Exploring the Origin and Physiological Significance of DNA Double Strand Breaks in the Developing Neuroretina
Abstract
1. Introduction
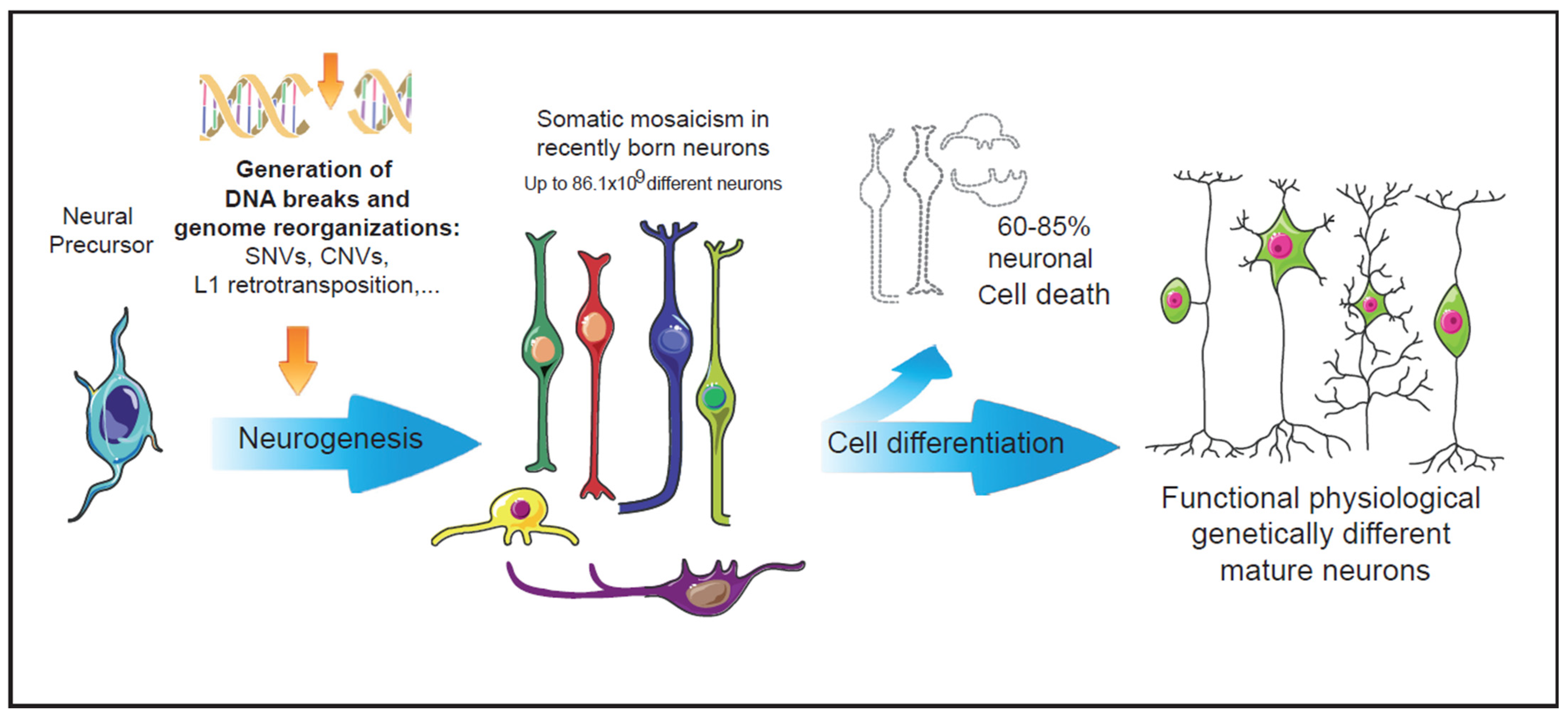
2. DSBs and Neural Development
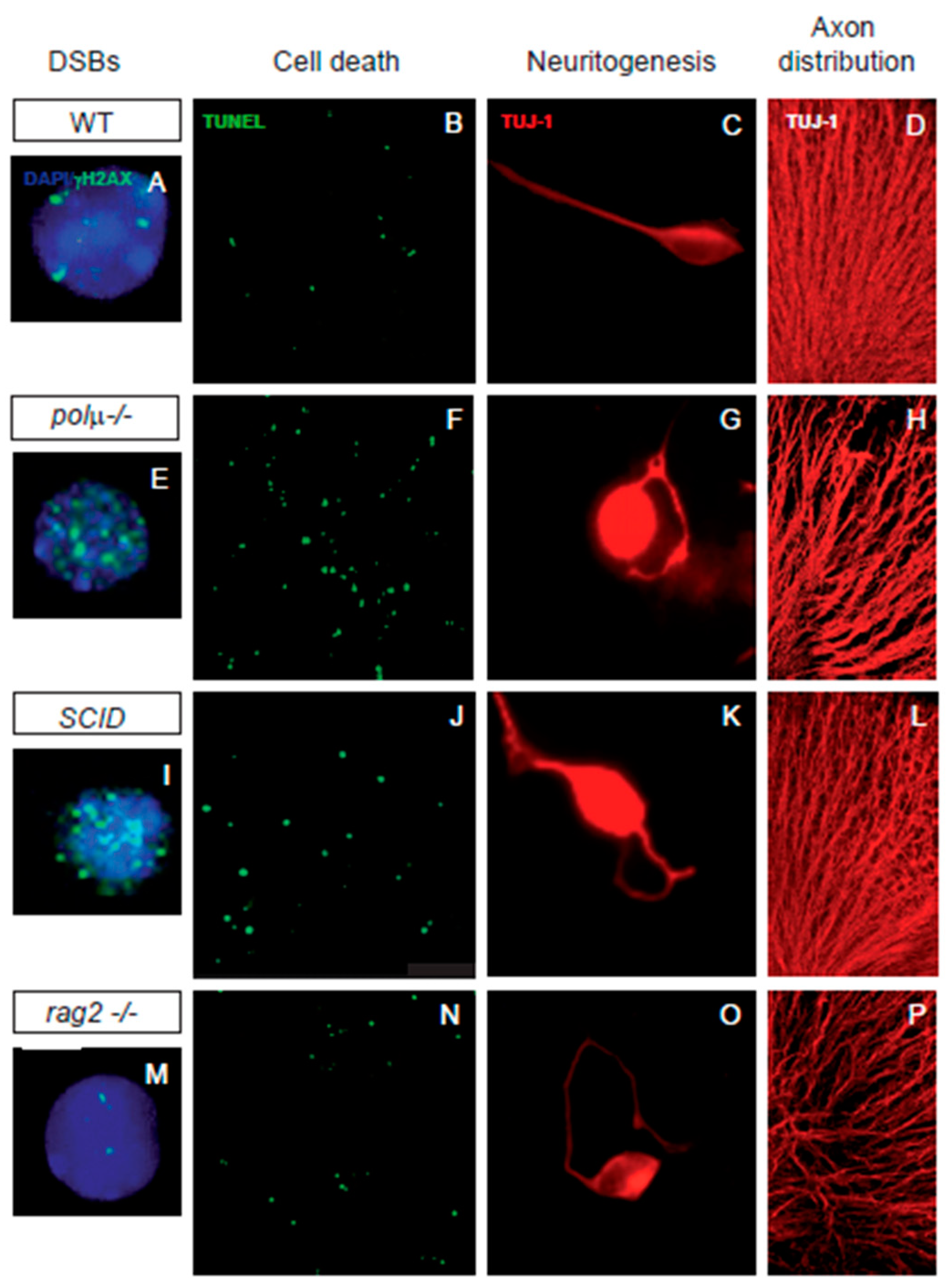
3. DSBs and Early Neural Cell Death
4. Mechanisms Underlying Specific DSBs: A Potential role for RAG-1,2 in Neural Development?
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Evrony, G.D.; Cai, X.; Lee, E.; Hills, L.B.; Elhosary, P.C.; Lehmann, H.S.; Parker, J.J.; Atabay, K.D.; Gilmore, E.C.; Poduri, A.; et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 2012, 151, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Evrony, G.D.; Lee, E.; Mehta, B.K.; Benjamini, Y.; Johnson, R.M.; Cai, X.; Yang, L.; Haseley, P.; Lehmann, H.S.; Park, P.J.; et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron 2015, 85, 49–59. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Lindberg, M.R.; Brennand, K.J.; Piper, J.C.; Voet, T.; Cowing-Zitron, C.; Shumilina, S.; Lasken, R.S.; Vermeesch, J.R.; Hall, I.M.; et al. Mosaic copy number variation in human neurons. Science 2013, 342, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Evrony, G.D.; Lehmann, H.S.; Elhosary, P.C.; Mehta, B.K.; Poduri, A.; Walsh, C.A. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2015, 10, 645. [Google Scholar] [CrossRef] [PubMed]
- Bushman, D.M.; Chun, J. The genomically mosaic brain: Aneuploidy and more in neural diversity and disease. Semin. Cell Dev. Biol. 2013, 24, 357–369. [Google Scholar] [CrossRef]
- Johnson, M.B.; Walsh, C.A. Cerebral cortical neuron diversity and development at single-cell resolution. Curr. Opin. Neurobiol. 2017, 42, 9–16. [Google Scholar] [CrossRef][Green Version]
- Westra, J.W.; Rivera, R.R.; Bushman, D.M.; Yung, Y.C.; Peterson, S.E.; Barral, S.; Chun, J. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J. Comp. Neurol. 2010, 518, 3981–4000. [Google Scholar] [CrossRef]
- Hazen, J.L.; Faust, G.G.; Rodriguez, A.R.; Ferguson, W.C.; Shumilina, S.; Clark, R.A.; Boland, M.J.; Martin, G.; Chubukov, P.; Tsunemoto, R.K.; et al. The Complete Genome Sequences, Unique Mutational Spectra, and Developmental Potency of Adult Neurons Revealed by Cloning. Neuron 2016, 89, 1223–1236. [Google Scholar] [CrossRef]
- Rohrback, S.; Siddoway, B.; Liu, C.S.; Chun, J. Genomic mosaicism in the developing and adult brain. Dev. Neurobiol. 2018, 78, 1026–1048. [Google Scholar] [CrossRef]
- Rohrback, S.; April, C.; Kaper, F.; Rivera, R.R.; Liu, C.S.; Siddoway, B.; Chun, J. Submegabase copy number variations arise during cerebral cortical neurogenesis as revealed by single-cell whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2018, 115, 10804–10809. [Google Scholar] [CrossRef]
- Kaushal, D.; Contos, J.J.A.; Treuner, K.; Yang, A.H.; Kingsbury, M.A.; Rehen, S.; McConnell, M.J.; Okabe, M.; Barlow, C.; Chun, J. Alteration of Gene Expression by Chromosome Loss in the Postnatal Mouse Brain. J. Neurosci. 2003, 23, 5599–5606. [Google Scholar] [CrossRef] [PubMed]
- Kingsbury, M.A.; Friedman, B.; McConnell, M.J.; Rehen, S.K.; Yang, A.H.; Kaushal, D.; Chun, J. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc. Natl. Acad. Sci. USA 2005, 102, 6143–6147. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.H.; Kaushal, D.; Rehen, S.; Kriedt, K.; Kingsbury, M.A.; McConnell, M.J.; Chun, J. Chromosome Segregation Defects Contribute to Aneuploidy in Normal Neural Progenitor Cells. J. Neurosci. 2003, 23, 10454–10462. [Google Scholar] [CrossRef] [PubMed]
