3.1. Midbrain and Isthmic Gene Patterns in E11.5 Mouse Embryos
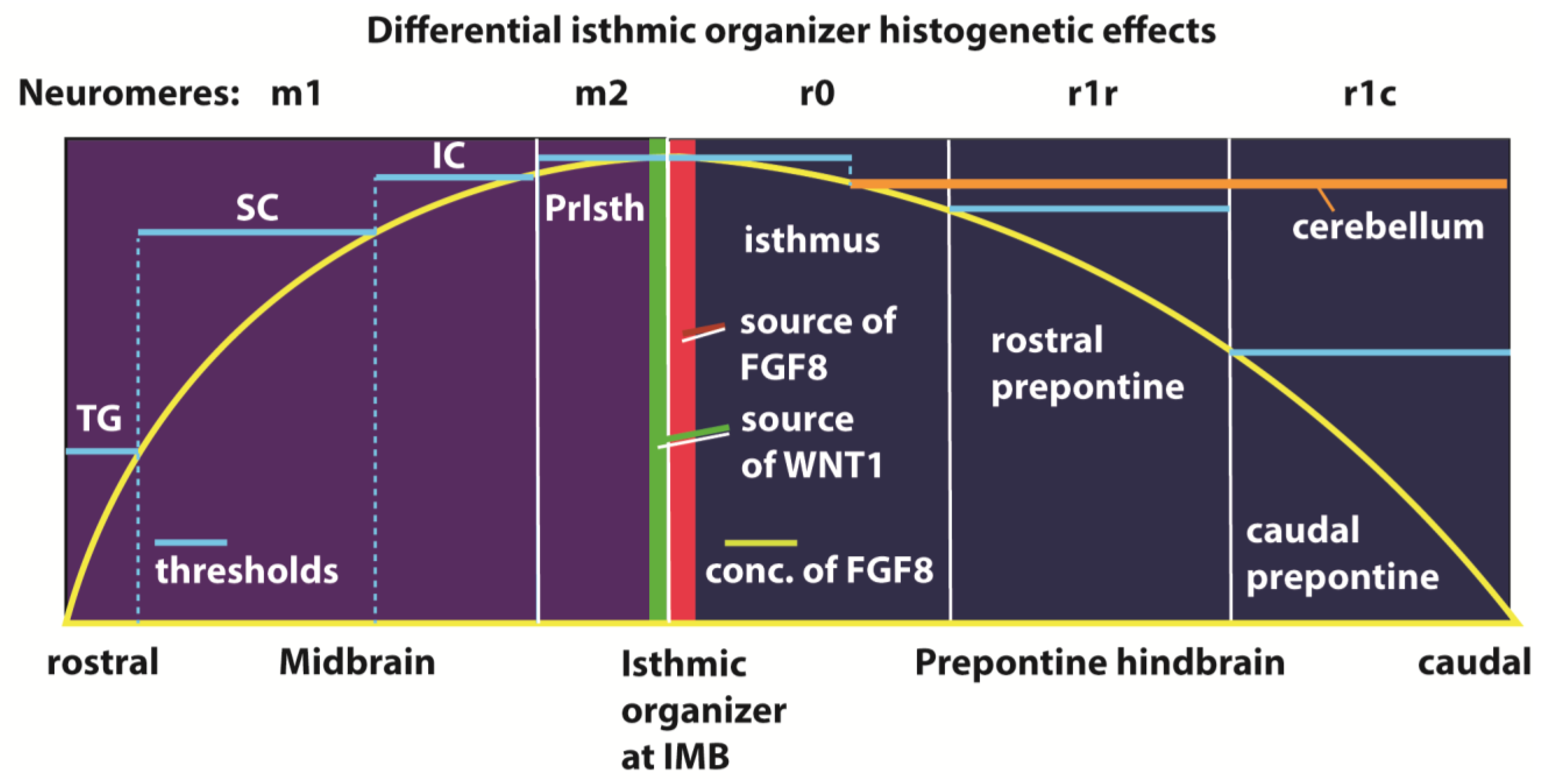
When cut in standard coronal sections, the isthmo-mesencephalic region is cut in fact horizontally; the axial dimension goes from the isthmus (bottom) up into the midbrain; in the sagittal sections shown here the ventral region lies to the left and the dorsal region to the right.
