Lmx1a-Dependent Activation of miR-204/211 Controls the Timing of Nurr1-Mediated Dopaminergic Differentiation
Abstract
1. Introduction
2. Results
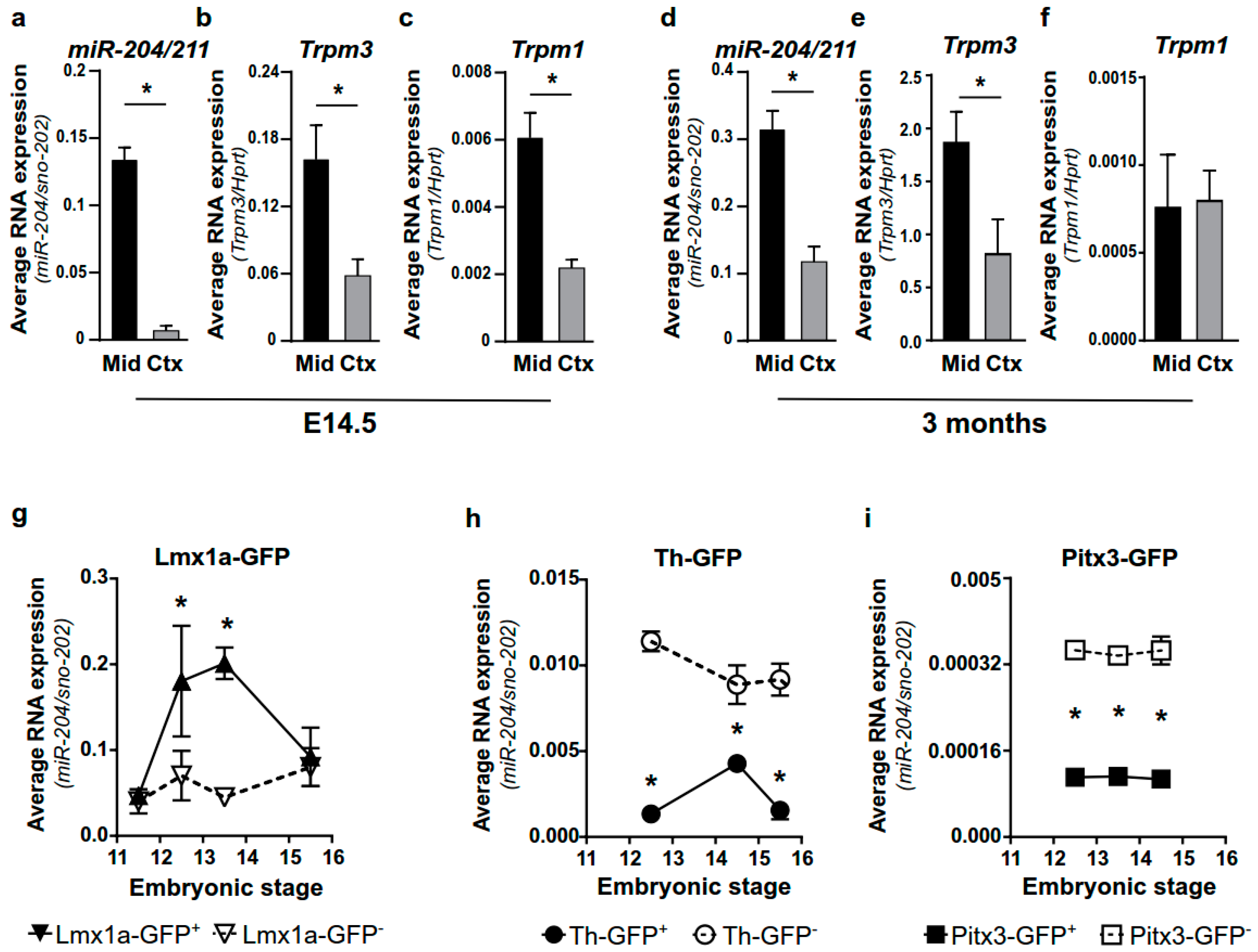
2.1. miR-204/211 Is Expressed in Mesencephalic DA Progenitor Cells
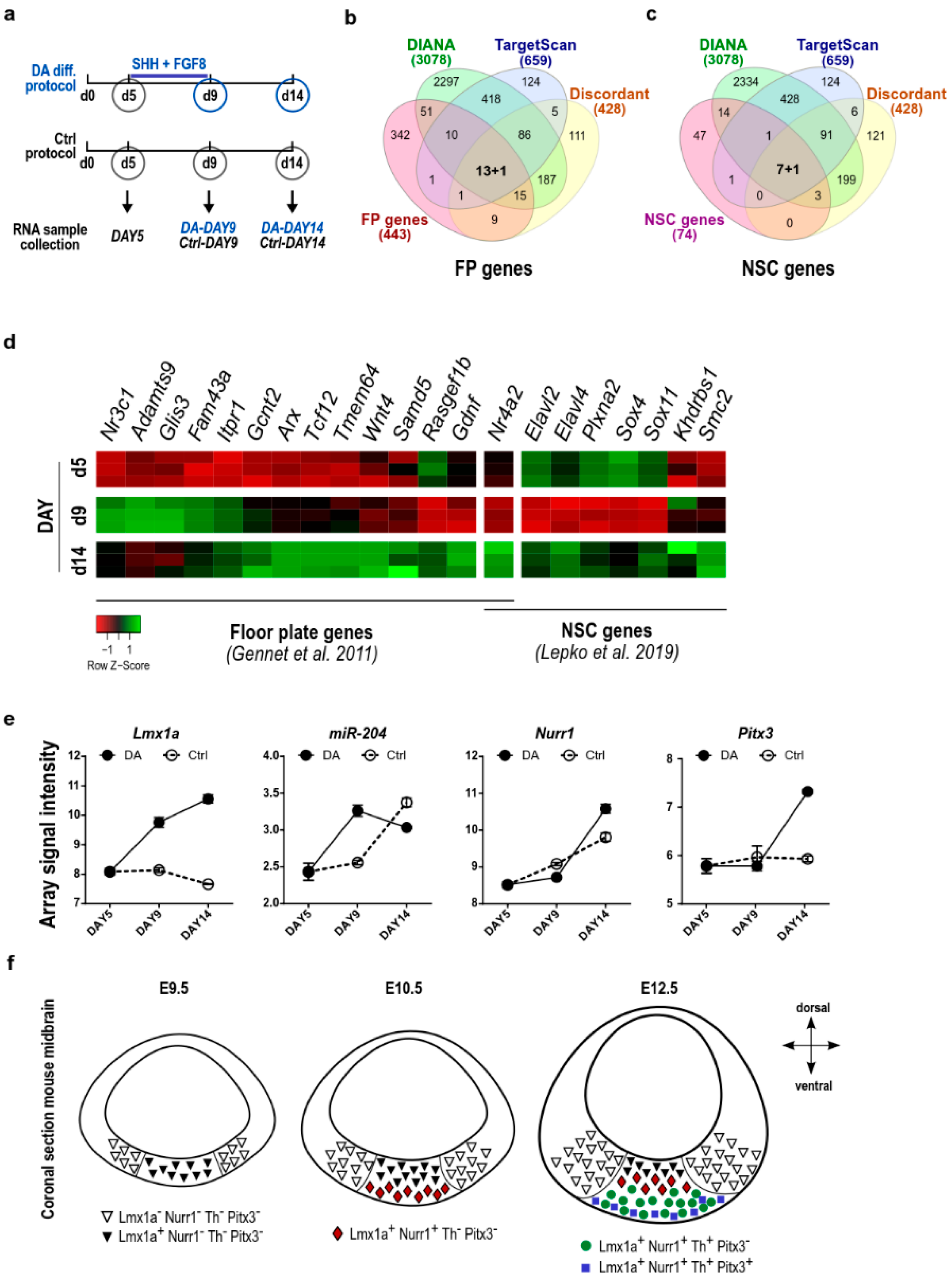
2.2. The Levels of Lmx1a Influence the Expression of miR-204/211 and Its Predicted Target Genes
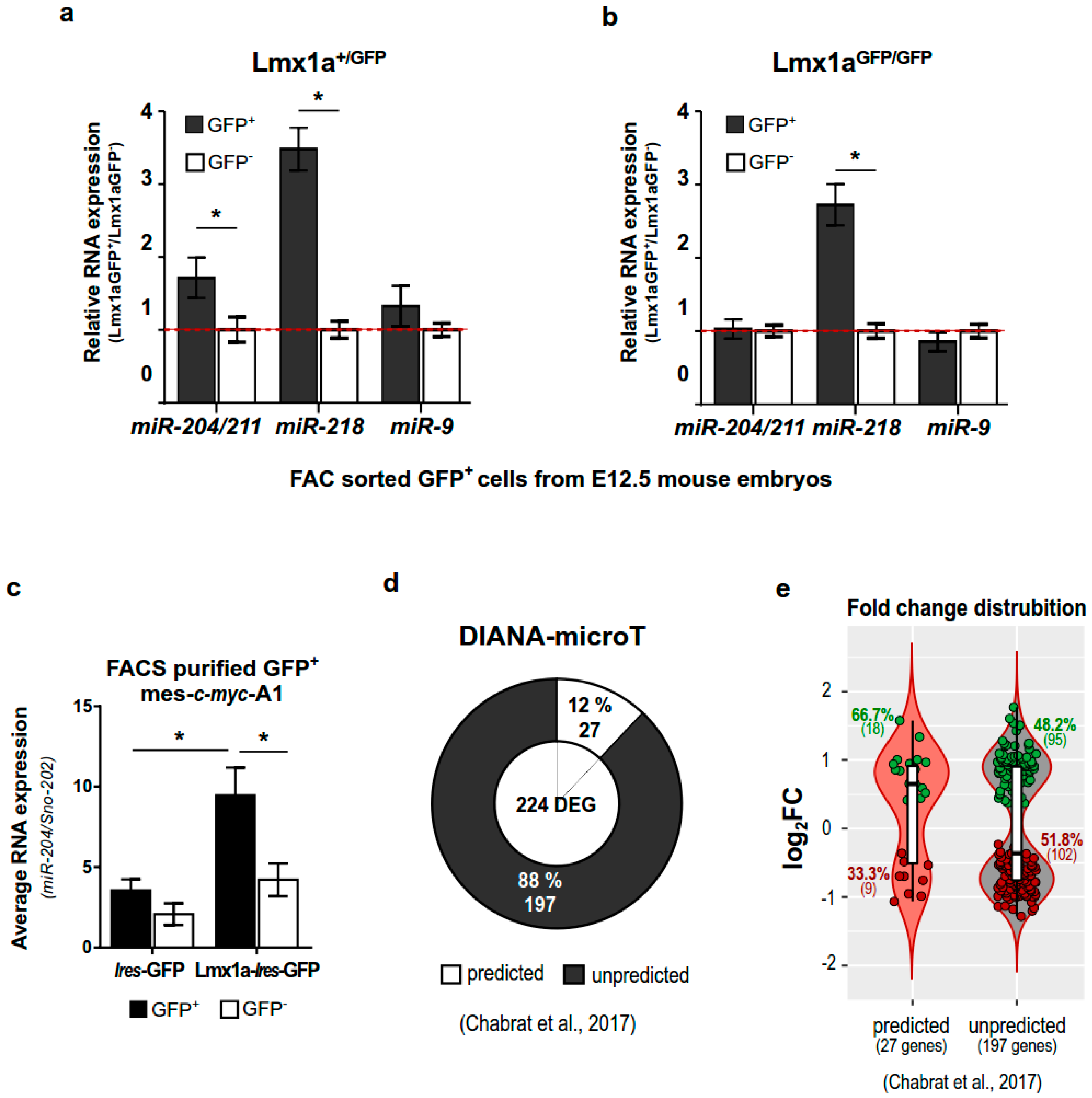
2.3. miR-204/211 Expression Is Regulated by Lmx1a
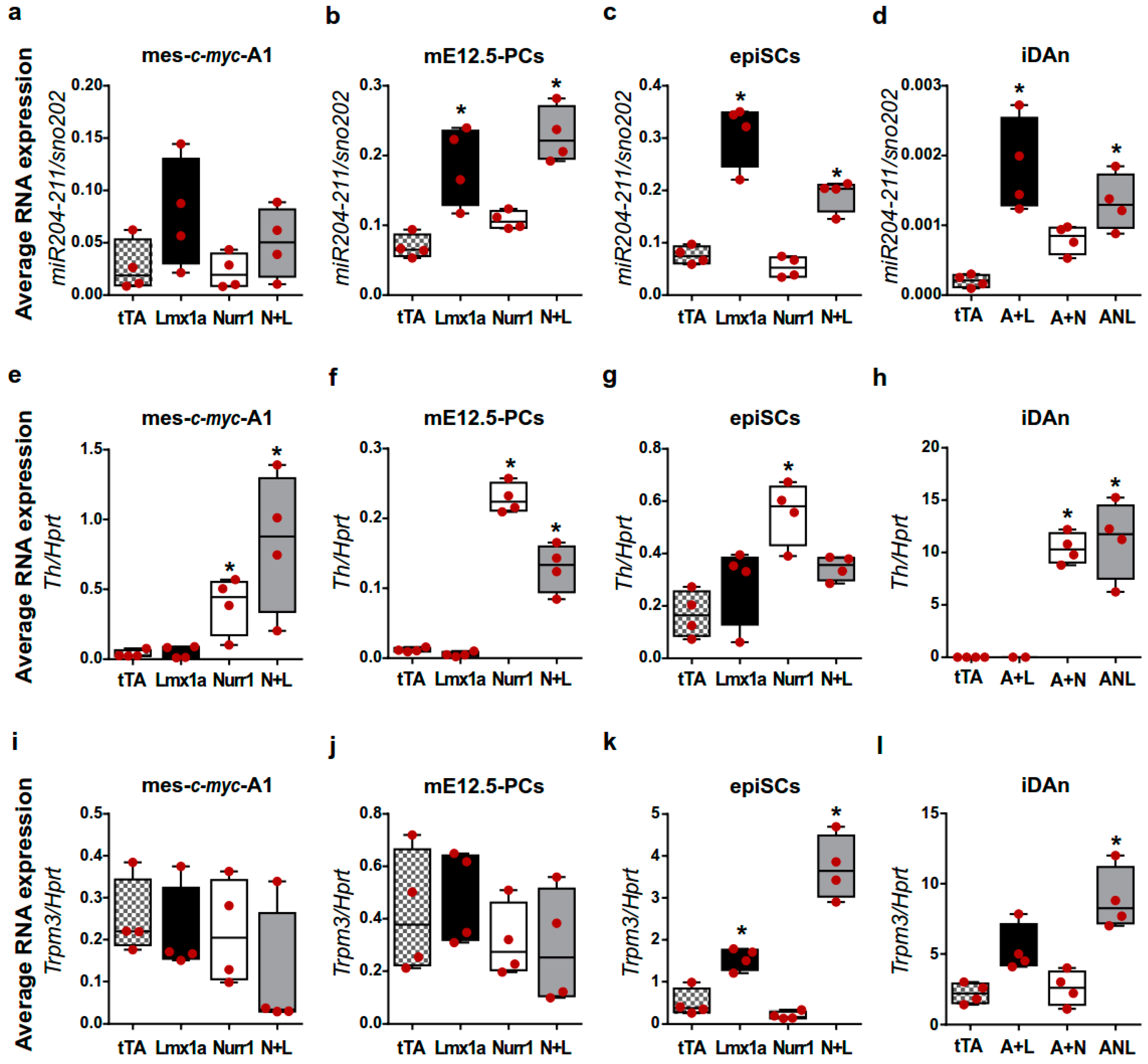
2.4. The Lmx1a-miR-204/211 Axis Controls the Timing of Midbrain Precursors Differentiation
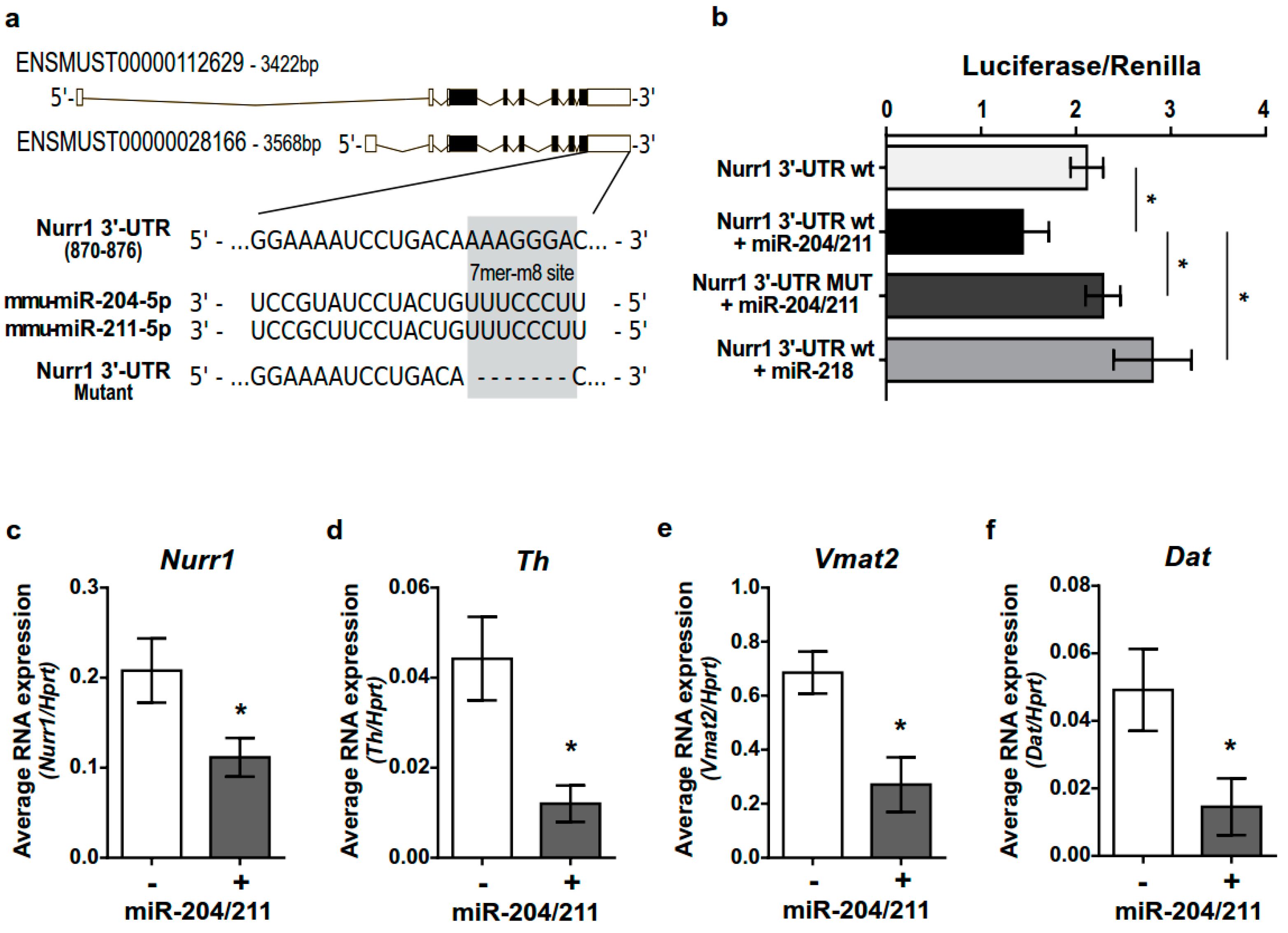
2.5. miR-204/211 Regulates Nurr1 Expression and Influences DA Differentiation
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Tissues Collection
4.3. GFP+ Cell Sorting
4.4. RNA Extraction and Real-Time qPCR
4.5. TaqMan MicroRNA Assays
4.6. Lentivirus Preparation and Viral Infection
4.7. Mesencephalic Primary Cultures (mE12.5-PCs)
4.8. mes-c-myc-A1
4.9. Fluorescence-Activated Cell Sorting
4.10. epiSC Differentiation
4.11. MEF and iDA Reprogramming
4.12. Luciferase
4.13. Putative miRNA-mRNA Interaction and Microarray Data Analysis
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Volpicelli, F.; Perrone-Capano, C.; Bellenchi, G.C.; Colucci-D’Amato, L.; di Porzio, U. Molecular Regulation in Dopaminergic Neuron Development. Cues to Unveil Molecular Pathogenesis and Pharmacological Targets of Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 3995. [Google Scholar] [CrossRef] [PubMed]
- Speranza, L.; di Porzio, U.; Viggiano, D.; de Donato, A.; Volpicelli, F. Dopamine: The Neuromodulator of Long-Term Synaptic Plasticity, Reward and Movement Control. Cells 2021, 10, 735. [Google Scholar] [CrossRef] [PubMed]
- Krashia, P.; Martini, A.; Nobili, A.; Aversa, D.; D’Amelio, M.; Berretta, N.; Guatteo, E.; Mercuri, N.B. On the Properties of Identified Dopaminergic Neurons in the Mouse Substantia Nigra and Ventral Tegmental Area. Eur. J. Neurosci. 2017, 45, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Lo Iacono, L.; Catale, C.; Martini, A.; Valzania, A.; Viscomi, M.T.; Chiurchiù, V.; Guatteo, E.; Bussone, S.; Perrone, F.; Di Sabato, P.; et al. From Traumatic Childhood to Cocaine Abuse: The Critical Function of the Immune System. Biol. Psychiatry 2018, 84, 905–916. [Google Scholar] [CrossRef]
- D’Addario, S.L.; Di Segni, M.; Ledonne, A.; Piscitelli, R.; Babicola, L.; Martini, A.; Spoleti, E.; Mancini, C.; Ielpo, D.; D’Amato, F.R.; et al. Resilience to Anhedonia-Passive Coping Induced by Early Life Experience Is Linked to a Long-Lasting Reduction of Ih Current in VTA Dopaminergic Neurons. Neurobiol. Stress 2021, 14, 100324. [Google Scholar] [CrossRef]
- Andersson, E.; Tryggvason, U.; Deng, Q.; Friling, S.; Alekseenko, Z.; Robert, B.; Perlmann, T.; Ericson, J. Identification of Intrinsic Determinants of Midbrain Dopamine Neurons. Cell 2006, 124, 393–405. [Google Scholar] [CrossRef]
- Yan, C.H.; Levesque, M.; Claxton, S.; Johnson, R.L.; Ang, S.-L. Lmx1a and Lmx1b Function Cooperatively to Regulate Proliferation, Specification, and Differentiation of Midbrain Dopaminergic Progenitors. J. Neurosci. 2011, 31, 12413–12425. [Google Scholar] [CrossRef]