- Muotri, A.R.; Gage, F.H. Generation of neuronal variability and complexity. Nature 2006, 441, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Boya, P.; de la Rosa, E.J. Cell death in early neural life. Birth Defects Res. C Embryo Today 2005, 75, 281–293. [Google Scholar] [CrossRef]
- Alt, F.W.; Schwer, B. DNA double-strand breaks as drivers of neural genomic change, function, and disease. DNA Repair 2018, 71, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Alt, F.W.; Wei, P.C.; Schwer, B. Recurrently breaking genes in neural progenitors: Potential roles of DNA breaks in neuronal function, degeneration and cancer. In Genome Editing in Neurosciences; Jaenisch, R., Zhang, F., Gage, F., Eds.; Springer: Cham, Switzerland, 2017; pp. 63–72. [Google Scholar]
- Jourdon, A.; Fasching, L.; Scuderi, S.; Abyzov, A.; Vaccarino, F.M. The role of somatic mosaicism in brain disease. Curr. Opin. Genet. Dev. 2020, 65, 84–90. [Google Scholar] [CrossRef]
- Kuzminov, A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. USA 2001, 98, 8241–8246. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef]
- Deriano, L.; Roth, D.B. Modernizing the nonhomologous end-joining repertoire: Alternative and classical NHEJ share the stage. Annu. Rev. Genet. 2013, 47, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Lescale, C.; Deriano, L. The RAG recombinase: Beyond breaking. Mech. Ageing Dev. 2017, 165, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Alt, F.W.; Zhang, Y.; Meng, F.L.; Guo, C.; Schwer, B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 2013, 152, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.A.; Paquola, A.C.; Muotri, A.R. LINE-1 retrotransposition in the nervous system. Annu. Rev. Cell Dev. Biol. 2012, 28, 555–573. [Google Scholar] [CrossRef] [PubMed]
- Suarez, N.A.; Macia, A.; Muotri, A.R. LINE-1 retrotransposons in healthy and diseased human brain. Dev. Neurobiol. 2017, 78, 434–455. [Google Scholar] [CrossRef]
- Lee, M.H.; Siddoway, B.; Kaeser, G.E.; Segota, I.; Rivera, R.; Romanow, W.J.; Liu, C.S.; Park, C.; Kennedy, G.; Long, T.; et al. Somatic APP gene recombination in Alzheimer’s disease and normal neurons. Nature 2018, 563, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef]
- Puc, J.; Aggarwal, A.K.; Rosenfeld, M.G. Physiological functions of programmed DNA breaks in signal-induced transcription. Nat. Rev. Mol. Cell Biol. 2017, 18, 471–476. [Google Scholar] [CrossRef]
- Suberbielle, E.; Sanchez, P.E.; Kravitz, A.V.; Wang, X.; Ho, K.; Eilertson, K.; Devidze, N.; Kreitzer, A.C.; Mucke, L. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 2013, 16, 613. [Google Scholar] [CrossRef]
- Frappart, P.O.; McKinnon, P.J. Mouse models of DNA double-strand break repair and neurological disease. DNA Repair 2008, 7, 1051–1060. [Google Scholar] [CrossRef]
- McKinnon, P.J.; Caldecott, K.W. DNA strand break repair and human genetic disease. Annu. Rev. Genom. Hum. Genet. 2007, 8, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; McKinnon, P.J. Responding to DNA double strand breaks in the nervous system. Neuroscience 2007, 145, 1365–1374. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, P.J. Maintaining genome stability in the nervous system. Nat. Neurosci. 2013, 16, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Valenciano, A.I.; Boya, P.; de la Rosa, E.J. Early neural cell death: Numbers and cues from the developing neuroretina. Int. J. Dev. Biol. 2009, 53, 1515–1528. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Sun, Y.; Frank, K.M.; Dikkes, P.; Fujiwara, Y.; Seidl, K.J.; Sekiguchi, J.M.; Rathbun, G.A.; Swat, W.; Wang, J.; et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 1998, 95, 891–902. [Google Scholar] [CrossRef]