The isthmo-mesencephalic boundary (IMB) appears clearly defined by the caudal limit of midbrain
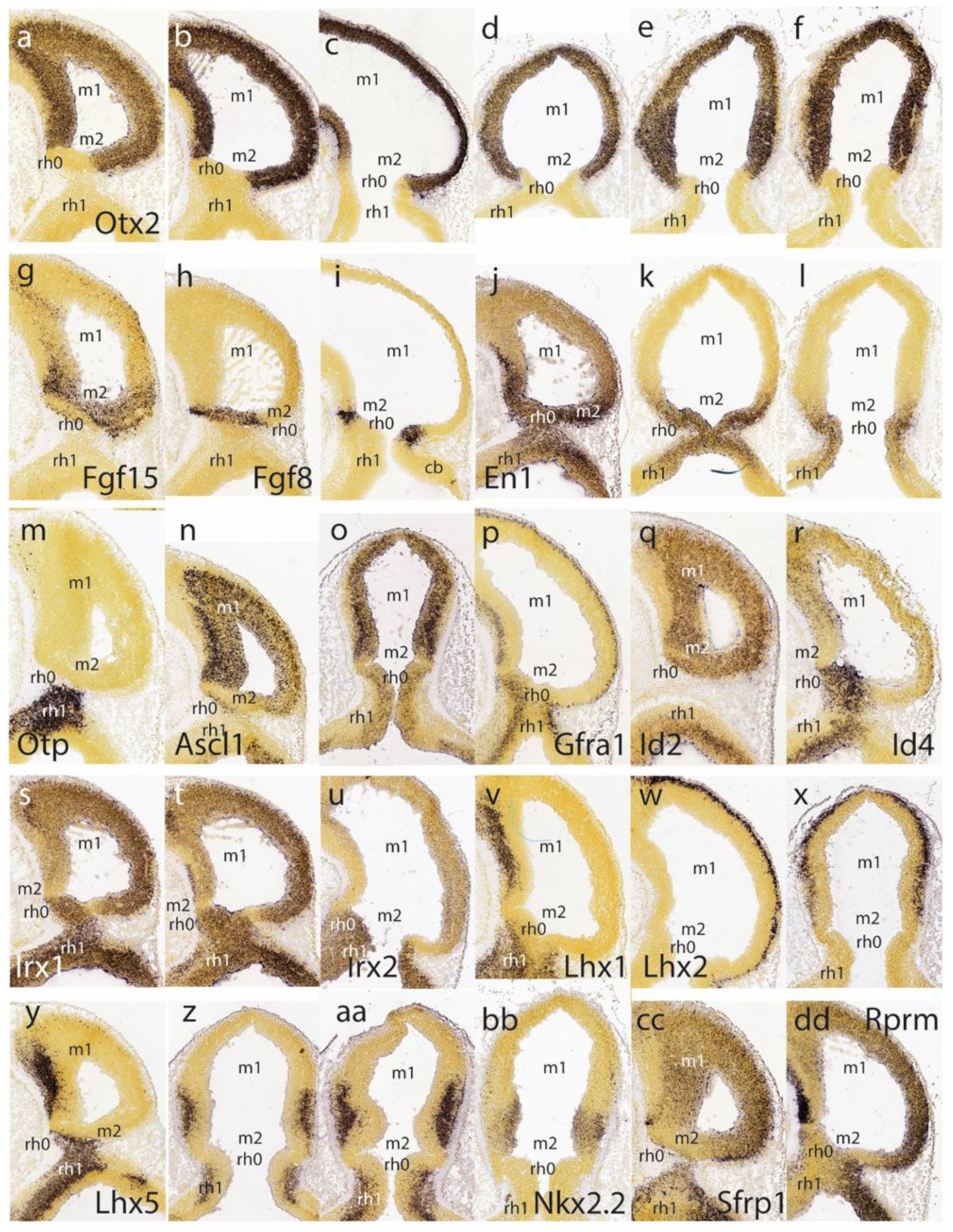
Otx2 expression in the largely undifferentiated neuroepithelium; the incipient mantle layer appears negative (
Figure 2a–f). It is clearly observed that this molecular fate boundary does not coincide with the macroscopic isthmic constriction, so that the rostralmost isthmus (r0) participates in the mesencephalic caudal vesicular evagination (
Figure 2a,b,d–f). Expression of
Fgf15 is weak at the rostral half of the midbrain, but it increases gradientally caudalwards to a maximum just in front of the IMB, at the expected locus of the m2 mesomere (
Figure 2g). In contrast, the
Fgf8 signal is already restricted at this stage to the rostral evaginated part of the isthmus, behind the IMB, at the caudal pole of the midbrain vesicle (
Figure 2h,i). The
engrailed genes are held to be activated downstream of
Fgf8. A double gradiental expression of
En1 overlaps with the isthmic organizer and characterizes the neighboring areas rostral and caudal to the IMB, penetrating maximally about one-third into the caudal basal midbrain (
Figure 2j) and less so dorsally (
Figure 2j–l). There is a specular
En1 gradient extending into the rostral hindbrain, possibly reaching the rh1 rhombomere (
Figure 2j–l). The latter is labeled differentially by the
Otp probe, contrasting with the
Otp-negative isthmus domain (rh0, rh1;
Figure 2m).