- Deng, Q.; Andersson, E.; Hedlund, E.; Alekseenko, Z.; Coppola, E.; Panman, L.; Millonig, J.H.; Brunet, J.-F.; Ericson, J.; Perlmann, T. Specific and Integrated Roles of Lmx1a, Lmx1b and Phox2a in Ventral Midbrain Development. Development 2011, 138, 3399–3408. [Google Scholar] [CrossRef]
- Ono, Y.; Nakatani, T.; Sakamoto, Y.; Mizuhara, E.; Minaki, Y.; Kumai, M.; Hamaguchi, A.; Nishimura, M.; Inoue, Y.; Hayashi, H.; et al. Differences in Neurogenic Potential in Floor Plate Cells along an Anteroposterior Location: Midbrain Dopaminergic Neurons Originate from Mesencephalic Floor Plate Cells. Development 2007, 134, 3213–3225. [Google Scholar] [CrossRef]
- Failli, V.; Bachy, I.; Rétaux, S. Expression of the LIM-Homeodomain Gene Lmx1a (Dreher) during Development of the Mouse Nervous System. Mech. Dev. 2002, 118, 225–228. [Google Scholar] [CrossRef]
- Millen, K.J.; Millonig, J.H.; Hatten, M.E. Roof Plate and Dorsal Spinal Cord Dl1 Interneuron Development in the Dreher Mutant Mouse. Dev. Biol. 2004, 270, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Chung, S.; Leung, K.; Hwang, I.; Moon, J.; Kim, K.-S. Functional Roles of Nurr1, Pitx3, and Lmx1a in Neurogenesis and Phenotype Specification of Dopamine Neurons During In Vitro Differentiation of Embryonic Stem Cells. Stem Cells Dev. 2014, 23, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Roybon, L.; Hjalt, T.; Christophersen, N.S.; Li, J.-Y.; Brundin, P. Effects on Differentiation of Embryonic Ventral Midbrain Progenitors by Lmx1a, Msx1, Ngn2, and Pitx3. J. Neurosci. 2008, 28, 3644–3656. [Google Scholar] [CrossRef] [PubMed]
- Gale, E.; Li, M. Midbrain Dopaminergic Neuron Fate Specification: Of Mice and Embryonic Stem Cells. Mol. Brain 2008, 1, 8. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Kim, C.-H.; Kim, K.-S. Lmx1a Regulates Dopamine Transporter Gene Expression during ES Cell Differentiation and Mouse Embryonic Development: Lmx1a Regulates Dopamine Transporter Expression. J. Neurochem. 2012, 122, 244–250. [Google Scholar] [CrossRef][Green Version]
- Hoekstra, E.J.; von Oerthel, L.; van der Linden, A.J.A.; Schellevis, R.D.; Scheppink, G.; Holstege, F.C.P.; Groot-Koerkamp, M.J.; van der Heide, L.P.; Smidt, M.P. Lmx1a Is an Activator of Rgs4 and Grb10 and Is Responsible for the Correct Specification of Rostral and Medial MdDA Neurons. Eur. J. Neurosci. 2013, 37, 23–32. [Google Scholar] [CrossRef]
- Perrone-Capano, C.; Tino, A.; Amadoro, G.; Pernas-Alonso, R.; di Porzio, U. Dopamine Transporter Gene Expression in Rat Mesencephalic Dopaminergic Neurons Is Increased by Direct Interaction with Target Striatal Cells in Vitro. Mol. Brain Res. 1996, 39, 160–166. [Google Scholar] [CrossRef]
- Doucet-Beaupré, H.; Gilbert, C.; Profes, M.S.; Chabrat, A.; Pacelli, C.; Giguère, N.; Rioux, V.; Charest, J.; Deng, Q.; Laguna, A.; et al. Lmx1a and Lmx1b Regulate Mitochondrial Functions and Survival of Adult Midbrain Dopaminergic Neurons. Proc. Natl. Acad. Sci. USA 2016, 113, E4387–E4396. [Google Scholar] [CrossRef]