- Frank, K.M.; Sharpless, N.E.; Gao, Y.; Sekiguchi, J.M.; Ferguson, D.O.; Zhu, C.; Manis, J.P.; Horner, J.; DePinho, R.A.; Alt, F.W. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell 2000, 5, 993–1002. [Google Scholar] [CrossRef]
- Barnes, D.E.; Stamp, G.; Rosewell, I.; Denzel, A.; Lindahl, T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998, 8, 1395–1398. [Google Scholar] [CrossRef]
- Woodbine, L.; Gennery, A.R.; Jeggo, P.A. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair 2014, 16, 84–96. [Google Scholar] [CrossRef]
- Zha, S.; Alt, F.W.; Cheng, H.L.; Brush, J.W.; Li, G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc. Natl. Acad. Sci. USA 2007, 104, 4518–4523. [Google Scholar] [CrossRef]
- El Waly, B.; Buhler, E.; Haddad, M.R.; Villard, L. Nhej1 Deficiency Causes Abnormal Development of the Cerebral Cortex. Mol. Neurobiol. 2014, 52, 771–782. [Google Scholar] [CrossRef]
- Buck, D.; Malivert, L.; de Chasseval, R.; Barraud, A.; Fondaneche, M.C.; Sanal, O.; Plebani, A.; Stephan, J.L.; Hufnagel, M.; le Deist, F.; et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 2006, 124, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Cipe, F.E.; Aydogmus, C.; Babayigit Hocaoglu, A.; Kilic, M.; Kaya, G.D.; Yilmaz Gulec, E. Cernunnos/XLF Deficiency: A Syndromic Primary Immunodeficiency. Case Rep. Pediatr. 2014, 2014, 614238. [Google Scholar] [PubMed]
- Rosin, N.; Elcioglu, N.H.; Beleggia, F.; Isguven, P.; Altmuller, J.; Thiele, H.; Steindl, K.; Joset, P.; Rauch, A.; Nurnberg, P.; et al. Mutations in XRCC4 cause primary microcephaly, short stature and increased genomic instability. Hum. Mol. Genet. 2015, 24, 3708–3717. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.E.; van der Burg, M.; IJspeert, I.J.; Carroll, P.; Wu, Q.; Ochi, T.; Leitch, A.; Miller, E.S.; Kysela, B.; Jawad, A.; et al. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am. J. Hum. Genet. 2015, 96, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Sobol, R.W. Genome instability caused by a germline mutation in the human DNA repair gene POLB. PLoS Genet. 2012, 8, e1003086. [Google Scholar] [CrossRef] [PubMed]
- Weissman, L.; Jo, D.G.; Sorensen, M.M.; de Souza-Pinto, N.C.; Markesbery, W.R.; Mattson, M.P.; Bohr, V.A. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007, 35, 5545–5555. [Google Scholar] [CrossRef]
- Baleriola, J.; Suarez, T.; de la Rosa, E.J. DNA-PK promotes the survival of young neurons in the embryonic mouse retina. Cell Death Differ. 2010, 17, 1697–1706. [Google Scholar] [CrossRef]
- Alvarez-Lindo, N.; Baleriola, J.; de Los Rios, V.; Suarez, T.; de la Rosa, E.J. RAG-2 deficiency results in fewer phosphorylated histone H2AX foci, but increased retinal ganglion cell death and altered axonal growth. Sci. Rep. 2019, 9, 18486. [Google Scholar] [CrossRef]
- Niimi, N.; Sugo, N.; Aratani, Y.; Koyama, H. Genetic interaction between DNA polymerase beta and DNA-PKcs in embryogenesis and neurogenesis. Cell Death Differ. 2005, 12, 184–191. [Google Scholar] [CrossRef]
- Fulop, G.M.; Phillips, R.A. The scid mutation in mice causes a general defect in DNA repair. Nature 1990, 347, 479–482. [Google Scholar] [CrossRef]
- Shultz, L.D.; Schweitzer, P.A.; Christianson, S.W.; Gott, B.; Schweitzer, I.B.; Tennent, B.; McKenna, S.; Mobraaten, L.; Rajan, T.V.; Greiner, D.L.; et al. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J. Immunol. 1995, 154, 180–191. [Google Scholar] [PubMed]