Ascl1 shows rather widespread expression in the E11.5 midbrain ventricular zone, with a decreasing dorsocaudal gradient and some positive mantle ventrally (
Figure 2n,o). Remarkably, its signal is sharply absent at a slightly evaginated part of the caudal midbrain neuroepithelium that covers the classic transverse ‘sulcus intraencephalicus posterior’ (
Figure 2n,o). This sulcal area lies just rostral to the
Fgf8-positive isthmic domain (compare
Figure 2h,i), and exhibits a weak
Ascl1 signal in its ventricular zone (
Figure 2o; note this is a topological horizontal section). If we check the
Otx2 signal again in similar horizontal sections (
Figure 2d–f), it seems to occupy integrally the sulcal radial domain next to the IMB, thus making it a candidate for the postulated m2 unit at E11.5 (note Palmgren [
17] used this landmark to identify the atrophic outpouching of m2, all the rest being m1). Nevertheless, we will see below that at later stages
Ascl1 comes to positively label the m2 domain.
A transverse negative gap apparently coinciding with the m2 locus and the associated sulcus at the caudal end of the midbrain vesicle is shown as well by
Gfra1 and
Id2 (
Figure 2p,q). Other expression patterns, such as those of
Id4,
Irx1 and
Irx2, occupy the rostral midbrain and the rostral hindbrain (including the isthmus), but leave an interposed caudal midbrain gap devoid of signal, which may correspond to the prospective m2 (
Figure 2r–u). Several
Lhx genes (e.g.,
Lhx1,
Lhx2,
Lhx5 in
Figure 2v–z,aa) as well as the genes
Nkx2.2,
Sfrp1, and
Rprm (
Figure 2bb,cc,dd) label either large or limited parts of prospective m1 (in the mantle or ventricular zone) but leave out the caudal sulcal midbrain domain (
Lhx1 and
Lhx5 both distinctly label the isthmus;
Figure 2v,y,z,aa). Interestingly,
Lhx5 labels the midbrain mantle at an intermediate rostrocaudal level, leaving negative both the rostral pole and the caudal sulcal area (
Figure 2y,z,aa). It is unclear whether this early labeling is restricted to the basal plate (tegmentum), as is suggested in
Figure 2y. In any case, at later stages this marker also becomes a selective alar m2 marker. The
Nkx2.2 gene, which otherwise is expressed along the whole forebrain (hypothalamus, diencephalon, and midbrain) as a thin longitudinal band approximating the alar-basal boundary [
28], ends its longitudinal route at the caudal midbrain, where the expression domain is deflected dorsalward into the illustrated area (
Figure 2bb) just in front of the caudal sulcal domain. Derived cells will later appear in the inferior colliculus.