- Jiménez-Moreno, N.; Stathakos, P.; Antón, Z.; Shoemark, D.K.; Sessions, R.B.; Witzgall, R.; Caldwell, M.; Lane, J.D. LIR-Dependent LMX1A/LMX1B Autophagy Crosstalk Shapes Human Midbrain Dopaminergic Neuronal Resilience. Cell Biol. 2019. [Google Scholar] [CrossRef]
- Cai, J.; Donaldson, A.; Yang, M.; German, M.S.; Enikolopov, G.; Iacovitti, L. The Role of Lmx1a in the Differentiation of Human Embryonic Stem Cells into Midbrain Dopamine Neurons in Culture and After Transplantation into a Parkinson’s Disease Model. Stem Cells 2009, 27, 220–229. [Google Scholar] [CrossRef]
- Caiazzo, M.; Dell’Anno, M.T.; Dvoretskova, E.; Lazarevic, D.; Taverna, S.; Leo, D.; Sotnikova, T.D.; Menegon, A.; Roncaglia, P.; Colciago, G.; et al. Direct Generation of Functional Dopaminergic Neurons from Mouse and Human Fibroblasts. Nature 2011, 476, 224–227. [Google Scholar] [CrossRef] [PubMed]
- De Luzy, I.R.; Niclis, J.C.; Gantner, C.W.; Kauhausen, J.A.; Hunt, C.P.J.; Ermine, C.; Pouton, C.W.; Thompson, L.H.; Parish, C.L. Isolation of LMX1a Ventral Midbrain Progenitors Improves the Safety and Predictability of Human Pluripotent Stem Cell-Derived Neural Transplants in Parkinsonian Disease. J. Neurosci. 2019, 39, 9521–9531. [Google Scholar] [CrossRef] [PubMed]
- Ng, Y.H.; Chanda, S.; Janas, J.A.; Yang, N.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Efficient Generation of Dopaminergic Induced Neuronal Cells with Midbrain Characteristics. Stem Cell Rep. 2021, 16, 1763–1776. [Google Scholar] [CrossRef] [PubMed]
- Rouillard, C.; Baillargeon, J.; Paquet, B.; St-Hilaire, M.; Maheux, J.; Lévesque, C.; Darlix, N.; Majeur, S.; Lévesque, D. Genetic Disruption of the Nuclear Receptor Nur77 (Nr4a1) in Rat Reduces Dopamine Cell Loss and l-Dopa-Induced Dyskinesia in Experimental Parkinson’s Disease. Exp. Neurol. 2018, 304, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Colebrooke, R.E.; Humby, T.; Lynch, P.J.; McGowan, D.P.; Xia, J.; Emson, P.C. Age-Related Decline in Striatal Dopamine Content and Motor Performance Occurs in the Absence of Nigral Cell Loss in a Genetic Mouse Model of Parkinson’s Disease: Age-Dependent Manifestations in a Model of Parkinsonism. Eur. J. Neurosci. 2006, 24, 2622–2630. [Google Scholar] [CrossRef]
- Giros, B.; Jaber, M.; Jones, S.R.; Wightman, R.M.; Caron, M.G. Hyperlocomotion and Indifference to Cocaine and Amphetamine in Mice Lacking the Dopamine Transporter. Nature 1996, 379, 606–612. [Google Scholar] [CrossRef]
- Jain, S.; Golden, J.P.; Wozniak, D.; Pehek, E.; Johnson, E.M.; Milbrandt, J. RET Is Dispensable for Maintenance of Midbrain Dopaminergic Neurons in Adult Mice. J. Neurosci. 2006, 26, 11230–11238. [Google Scholar] [CrossRef]
- Joseph, B.; Wallén-Mackenzie, Å.; Benoit, G.; Murata, T.; Joodmardi, E.; Okret, S.; Perlmann, T. P57 Kip2 Cooperates with Nurr1 in Developing Dopamine Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 15619–15624. [Google Scholar] [CrossRef]
- Kramer, E.R.; Aron, L.; Ramakers, G.M.J.; Seitz, S.; Zhuang, X.; Beyer, K.; Smidt, M.P.; Klein, R. Absence of Ret Signaling in Mice Causes Progressive and Late Degeneration of the Nigrostriatal System. PLoS Biol. 2007, 5, e39. [Google Scholar] [CrossRef]