- van der Burg, M.; Ijspeert, H.; Verkaik, N.S.; Turul, T.; Wiegant, W.W.; Morotomi-Yano, K.; Mari, P.O.; Tezcan, I.; Chen, D.J.; Zdzienicka, M.Z.; et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J. Clin. Invest. 2009, 119, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Rooney, S.; Alt, F.W.; Lombard, D.; Whitlow, S.; Eckersdorff, M.; Fleming, J.; Fugmann, S.; Ferguson, D.O.; Schatz, D.G.; Sekiguchi, J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 2003, 197, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Petersen, S.; Tessarollo, L.; Nussenzweig, A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 2001, 11, 105–109. [Google Scholar] [CrossRef]
- Buis, J.; Wu, Y.; Deng, Y.; Leddon, J.; Westfield, G.; Eckersdorff, M.; Sekiguchi, J.M.; Chang, S.; Ferguson, D.O. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 2008, 135, 85–96. [Google Scholar] [CrossRef]
- Gu, Y.; Sekiguchi, J.; Gao, Y.; Dikkes, P.; Frank, K.; Ferguson, D.; Hasty, P.; Chun, J.; Alt, F.W. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc. Natl. Acad. Sci. USA 2000, 97, 2668–2673. [Google Scholar] [CrossRef]
- Wang, Y.; Ghosh, G.; Hendrickson, E.A. Ku86 represses lethal telomere deletion events in human somatic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 12430–12435. [Google Scholar] [CrossRef]
- Korabiowska, M.; Konig, F.; Schlott, T.; Slotty, P.; Romeike, B.; Cordon-Cardo, C.; Brinck, U. Quantitative Analysis of Ku70 and Ku80 mRNA Gene Expression in Melanoma Brain Metastases. Correlation with Immunohistochemistry and In Situ Hybridization. Cancer Genom. Proteom. 2004, 1, 225–230. [Google Scholar]
- Eilam, R.; Peter, Y.; Elson, A.; Rotman, G.; Shiloh, Y.; Groner, Y.; Segal, M. Selective loss of dopaminergic nigro-striatal neurons in brains of Atm-deficient mice. Proc. Natl. Acad. Sci. USA 1998, 95, 12653–12656. [Google Scholar] [CrossRef]
- Barlow, C.; Hirotsune, S.; Paylor, R.; Liyanage, M.; Eckhaus, M.; Collins, F.; Shiloh, Y.; Crawley, J.N.; Ried, T.; Tagle, D.; et al. Atm-deficient mice: A paradigm of ataxia telangiectasia. Cell 1996, 86, 159–171. [Google Scholar] [CrossRef]
- Baleriola, J.; Alvarez-Lindo, N.; de la Villa, P.; Bernad, A.; Blanco, L.; Suarez, T.; de la Rosa, E.J. Increased neuronal death and disturbed axonal growth in the Polmu-deficient mouse embryonic retina. Sci. Rep. 2016, 6, 25928. [Google Scholar] [CrossRef] [PubMed]
- Lucas, D.; Delgado-Garcia, J.M.; Escudero, B.; Albo, C.; Aza, A.; Acin-Perez, R.; Torres, Y.; Moreno, P.; Enriquez, J.A.; Samper, E.; et al. Increased learning and brain long-term potentiation in aged mice lacking DNA polymerase mu. PLoS ONE 2013, 8, e53243. [Google Scholar] [CrossRef] [PubMed]
- Lucas, D.; Escudero, B.; Ligos, J.M.; Segovia, J.C.; Estrada, J.C.; Terrados, G.; Blanco, L.; Samper, E.; Bernad, A. Altered hematopoiesis in mice lacking DNA polymerase mu is due to inefficient double-strand break repair. PLoS Genet. 2009, 5, e1000389. [Google Scholar] [CrossRef] [PubMed]
- Gatz, S.A.; Ju, L.; Gruber, R.; Hoffmann, E.; Carr, A.M.; Wang, Z.Q.; Liu, C.; Jeggo, P.A. Requirement for DNA ligase IV during embryonic neuronal development. J. Neurosci. 2011, 31, 10088–10100. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.C.; Chang, A.N.; Kao, J.; Du, Z.; Meyers, R.M.; Alt, F.W.; Schwer, B. Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells. Cell 2016, 164, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Rehen, S.K.; McConnell, M.J.; Kaushal, D.; Kingsbury, M.A.; Yang, A.H.; Chun, J. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc. Natl. Acad. Sci. USA 2001, 98, 13361–13366. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Kaushal, D.; Yang, A.H.; Kingsbury, M.A.; Rehen, S.K.; Treuner, K.; Helton, R.; Annas, E.G.; Chun, J.; Barlow, C. Failed clearance of aneuploid embryonic neural progenitor cells leads to excess aneuploidy in the Atm-deficient but not the Trp53-deficient adult cerebral cortex. J. Neurosci. 2004, 24, 8090–8096. [Google Scholar] [CrossRef]
