.74 | 7.75 |
| 2 | EF-1α | 0.80 | 4.78 | |
| 3 | Actin | 0.89 | 4.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, S.-W.; Hang, L.-F.; Ali, S.; Xu, X.-Y.; Liu, Y.-J.; Yan, Q.-Q.; Luo, Q.-Y.; Li, Y.; Lin, L.-J.; Li, H.-X.; et al. Biosynthesis and Metabolism of Garlic Odor Compounds in Cultivated Chinese Chives (Allium tuberosum) and Wild Chinese Chives (Allium hookeri). Int. J. Mol. Sci. 2022, 23, 7013. https://doi.org/10.3390/ijms23137013
Xia S-W, Hang L-F, Ali S, Xu X-Y, Liu Y-J, Yan Q-Q, Luo Q-Y, Li Y, Lin L-J, Li H-X, et al. Biosynthesis and Metabolism of Garlic Odor Compounds in Cultivated Chinese Chives (Allium tuberosum) and Wild Chinese Chives (Allium hookeri). International Journal of Molecular Sciences. 2022; 23(13):7013. https://doi.org/10.3390/ijms23137013
Chicago/Turabian StyleXia, Shi-Wei, Lin-Feng Hang, Siyad Ali, Xiao-Yu Xu, Yan-Jun Liu, Qian-Qian Yan, Qiu-Yu Luo, Yu Li, Li-Jing Lin, Huan-Xiu Li, and et al. 2022. "Biosynthesis and Metabolism of Garlic Odor Compounds in Cultivated Chinese Chives (Allium tuberosum) and Wild Chinese Chives (Allium hookeri)" International Journal of Molecular Sciences 23, no. 13: 7013. https://doi.org/10.3390/ijms23137013
APA StyleXia, S.-W., Hang, L.-F., Ali, S., Xu, X.-Y., Liu, Y.-J., Yan, Q.-Q., Luo, Q.-Y., Li, Y., Lin, L.-J., Li, H.-X., Zhang, X.-A., Huang, L.-K., Ma, X., & Lai, Y.-S. (2022). Biosynthesis and Metabolism of Garlic Odor Compounds in Cultivated Chinese Chives (Allium tuberosum) and Wild Chinese Chives (Allium hookeri). International Journal of Molecular Sciences, 23(13), 7013. https://doi.org/10.3390/ijms23137013