- Volpicelli, F.; Caiazzo, M.; Greco, D.; Consales, C.; Leone, L.; Perrone-Capano, C.; D’Amato, L.C.; Porzio, U. di Bdnf Gene Is a Downstream Target of Nurr1 Transcription Factor in Rat Midbrain Neurons in Vitro: Nurr1 Regulates Bdnf Expression in DA Neurons. J. Neurochem. 2007, 102, 441–453. [Google Scholar] [CrossRef]
- Volpicelli, F.; De Gregorio, R.; Pulcrano, S.; Perrone-Capano, C.; di Porzio, U.; Bellenchi, G.C. Direct Regulation of Pitx3 Expression by Nurr1 in Culture and in Developing Mouse Midbrain. PLoS ONE 2012, 7, e30661. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.-Y.; Quaife, C.J.; Palmiter, R.D. Targeted Disruption of the Tyrosine Hydroxylase Gene Reveals That Catecholamines Are Required for Mouse Fetal Development. Nature 1995, 374, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Zetterström, R.H.; Solomin, L.; Jansson, L.; Hoffer, B.J.; Olson, L.; Perlmann, T. Dopamine Neuron Agenesis in Nurr1-Deficient Mice. Science 1997, 276, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P.; Chen, C.-Z. Micromanagers of Gene Expression: The Potentially Widespread Influence of Metazoan MicroRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Inoue, K.; Ishii, J.; Vanti, W.B.; Voronov, S.V.; Murchison, E.; Hannon, G.; Abeliovich, A. A MicroRNA Feedback Circuit in Midbrain Dopamine Neurons. Science 2007, 317, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, R.; Pulcrano, S.; De Sanctis, C.; Volpicelli, F.; Guatteo, E.; von Oerthel, L.; Latagliata, E.C.; Esposito, R.; Piscitelli, R.M.; Perrone-Capano, C.; et al. MiR-34b/c Regulates Wnt1 and Enhances Mesencephalic Dopaminergic Neuron Differentiation. Stem Cell Rep. 2018, 10, 1237–1250. [Google Scholar] [CrossRef]
- Rivetti di Val Cervo, P.; Romanov, R.A.; Spigolon, G.; Masini, D.; Martín-Montañez, E.; Toledo, E.M.; La Manno, G.; Feyder, M.; Pifl, C.; Ng, Y.-H.; et al. Induction of Functional Dopamine Neurons from Human Astrocytes in Vitro and Mouse Astrocytes in a Parkinson’s Disease Model. Nat. Biotechnol. 2017, 35, 444–452. [Google Scholar] [CrossRef]
- Anderegg, A.; Lin, H.-P.; Chen, J.-A.; Caronia-Brown, G.; Cherepanova, N.; Yun, B.; Joksimovic, M.; Rock, J.; Harfe, B.D.; Johnson, R.; et al. An Lmx1b-MiR135a2 Regulatory Circuit Modulates Wnt1/Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool. PLoS Genet. 2013, 9, e1003973. [Google Scholar] [CrossRef]
- Chiu, C.-C.; Yeh, T.-H.; Chen, R.-S.; Chen, H.-C.; Huang, Y.-Z.; Weng, Y.-H.; Cheng, Y.-C.; Liu, Y.-C.; Cheng, A.-J.; Lu, Y.-C.; et al. Upregulated Expression of MicroRNA-204-5p Leads to the Death of Dopaminergic Cells by Targeting DYRK1A-Mediated Apoptotic Signaling Cascade. Front. Cell. Neurosci. 2019, 13, 399. [Google Scholar] [CrossRef]
- Nair, V.D.; Ge, Y. Alterations of MiRNAs Reveal a Dysregulated Molecular Regulatory Network in Parkinson’s Disease Striatum. Neurosci. Lett. 2016, 629, 99–104. [Google Scholar] [CrossRef]
- Talepoor Ardakani, M.; Rostamian Delavar, M.; Baghi, M.; Nasr-Esfahani, M.H.; Kiani-Esfahani, A.; Ghaedi, K. Upregulation of MiR-200a and MiR-204 in MPP + -treated Differentiated PC12 Cells as a Model of Parkinson’s Disease. Mol. Genet. Genom. Med. 2019, 7, e548. [Google Scholar] [CrossRef] [PubMed]