- Rehen, S.K.; Yung, Y.C.; McCreight, M.P.; Kaushal, D.; Yang, A.H.; Almeida, B.S.; Kingsbury, M.A.; Cabral, K.M.; McConnell, M.J.; Anliker, B.; et al. Constitutional aneuploidy in the normal human brain. J. Neurosci. 2005, 25, 2176–2180. [Google Scholar] [CrossRef]
- Strasser, A. The role of BH3-only proteins in the immune system. Nat. Rev. Immunol. 2005, 5, 189–200. [Google Scholar] [CrossRef]
- Yeo, W.; Gautier, J. Early neural cell death: Dying to become neurons. Dev. Biol. 2004, 274, 233–244. [Google Scholar] [CrossRef]
- Blaschke, A.J.; Staley, K.; Chun, J. Widespread programmed cell death in proliferative and postmitotic regions of the fetal cerebral cortex. Development 1996, 122, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- de la Rosa, E.J.; de Pablo, F. Cell death in early neural development: Beyond the neurotrophic theory. Trends Neurosci. 2000, 23, 454–458. [Google Scholar] [CrossRef]
- Chavarria, T.; Baleriola, J.; Mayordomo, R.; de Pablo, F.; de la Rosa, E.J. Early neural cell death is an extensive, dynamic process in the embryonic chick and mouse retina. Sci. World J. 2013, 2013, 627240. [Google Scholar] [CrossRef] [PubMed]
- Buss, R.R.; Sun, W.; Oppenheim, R.W. Adaptive roles of programmed cell death during nervous system development. Annu. Rev. Neurosci. 2006, 29, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Yankner, B.A. Apoptosis in the nervous system. Nature 2000, 407, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.; Hengartner, M.O. Programmed cell death: Alive and well in the new millennium. Trends Cell Biol. 2001, 11, 526–534. [Google Scholar] [CrossRef]
- Mayordomo, R.; Valenciano, A.I.; de la Rosa, E.J.; Hallbook, F. Generation of retinal ganglion cells is modulated by caspase-dependent programmed cell death. Eur. J. Neurosci. 2003, 18, 1744–1750. [Google Scholar] [CrossRef]
- Galli-Resta, L.; Ensini, M. An intrinsic time limit between genesis and death of individual neurons in the developing retinal ganglion cell layer. J. Neurosci. 1996, 16, 2318–2324. [Google Scholar] [CrossRef]
- Southwell, D.G.; Paredes, M.F.; Galvao, R.P.; Jones, D.L.; Froemke, R.C.; Sebe, J.Y.; Alfaro-Cervello, C.; Tang, Y.; Garcia-Verdugo, J.M.; Rubenstein, J.L.; et al. Intrinsically determined cell death of developing cortical interneurons. Nature 2012, 491, 109–113. [Google Scholar] [CrossRef]
- Clarke, P.G.; Posada, A.; Primi, M.P.; Castagne, V. Neuronal death in the central nervous system during development. Biomed. Pharmacother. 1998, 52, 356–362. [Google Scholar] [CrossRef]
- Cellerino, A.; Galli-Resta, L.; Colombaioni, L. The dynamics of neuronal death: A time-lapse study in the retina. J. Neurosci. 2000, 20, RC92. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, R.W.; Flavell, R.A.; Vinsant, S.; Prevette, D.; Kuan, C.Y.; Rakic, P. Programmed cell death of developing mammalian neurons after genetic deletion of caspases. J. Neurosci. 2001, 21, 4752–4760. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yu, Y.; Hamrick, H.E.; Duerksen-Hughes, P.J. ATM, ATR and DNA-PK: Initiators of the cellular genotoxic stress responses. Carcinogenesis 2003, 24, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Lindo, N. Implicación de la Generación y Reparación de Roturas del DNA en el Desarrollo de la Retina de Ratón. Ph.D. Thesis, Universidad Complutense de Madrid, Madrid, Spain, 2015. [Google Scholar]