The caudal sulcal ventricular zone of the midbrain appears densely labeled by the
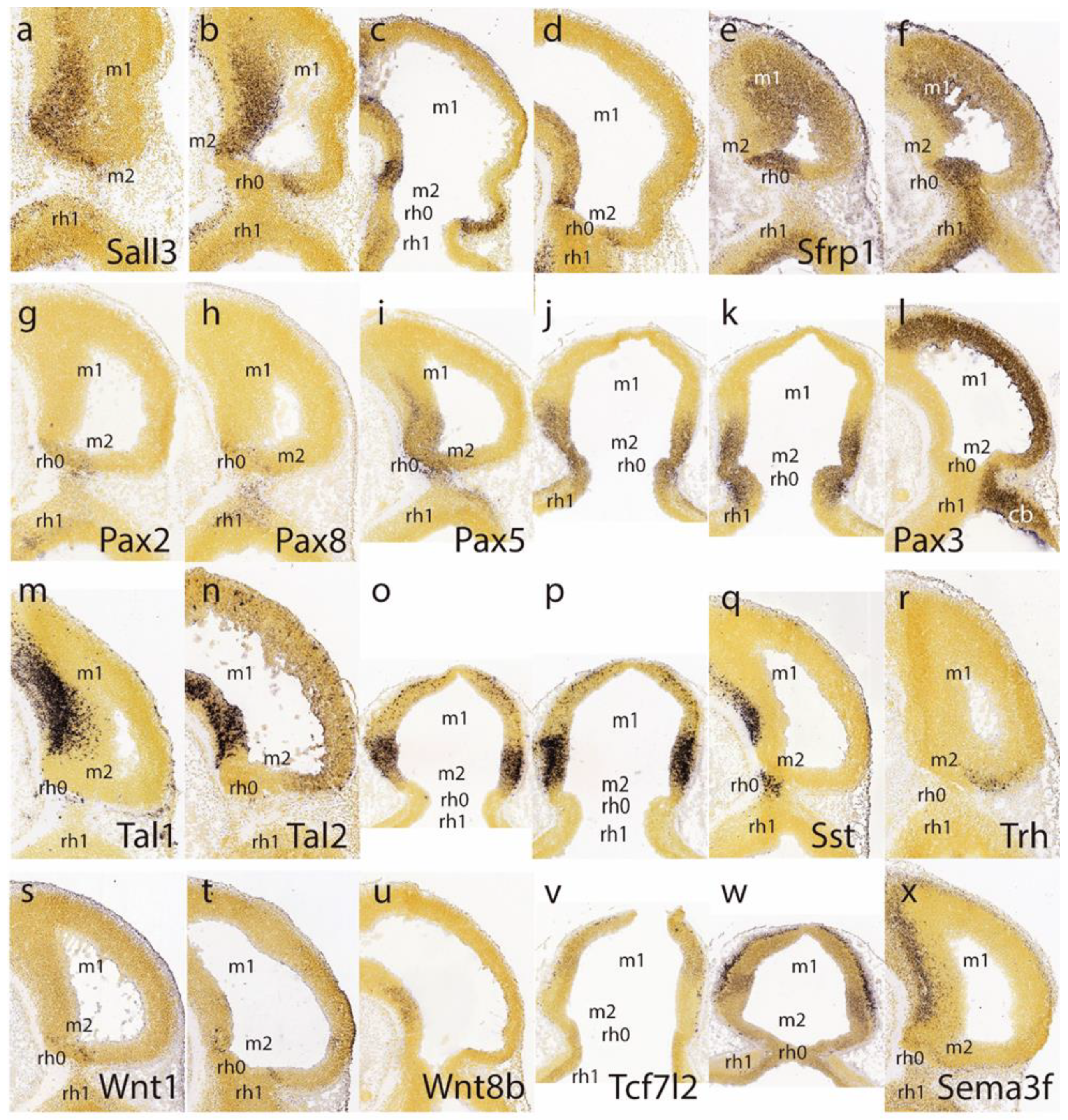
Sall3 signal at several lateromedial parasagittal section levels (
Figure 3a–d); this marker is also expressed in more rostral basal mantle elements in m1, but not in the adjoining isthmus domain. The latter is strongly labeled by
Sfrp1, clearly leaving negative the adjacent caudal sulcal midbrain area (
Figure 3e,f).
Pax2 and
Pax8 both seem weakly expressed in the isthmic domain, stopping rostrally at the IMB, just caudal to the negative sulcal area (
Figure 3g,h). In contrast to findings in the chick [
15], m2 seems devoid of
Pax2 in the mouse, at least at this stage. In contrast, the
Pax5 signal extends rostralward from the positive isthmus and penetrates the caudal part of the midbrain (
Figure 2i–k) analogously, as observed above for En1 (
Figure 2j; note the positive sulcal area).