- Avellino, R.; Carrella, S.; Pirozzi, M.; Risolino, M.; Salierno, F.G.; Franco, P.; Stoppelli, P.; Verde, P.; Banfi, S.; Conte, I. MiR-204 Targeting of Ankrd13A Controls Both Mesenchymal Neural Crest and Lens Cell Migration. PLoS ONE 2013, 8, e61099. [Google Scholar] [CrossRef]
- Conte, I.; Carrella, S.; Avellino, R.; Karali, M.; Marco-Ferreres, R.; Bovolenta, P.; Banfi, S. MiR-204 Is Required for Lens and Retinal Development via Meis2 Targeting. Proc. Natl. Acad. Sci. USA 2010, 107, 15491–15496. [Google Scholar] [CrossRef] [PubMed]
- Conte, I.; Merella, S.; Garcia-Manteiga, J.M.; Migliore, C.; Lazarevic, D.; Carrella, S.; Marco-Ferreres, R.; Avellino, R.; Davidson, N.P.; Emmett, W.; et al. The Combination of Transcriptomics and Informatics Identifies Pathways Targeted by MiR-204 during Neurogenesis and Axon Guidance. Nucleic Acids Res. 2014, 42, 7793–7806. [Google Scholar] [CrossRef]
- Lepko, T.; Pusch, M.; Müller, T.; Schulte, D.; Ehses, J.; Kiebler, M.; Hasler, J.; Huttner, H.B.; Vandenbroucke, R.E.; Vandendriessche, C.; et al. Choroid Plexus-derived MiR-204 Regulates the Number of Quiescent Neural Stem Cells in the Adult Brain. EMBO J. 2019, 38, e100481. [Google Scholar] [CrossRef]
- Danka Mohammed, C.P.; Rhee, H.; Phee, B.; Kim, K.; Kim, H.; Lee, H.; Park, J.H.; Jung, J.H.; Kim, J.Y.; Kim, H.; et al. MiR-204 Downregulates EphB2 in Aging Mouse Hippocampal Neurons. Aging Cell 2016, 15, 380–388. [Google Scholar] [CrossRef]
- Venø, M.T.; Venø, S.T.; Rehberg, K.; van Asperen, J.V.; Clausen, B.H.; Holm, I.E.; Pasterkamp, R.J.; Finsen, B.; Kjems, J. Cortical Morphogenesis during Embryonic Development Is Regulated by MiR-34c and MiR-204. Front. Mol. Neurosci. 2017, 10, 31. [Google Scholar] [CrossRef]
- Pereira, L.A.; Munita, R.; González, M.P.; Andrés, M.E. Long 3′UTR of Nurr1 MRNAs Is Targeted by MiRNAs in Mesencephalic Dopamine Neurons. PLoS ONE 2017, 12, e0188177. [Google Scholar] [CrossRef]
- Wang, X.; Liu, L.; Zhang, L.; Guo, J.; Yu, L.; Li, T. Circ_0057583 Facilitates Brain Microvascular Endothelial Cell Injury through Modulating MiR-204-5p/NR4A1 Axis. Metab. Brain Dis. 2022, 37, 501–511. [Google Scholar] [CrossRef]
- Sawamoto, K.; Nakao, N.; Kobayashi, K.; Matsushita, N.; Takahashi, H.; Kakishita, K.; Yamamoto, A.; Yoshizaki, T.; Terashima, T.; Murakami, F.; et al. Visualization, Direct Isolation, and Transplantation of Midbrain Dopaminergic Neurons. Proc. Natl. Acad. Sci. USA 2001, 98, 6423–6428. [Google Scholar] [CrossRef]
- Zhao, S.; Maxwell, S.; Jimenez-Beristain, A.; Vives, J.; Kuehner, E.; Zhao, J.; O’Brien, C.; de Felipe, C.; Semina, E.; Li, M. Generation of Embryonic Stem Cells and Transgenic Mice Expressing Green Fluorescence Protein in Midbrain Dopaminergic Neurons. Eur. J. Neurosci. 2004, 19, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Liu, Y.; Huang, M.; Zhao, X.; Cheng, L. Wnt1-Cre-Mediated Conditional Loss of Dicer Results in Malformation of the Midbrain and Cerebellum and Failure of Neural Crest and Dopaminergic Differentiation in Mice. J. Mol. Cell Biol. 2010, 2, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Bak, M.; Silahtaroglu, A.; Møller, M.; Christensen, M.; Rath, M.F.; Skryabin, B.; Tommerup, N.; Kauppinen, S. MicroRNA Expression in the Adult Mouse Central Nervous System. RNA 2008, 14, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A Mammalian MicroRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Colucci-D’Amato, G. Neuronal and Glial Properties Coexist in a Novel Mouse CNS Immortalized Cell Line. Exp. Cell Res. 1999, 252, 383–391. [Google Scholar] [CrossRef]