- Chun, J.J.; Schatz, D.G.; Oettinger, M.A.; Jaenisch, R.; Baltimore, D. The recombination activating gene-1 (RAG-1) transcript is present in the murine central nervous system. Cell 1991, 64, 189–200. [Google Scholar] [CrossRef]
- Gilmore, E.C.; Nowakowski, R.S.; Caviness, V.S., Jr.; Herrup, K. Cell birth, cell death, cell diversity and DNA breaks: How do they all fit together? Trends Neurosci. 2000, 23, 100–105. [Google Scholar] [CrossRef]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Yeo, G.W.; Mu, Y.; Lovci, M.T.; Morell, M.; O’Shea, K.S.; Moran, J.V.; Gage, F.H. L1 retrotransposition in human neural progenitor cells. Nature 2009, 460, 1127–1131. [Google Scholar] [CrossRef]
- Muotri, A.R. L1 Retrotransposition in Neural Progenitor Cells. Methods Mol. Biol. 2016, 1400, 157–163. [Google Scholar]
- Gomez-Herreros, F. DNA Double Strand Breaks and Chromosomal Translocations Induced by DNA Topoisomerase II. Front. Mol. Biosci. 2019, 6, 141. [Google Scholar] [CrossRef]
- Madabhushi, R.; Gao, F.; Pfenning, A.R.; Pan, L.; Yamakawa, S.; Seo, J.; Rueda, R.; Phan, T.X.; Yamakawa, H.; Pao, P.C.; et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell 2015, 161, 1592–1605. [Google Scholar] [CrossRef]
- Dreyer, W.J.; Gray, W.R.; Hood, L. The genetic, molecular and cellular basis of antibody formation: Some facts and a unifying hipothesis. Cold Spring Harb. Symp. Quant. Biol. 1967, 32, 353–367. [Google Scholar] [CrossRef]
- Edelman, G.M.; Gally, J.A. Somatic recombination of duplicated genes: An hypothesis on the origin of antibody diversity. Proc. Natl. Acad. Sci. USA 1967, 57, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Schatz, D.G. Rearranging views on neurogenesis: Neuronal death in the absence of DNA end-joining proteins. Neuron 1999, 22, 7–10. [Google Scholar] [CrossRef][Green Version]
- Kaeser, G.; Chun, J. Brain cell somatic gene recombination and its phylogenetic foundations. J. Biol. Chem. 2020, 295, 12786–12795. [Google Scholar] [CrossRef]
- Helmink, B.A.; Sleckman, B.P. The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol. 2012, 30, 175–202. [Google Scholar] [CrossRef] [PubMed]
- Gigi, V.; Lewis, S.; Shestova, O.; Mijuskovic, M.; Deriano, L.; Meng, W.; Luning Prak, E.T.; Roth, D.B. RAG2 mutants alter DSB repair pathway choice in vivo and illuminate the nature of ’alternative NHEJ’. Nucleic Acids Res. 2014, 42, 6352–6364. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.M.; Agard, E.; Suh, S.; Czyzyk, L. Cryptic signals and the fidelity of V(D)J joining. Mol. Cell Biol. 1997, 17, 3125–3136. [Google Scholar] [CrossRef] [PubMed]
- Teng, G.; Maman, Y.; Resch, W.; Kim, M.; Yamane, A.; Qian, J.; Kieffer-Kwon, K.R.; Mandal, M.; Ji, Y.; Meffre, E.; et al. RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell 2015, 162, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Laszkiewicz, A.; Sniezewski, L.; Kasztura, M.; Bzdzion, L.; Cebrat, M.; Kisielow, P. Bidirectional activity of the NWC promoter is responsible for RAG-2 transcription in non-lymphoid cells. PLoS ONE 2012, 7, e44807. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jessen, J.R.; Jessen, T.N.; Vogel, S.S.; Lin, S. Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons. Genesis 2001, 29, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Zhang, J.; Sun, Y.; Wang, S.; Korteweg, C.; Gao, W.; Gu, J. Expression and distribution of immunoglobulin G and its receptors in an immune privileged site: The eye. Cell Mol. Life Sci. 2011, 68, 2481–2492. [Google Scholar] [CrossRef]