Pax3 shows alar expression in the midbrain and cerebellar hindbrain but leaves an area devoid of expression coinciding with the caudal sulcal locus (
Figure 3l).
Tal1 and
Tal2 show at E11.5 strong expression in the midbrain basal domain (ventricular zone and mantle;
Tal2 also has rather weak expression at the alar domain), but neither is expressed at the caudal sulcal area (
Figure 3m–p).
Sst-positive postmitotic cells are observed rostrally in the basal midbrain as well as in the basal isthmus, but are lacking at the caudal sulcal area (
Figure 3q). The
Trh signal appears selectively in a semilunar postmitotic mantle area that might correspond to the caudal sulcal area (
Figure 3r).
Ventricular zone expression of
Wnt1 appears at E11.5 along a transverse line aligned precisely with the IMB, that is, in between the evaginated
Fgf8-positive isthmus and the sulcal midbrain area (both being negative to
Wnt1;
Figure 3s,t). The
Wnt1 signal also extends rostralward along the midbrain roof plate, but not along the isthmic one. No relevant
Wnt8 signal was observed at the preisthmic domain (
Figure 3u; there is some basal signal in m1, though). There is little
Tcf7l2 signal at either the isthmus or midbrain at E11.5 (
Figure 3v), but at E12.5 positive mantle cells have started to aggregate subpially, particularly in an intermediate domain of the midbrain lying rostral to the sulcal area (
Figure 3w); this primordium later seems to transform into the inferior colliculus (see below).
Sema3f shows expression concentrated at the midbrain basal plate, but evades the caudal sulcal area (
Figure 3x).
3.2. Midbrain and Isthmic Gene Patterns in E13.5 Mouse Embryos
At E13.5 the major structural primordia of the alar midbrain mantle are clearly distinguishable. Sagittal sections that reacted in situ for
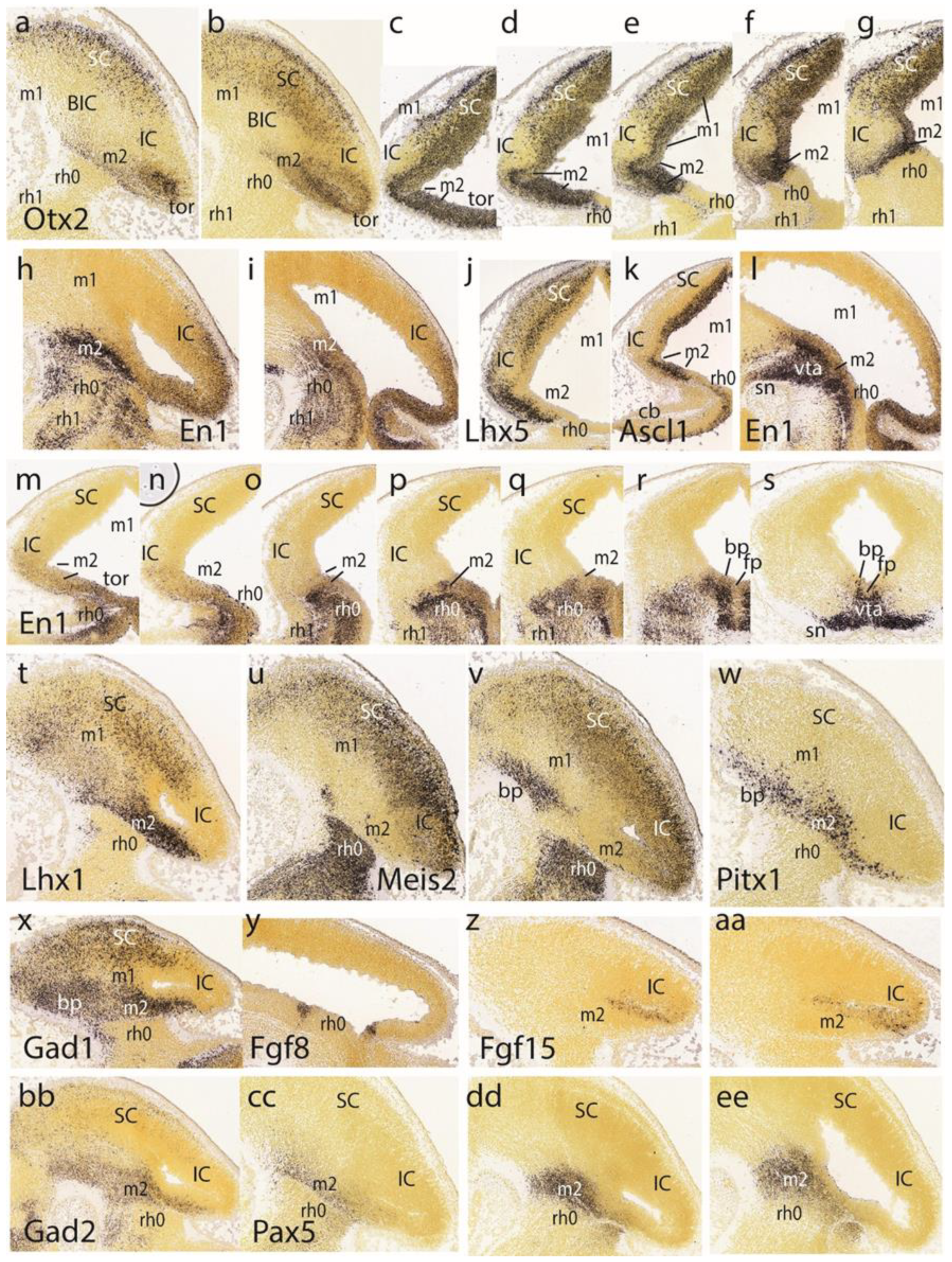
Otx2 show a positive and elongated dorsorostral area that corresponds to the superior colliculus (SC; this apparently includes the still more rostral ‘tectal gray’ formation–TG—which we will point out only occasionally) (
Figure 4a,b). The SC area limits ventrocaudally with a mantle domain devoid of an