- Chabrat, A.; Brisson, G.; Doucet-Beaupré, H.; Salesse, C.; Schaan Profes, M.; Dovonou, A.; Akitegetse, C.; Charest, J.; Lemstra, S.; Côté, D.; et al. Transcriptional Repression of Plxnc1 by Lmx1a and Lmx1b Directs Topographic Dopaminergic Circuit Formation. Nat. Commun. 2017, 8, 933. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-MicroT Web Server v5.0: Service Integration into MiRNA Functional Analysis Workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef]
- Gennet, N.; Gale, E.; Nan, X.; Farley, E.; Takacs, K.; Oberwallner, B.; Chambers, D.; Li, M. Doublesex and Mab-3–Related Transcription Factor 5 Promotes Midbrain Dopaminergic Identity in Pluripotent Stem Cells by Enforcing a Ventral-Medial Progenitor Fate. Proc. Natl. Acad. Sci. USA 2011, 108, 9131–9136. [Google Scholar] [CrossRef]
- Hedlund, E.; Belnoue, L.; Theofilopoulos, S.; Salto, C.; Bye, C.; Parish, C.; Deng, Q.; Kadkhodaei, B.; Ericson, J.; Arenas, E.; et al. Dopamine Receptor Antagonists Enhance Proliferation and Neurogenesis of Midbrain Lmx1a-Expressing Progenitors. Sci. Rep. 2016, 6, 26448. [Google Scholar] [CrossRef] [PubMed]
- Cantone, M.; Küspert, M.; Reiprich, S.; Lai, X.; Eberhardt, M.; Göttle, P.; Beyer, F.; Azim, K.; Küry, P.; Wegner, M.; et al. A Gene Regulatory Architecture That Controls Region-independent Dynamics of Oligodendrocyte Differentiation. Glia 2019, 67, 825–843. [Google Scholar] [CrossRef]
- Reiprich, S.; Cantone, M.; Weider, M.; Baroti, T.; Wittstatt, J.; Schmitt, C.; Küspert, M.; Vera, J.; Wegner, M. Transcription Factor Sox10 Regulates Oligodendroglial Sox9 Levels via MicroRNAs: Sox Proteins and MicroRNAs. Glia 2017, 65, 1089–1102. [Google Scholar] [CrossRef] [PubMed]
- Amin, N.D.; Bai, G.; Klug, J.R.; Bonanomi, D.; Pankratz, M.T.; Gifford, W.D.; Hinckley, C.A.; Sternfeld, M.J.; Driscoll, S.P.; Dominguez, B.; et al. Loss of Motoneuron-Specific MicroRNA-218 Causes Systemic Neuromuscular Failure. Science 2015, 350, 1525–1529. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Chen, D.; Xiao, T.; Lin, D.; Lin, D.; Lin, L.; Zhu, H.; Xu, J.; Huang, W.; Yang, T. DNA Methylation-mediated Silencing of MicroRNA-204 Enhances T Cell Acute Lymphoblastic Leukemia by Up-regulating MMP-2 and MMP-9 via NF-κB. J. Cell. Mol. Med. 2021, 25, 2365–2376. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Smibert, C.A.; Kaplan, D.R.; Miller, F.D. An EIF4E1/4E-T Complex Determines the Genesis of Neurons from Precursors by Translationally Repressing a Proneurogenic Transcription Program. Neuron 2014, 84, 723–739. [Google Scholar] [CrossRef]