- Aoki, T.; Tashiro, K.; Miyatake, S.; Nakano, T.; Oda, Y.; Kikuchi, H.; Honjo, T. Expression of the RAG-2 gene in murine central nervous system tumor cell lines. Biochem. Biophys. Res. Commun. 1991, 181, 151–158. [Google Scholar] [PubMed]
- Dhingra, N.; Yadav, S.P.; de Villartay, J.P.; Picard, C.; Sabharwal, R.K.; Dinand, V.; Ghuman, S.S.; Sachdeva, A. Severe combined immunodeficiency caused by a new homozygous RAG1 mutation with progressive encephalopathy. Hematol. Oncol. Stem Cell Ther. 2014, 7, 44–49. [Google Scholar] [CrossRef] [PubMed]
- McGowan, P.O.; Hope, T.A.; Meck, W.H.; Kelsoe, G.; Williams, C.L. Impaired social recognition memory in recombination activating gene 1-deficient mice. Brain Res. 2011, 1383, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Rattazzi, L.; Cariboni, A.; Poojara, R.; Shoenfeld, Y.; D’Acquisto, F. Impaired sense of smell and altered olfactory system in RAG-1(--) immunodeficient mice. Front. Neurosci. 2015, 9, 318. [Google Scholar] [CrossRef]
- Hirano, T.; Murata, T.; Hayashi, T. Physiological significance of recombination-activating gene 1 in neuronal death, especially optic neuropathy. FEBS J. 2015, 282, 129–141. [Google Scholar] [CrossRef]
- Karanjawala, Z.E.; Hinton, D.R.; Oh, E.; Hsieh, C.L.; Lieber, M.R. Developmental retinal apoptosis in Ku86-/- mice. DNA Repair 2003, 2, 1429–1434. [Google Scholar] [CrossRef] [PubMed]
- Gozalbo-Lopez, B.; Andrade, P.; Terrados, G.; de Andres, B.; Serrano, N.; Cortegano, I.; Palacios, B.; Bernad, A.; Blanco, L.; Marcos, M.A.; et al. A role for DNA polymerase mu in the emerging DJH rearrangements of the postgastrulation mouse embryo. Mol. Cell Biol. 2009, 29, 1266–1275. [Google Scholar] [CrossRef]
- Merelli, I.; Guffanti, A.; Fabbri, M.; Cocito, A.; Furia, L.; Grazini, U.; Bonnal, R.J.; Milanesi, L.; McBlane, F. RSSsite: A reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucleic Acids Res. 2010, 38, W262–W267. [Google Scholar] [CrossRef]
- Erskine, L.; Herrera, E. The retinal ganglion cell axon’s journey: Insights into molecular mechanisms of axon guidance. Dev. Biol. 2007, 308, 1–14. [Google Scholar] [CrossRef]
| NHEJ MUTATED GENE (and Function) | MURINE NEURAL PHENOTYPE | HUMAN PHENOTYPE Immune System and Genomic Stability | HUMAN PHENOTYPE Nervous System |
|---|---|---|---|
| LIG IV (DSB sealing) | Lethal in E14-E16 (depending on the study). Increased apoptosis in early postmitotic neurons. Acellularity in central and peripheral nervous system [15,37,38]. | Immunodeficiency (residual T and B cells), pancytopenia, lymphomas, leukemia [36]. | (Only hypomorphic mutants described). Microcephaly (non progressive after birth). Delayed development, primordial dwarfism and neurological abnormalities. Dubowitz syndrome, LIG4 syndrome [39]. |
| Nhej-1/XLF/Cernunnos (DSB sealing) | Viable. Frequent spontaneous genomic instability, including translocations [40]. Increased neuronal cell death and neuronal migration defects in brain cortex [15,41]. | Immunodeficiency (residual T and B cells), neutropenia, macrocytic anemia, autoimmunity [42,43]. | In hypomorphic mutants, microcephaly, delayed development, chromosomal translocations. Nijmegen breakage syndrome-like phenotype, polymicrogyria [39]. |