Otx2 signal, which represents the inferior colliculus—IC—and its rostral continuation in the interstitial bed nucleus of the brachium of the inferior colliculus—the BIC (
Figure 3a,b). Ventrocaudal to this band there appears again
Otx2 expression in what we interpret as the preisthmus or m2 derivative (
Figure 3a,b). This domain seems wider dorsally (i.e., label ‘tor’, close to the roof plate), where it seems to participate somehow in the toral area of the inferior colliculus (more on this below). As the domain thins out ventralwards (topologically) it displays a sharp boundary with the
Otx2-negative isthmus domain (across the IMB).
Examination of a dorsoventral series of horizontal
Otx2 sections (
Figure 4c–g) identifies again these three alar midbrain territories (positive SC, negative IC, and positive preisthmus) and the molecular limit with the isthmic hindbrain, which partly evaginates into the midbrain vesicle (as seen in
Figure 4d–g). We now notice that the IC is
Otx2-negative only in its mantle zone, whereas its underlying ventricular zone is actually positive, thus establishing the validity of the marker for all the midbrain (the same pattern was discovered in the avian auditory midbrain [
15]). These sections also show the sectioned caudal sulcal area. The posterior intraencephalic sulcus now forms dorsally a deep ventricular diverticulum that is not limited here by the isthmus domain (which lies more ventrally) and lies just
caudal to the IC; it gradually flattens out ventralwards as the IC acquires a larger mass (
Figure 4c–g). The sulcus is separated dorsally (sections in
Figure 4c,d) from the median roof plate by an undifferentiated stretch of positive neuroepithelium that diminishes in surface extent ventralwards, finishing as a narrow radial interstice (the isthmus starts to appear at level of
Figure 4d). This sequence corresponds to the change in dimensions of the alar preisthmus, which we already observed in the sagittal sections. We will see at later stages that the preisthmus seems to contribute to inferior collicular structure via this as yet undifferentiated dorsomedial ‘toral’ domain (the name alludes to its subsequent intraventricular bulge).