- Kambey, P.A.; Kanwore, K.; Ayanlaja, A.A.; Nadeem, I.; Du, Y.; Buberwa, W.; Liu, W.; Gao, D. Failure of Glial Cell-Line Derived Neurotrophic Factor (GDNF) in Clinical Trials Orchestrated By Reduced NR4A2 (NURR1) Transcription Factor in Parkinson’s Disease. A Systematic Review. Front. Aging Neurosci. 2021, 13, 645583. [Google Scholar] [CrossRef]
- Wu, M.; Deng, Q.; Lei, X.; Du, Y.; Shen, Y. Elavl2 Regulates Retinal Function Via Modulating the Differentiation of Amacrine Cells Subtype. Investig. Ophthalmol. Vis. Sci. 2021, 62, 1. [Google Scholar] [CrossRef]
- Jacobs, F.M.J.; van der Linden, A.J.A.; Wang, Y.; von Oerthel, L.; Sul, H.S.; Burbach, J.P.H.; Smidt, M.P. Identification of Dlk1, Ptpru and Klhl1 as Novel Nurr1 Target Genes in Meso-Diencephalic Dopamine Neurons. Development 2009, 136, 2363–2373. [Google Scholar] [CrossRef]
- Mesman, S.; Smidt, M.P. Tcf12 Is Involved in Early Cell-Fate Determination and Subset Specification of Midbrain Dopamine Neurons. Front. Mol. Neurosci. 2017, 10, 353. [Google Scholar] [CrossRef]
- Belle, M.; Parray, A.; Belle, M.; Chédotal, A.; Nguyen-Ba-Charvet, K.T. PlexinA2 and Sema6A Are Required for Retinal Progenitor Cell Migration. Dev. Growth Differ. 2016, 58, 492–502. [Google Scholar] [CrossRef]
- Epting, D.; Vorwerk, S.; Hageman, A.; Meyer, D. Expression of Rasgef1b in Zebrafish. Gene Expr. Patterns 2007, 7, 389–395. [Google Scholar] [CrossRef]
- Goodings, L.; He, J.; Wood, A.J.; Harris, W.A.; Currie, P.D.; Jusuf, P.R. In Vivo Expression of Nurr1/Nr4a2a in Developing Retinal Amacrine Subtypes in Zebrafish Tg(Nr4a2a:EGFP) Transgenics: GOODINGS et Al. J. Comp. Neurol. 2017, 525, 1962–1979. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.D.; Zhang, M.; Ferguson, J.W.; Koch, M.; Brunken, W.J. The Extracellular Matrix Component WIF-1 Is Expressed during, and Can Modulate, Retinal Development. Mol. Cell. Neurosci. 2004, 27, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Usui, A.; Mochizuki, Y.; Iida, A.; Miyauchi, E.; Satoh, S.; Sock, E.; Nakauchi, H.; Aburatani, H.; Murakami, A.; Wegner, M.; et al. The Early Retinal Progenitor-Expressed Gene Sox11 Regulates the Timing of the Differentiation of Retinal Cells. Development 2013, 140, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Fajol, A.; Tu, X.; Ren, B.; Shi, S. MiR-204 Suppresses the Development and Progression of Human Glioblastoma by Targeting ATF2. Oncotarget 2016, 7, 70058–70065. [Google Scholar] [CrossRef]
- Jacobs, F.M.J.; Veenvliet, J.V.; Almirza, W.H.; Hoekstra, E.J.; von Oerthel, L.; van der Linden, A.J.A.; Neijts, R.; Koerkamp, M.G.; van Leenen, D.; Holstege, F.C.P.; et al. Retinoic Acid-Dependent and -Independent Gene-Regulatory Pathways of Pitx3 in Meso-Diencephalic Dopaminergic Neurons. Development 2011, 138, 5213–5222. [Google Scholar] [CrossRef]
- Kee, N.; Volakakis, N.; Kirkeby, A.; Dahl, L.; Storvall, H.; Nolbrant, S.; Lahti, L.; Björklund, Å.K.; Gillberg, L.; Joodmardi, E.; et al. Single-Cell Analysis Reveals a Close Relationship between Differentiating Dopamine and Subthalamic Nucleus Neuronal Lineages. Cell Stem Cell 2017, 20, 29–40. [Google Scholar] [CrossRef]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct Conversion of Fibroblasts to Functional Neurons by Defined Factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef]