| XRCC4 (DSB sealing) | Lethal in E14,5. Increased apoptosis in early postmitotic neurons, acellularity in central and peripheral nervous system [15]. | Genomic instability, hypersensitivity to radiation and cancer predisposition [44]. | Microcephaly and delayed development. [44] Primordial dwarfism [45]. |
| Pol β (DSB gap filling)) | Neonatal lethality. Increased apoptosis in early postmitotic neurons, apoptosis in central and peripheral nervous system, genomic instability [15]. | Genomic instability [46]. | Reduced activity in patients with Alzheimer disease [47]. |
| DNA-PK (Nuclease. DSB end processing) | Viable. Increased apoptosis in early retina postmitotic neurons [48]. Altered axonal emission [49]. If combined with Pol β deficiency, lethal in E11,5, delayed embryonic development and massive neuronal apoptosis [15,50]. | Severe combined immunodeficiency, total loss of T and B cells [51,52]. | Microcephaly, delayed development, progressive neural degeneration and telomere shrinkage [39,53]. |
| Artemis (Nuclease. DSB end processing) | Viable. Hypersensitivity to radiation and genomic instability, including telomeric fusions [54]. | Progressive immunodeficiency, reaching total T and B cell loss, autoimmunity and Omenn Syndrome. Leukemia and non lymphoid carcinomas [39]. | Not described. |
| MRE11/NBS1-1/RAD50 (Sensor of DNA damage) | Lethal at E6. Elevated genomic instability [55,56]. | Predisposition to lymphomas, breast and ovary cancer [39]. | Nijmegen breakage syndrome (NBS), microcephaly and ataxia [39]. |
| KU 70/80 (Recognition of DNA lesions) | Viable. Increased apoptosis in early postmitotic neurons, especially in the retina [57]. | Suspected to induce embryonic lethality due to telomeric instability [58]. | Melanoma brain metastases with high genomic instability [59]. |
| ATM (Sensor of DNA damage) | Viable. Delayed embryonic development, with neurologic disfunction [15]. Specific loss of a subpopulation of dopaminergic neurons [60]. Hypersensitivity to radiation. [61]. | Reduced or absent levels of IgE, IgA and IgG2, genomic instability, telomere shrinkage and lymphoma predisposition [39]. | Ataxia, progressive neurodegeneration, ocular telangiectasia [39]. |
| Polymerase mu (DSB gap filling) | Viable. Increased apoptosis in early retina postmitotic neurons, ectopic neurons and axonal pathfinding cues, and altered axonal emission [62]. Increased learning and brain long term potentiation in aged mice [63]. | Not described in humans, but altered hematopoiesis has been detected in mice [64]. | Not described. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez-Lindo, N.; Suárez, T.; de la Rosa, E.J. Exploring the Origin and Physiological Significance of DNA Double Strand Breaks in the Developing Neuroretina. Int. J. Mol. Sci. 2022, 23, 6449. https://doi.org/10.3390/ijms23126449
Álvarez-Lindo N, Suárez T, de la Rosa EJ. Exploring the Origin and Physiological Significance of DNA Double Strand Breaks in the Developing Neuroretina. International Journal of Molecular Sciences. 2022; 23(12):6449. https://doi.org/10.3390/ijms23126449
Chicago/Turabian StyleÁlvarez-Lindo, Noemí, Teresa Suárez, and Enrique J. de la Rosa. 2022. "Exploring the Origin and Physiological Significance of DNA Double Strand Breaks in the Developing Neuroretina" International Journal of Molecular Sciences 23, no. 12: 6449. https://doi.org/10.3390/ijms23126449
APA StyleÁlvarez-Lindo, N., Suárez, T., & de la Rosa, E. J. (2022). Exploring the Origin and Physiological Significance of DNA Double Strand Breaks in the Developing Neuroretina. International Journal of Molecular Sciences, 23(12), 6449. https://doi.org/10.3390/ijms23126449