At the magnification represented it is hardly possible to distinguish that a stream of
Otx2-positive cells exits the caudalmost midbrain. These migrate tangentially caudalwards along a cell-poor intermediate stratum of the isthmus (tiny dark dots in
Figure 4e,f; see the Allen Developing Mouse Brain Atlas). We believe this represents the caudal migration of cells of the mesencephalic nucleus of the trigeminal nerve (mes5), which targets the isthmus and rh1 alar domains [
20]. Various other studied gene markers label these particular neurons. On the other hand, we cannot identify at this stage any of the diverse alar derivatives of the isthmic region, though their development seems quite advanced.
In the second and third rows of
Figure 4, we show the
En1 pattern at E13.5 (
Figure 4h,i,l–s). We intersected two relatively equivalent horizontal sections labeled for
Lhx5 and
Ascl1 (
Figure 4j,k respectively) because they are directly comparable to the overlying
Otx2 sections in
Figure 4d,e. Sagittal parasagittal sections at lateral, intermediate, and medial section levels minimally illustrate the complex
En1 pattern (
Figure 4h,i,l). The labeled preisthmus shows a strong subpial signal, a moderate to weak signal at intermediate levels, and a strong signal medially, the latter in apparent relation to the generation and migration of dopaminergic neurons of the ventral tegmental area and substantia nigra (compare
Figure 4r,s). The dorsoventral series of horizontal sections shown in
Figure 4m–s illustrate other details. The
En1 signal decreases gradientally across the preisthmus, barely reaching the caudal sulcal area (
Figure 4m,n).
Turning now to
Figure 4j,k, it will be noted that at E13.5
Lhx5 distinguishes (similarly to
Otx2) positive SC and preisthmic mantle domains (including the caudal sulcal area), whereas the IC mantle and ventricular zones are largely negative; the isthmus is also
Lhx5-negative (
Figure 4j). A similar pattern is shown by
Ascl1, whose signal is restricted to the SC and preisthmic ventricular zones, including the sulcal area (
Figure 4k). A much weaker signal slightly labels the IC and isthmic ventricular zone.
The fourth row in
Figure 4 shows similar lateral parasagittal sections illustrating, first, restricted preisthmic expression of
Lhx1 (
Figure 4t). Secondly, two sections showing massive complementary labeling by
Meis2 of the isthmus and neighboring hindbrain domains, as well as weaker labeling of the SC and IC/BIC midbrain domains; a distinct basal patch of mantle cells in m1 and the negative preisthmus stand out (
Figure 4u,v). Finally, we see selective preisthmic labeling (with separate signal in basal m1) with the
Pitx1 probe. Note the constancy and sharpness of the IMG boundary, irrespective of the side labeled.
Gad1 and
Gad2, both markers of GABAergic neurons, are strongly represented at the preisthmic level (not so at this stage at the IC primordium;
Figure 4x,bb). The
Gad1 signal also appears in the SC primordium. Dorsal and ventral intersections of the semicircular arch of
Fgf8 expression along the rostralmost isthmus (=the isthmic organizer) are seen in
Figure 4y. Note the implicit topographic obliquity of the
transverse IMG boundary. The
Fgf15 signal appears as a positive transverse line that in comparison to the
Gad1 pattern seems to separate the negative preisthmic domain from the IC domain (
Figure 4z,aa).