- De Risi, M.; Tufano, M.; Alvino, F.G.; Ferraro, M.G.; Torromino, G.; Gigante, Y.; Monfregola, J.; Marrocco, E.; Pulcrano, S.; Tunisi, L.; et al. Altered Heparan Sulfate Metabolism during Development Triggers Dopamine-Dependent Autistic-Behaviours in Models of Lysosomal Storage Disorders. Nat. Commun. 2021, 12, 3495. [Google Scholar] [CrossRef]
- Jaeger, I.; Arber, C.; Risner-Janiczek, J.R.; Kuechler, J.; Pritzsche, D.; Chen, I.-C.; Naveenan, T.; Ungless, M.A.; Li, M. Temporally Controlled Modulation of FGF/ERK Signaling Directs Midbrain Dopaminergic Neural Progenitor Fate in Mouse and Human Pluripotent Stem Cells. Development 2011, 138, 4363–4374. [Google Scholar] [CrossRef]
- Della Valle, F.; Thimma, M.P.; Caiazzo, M.; Pulcrano, S.; Celii, M.; Adroub, S.A.; Liu, P.; Alanis-Lobato, G.; Broccoli, V.; Orlando, V. Transdifferentiation of Mouse Embryonic Fibroblasts into Dopaminergic Neurons Reactivates LINE-1 Repetitive Elements. Stem Cell Rep. 2020, 14, 60–74. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective MicroRNA Target Sites in Mammalian MRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
| Gene | Forward | Reverse |
|---|---|---|
| Dat | TCTGGGTATCGACAGTGCCA | GCAGCTGGAACTCATCGACAA |
| Hprt | TGGGAGGCCATCACATTGT | AATCCAGCAGGTCAGCAAAGA |
| Lmx1a | AACCAGCGAGCCAAGATGAA | TGGGTGTTCTGTTGGTCCTGT |
| Nurr1 | CAACTACAGCACAGGCTACGA | GCATCTGAATGTCTTCTACTTAAT |
| Th | CCTTTGACCCAGACACAGCA | ATACGAGAGGCATAGTTCCTGAG |
| Trpm1 | CTGCCTTGCTCAAAGGAACCAA | GAGGGGCCAGGCGGCCCAG |
| Trpm3 | CATGCACTCCCACTTCATCC | TGGAACCCCTTGACCGATT |
| Vmat2 | TTGCTCATCTGTGGCTGGG | TGGCGTTACCCCTCTCTTCAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pulcrano, S.; De Gregorio, R.; De Sanctis, C.; Lahti, L.; Perrone-Capano, C.; Ponti, D.; di Porzio, U.; Perlmann, T.; Caiazzo, M.; Volpicelli, F.; et al. Lmx1a-Dependent Activation of miR-204/211 Controls the Timing of Nurr1-Mediated Dopaminergic Differentiation. Int. J. Mol. Sci. 2022, 23, 6961. https://doi.org/10.3390/ijms23136961
Pulcrano S, De Gregorio R, De Sanctis C, Lahti L, Perrone-Capano C, Ponti D, di Porzio U, Perlmann T, Caiazzo M, Volpicelli F, et al. Lmx1a-Dependent Activation of miR-204/211 Controls the Timing of Nurr1-Mediated Dopaminergic Differentiation. International Journal of Molecular Sciences. 2022; 23(13):6961. https://doi.org/10.3390/ijms23136961
Chicago/Turabian StylePulcrano, Salvatore, Roberto De Gregorio, Claudia De Sanctis, Laura Lahti, Carla Perrone-Capano, Donatella Ponti, Umberto di Porzio, Thomas Perlmann, Massimiliano Caiazzo, Floriana Volpicelli, and et al. 2022. "Lmx1a-Dependent Activation of miR-204/211 Controls the Timing of Nurr1-Mediated Dopaminergic Differentiation" International Journal of Molecular Sciences 23, no. 13: 6961. https://doi.org/10.3390/ijms23136961
APA StylePulcrano, S., De Gregorio, R., De Sanctis, C., Lahti, L., Perrone-Capano, C., Ponti, D., di Porzio, U., Perlmann, T., Caiazzo, M., Volpicelli, F., & Bellenchi, G. C. (2022). Lmx1a-Dependent Activation of miR-204/211 Controls the Timing of Nurr1-Mediated Dopaminergic Differentiation. International Journal of Molecular Sciences, 23(13), 6961. https://doi.org/10.3390/ijms23136961