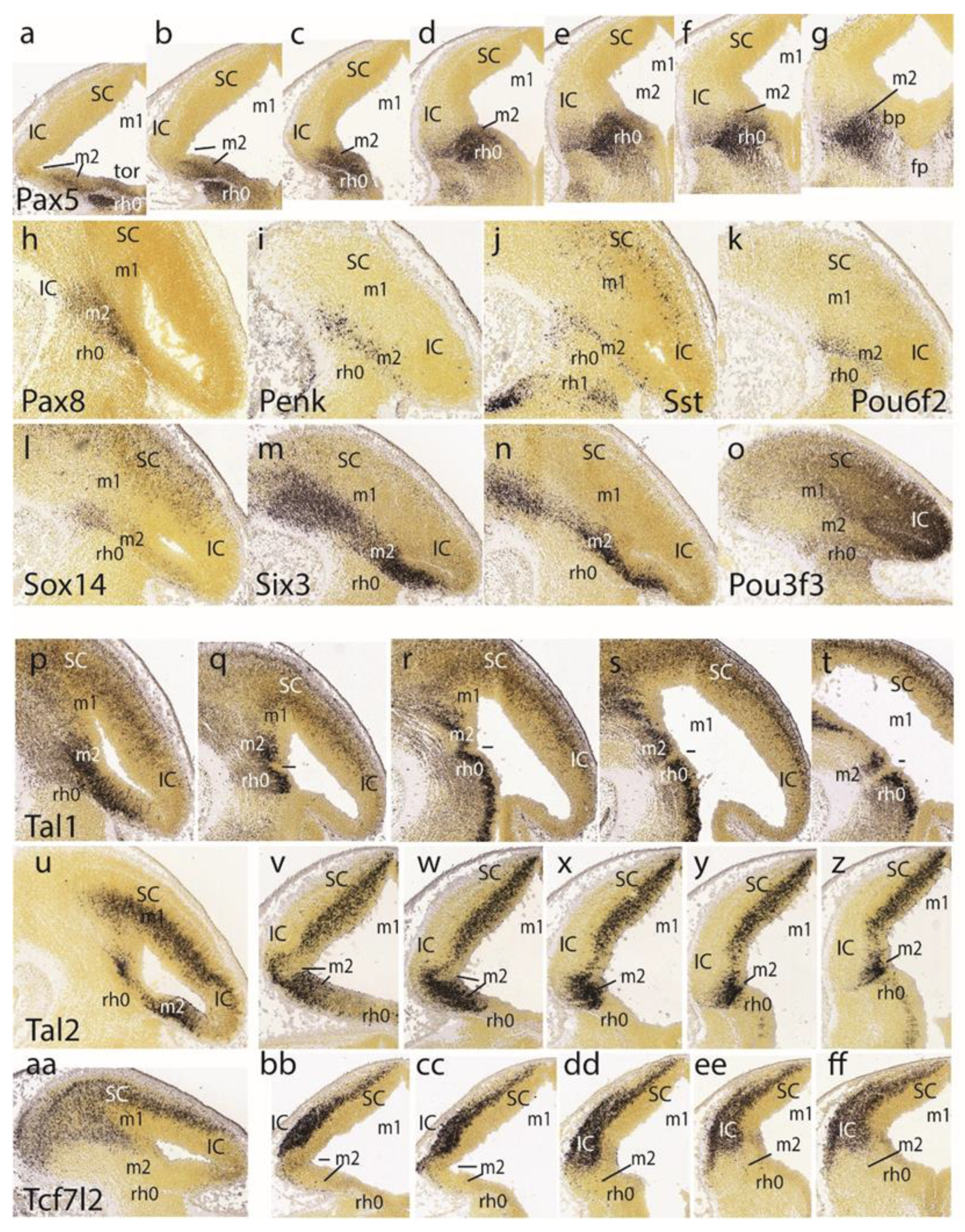
The preisthmic midbrain territory is clearly invaded by the expression of
Pax5 (
Figure 4cc,dd,ee). This pattern is also detailed in a series of horizontal sections (
Figure 4a–g). Note that both the isthmus and preisthmus are variously labeled, but the IC area remains negative. A similar preisthmic
Pax8 labeling was observed (
Figure 5h). Comparable selective positive patterns are likewise provided by
Penk,
Sst,
Pou6f2,
Sox14, and
Six3 (
Figure 5i–n, respectively). Note in contrast the selective preisthmic
negativity demonstrated by
Pou3f3 (
Figure 5o).
A series of parasagittal sections labeled with
Tal1 illustrates its marked expression laterally at the alar preisthmus (
Figure 5p), as well as more ventrally along the transition into the preisthmic basal domain, where it is separated distinctly from an analogous isthmic basal signal by a negative narrow tissue gap associated with the posterior intraencephalic sulcus (
Figure 5q–t). A parasagittal section (
Figure 4u) and a dorsoventral set of five horizontal sections (
Figure 5v–z) labeled with
Tal2 illustrate major labeling at the preisthmus and SC, and a minor periventricular signal at the IC primordium. There is only very weak ventricular expression at the isthmus.
The last row of
Figure 5 shows the
Tcf7l2 expression pattern in a parasagittal section (
Figure 5aa) and five dorsoventral horizontal sections. This marker selectively characterizes the SC and IC mantle primordia, leaving negative the preisthmus and isthmus. Note the lack of signal at the caudal sulcal area.
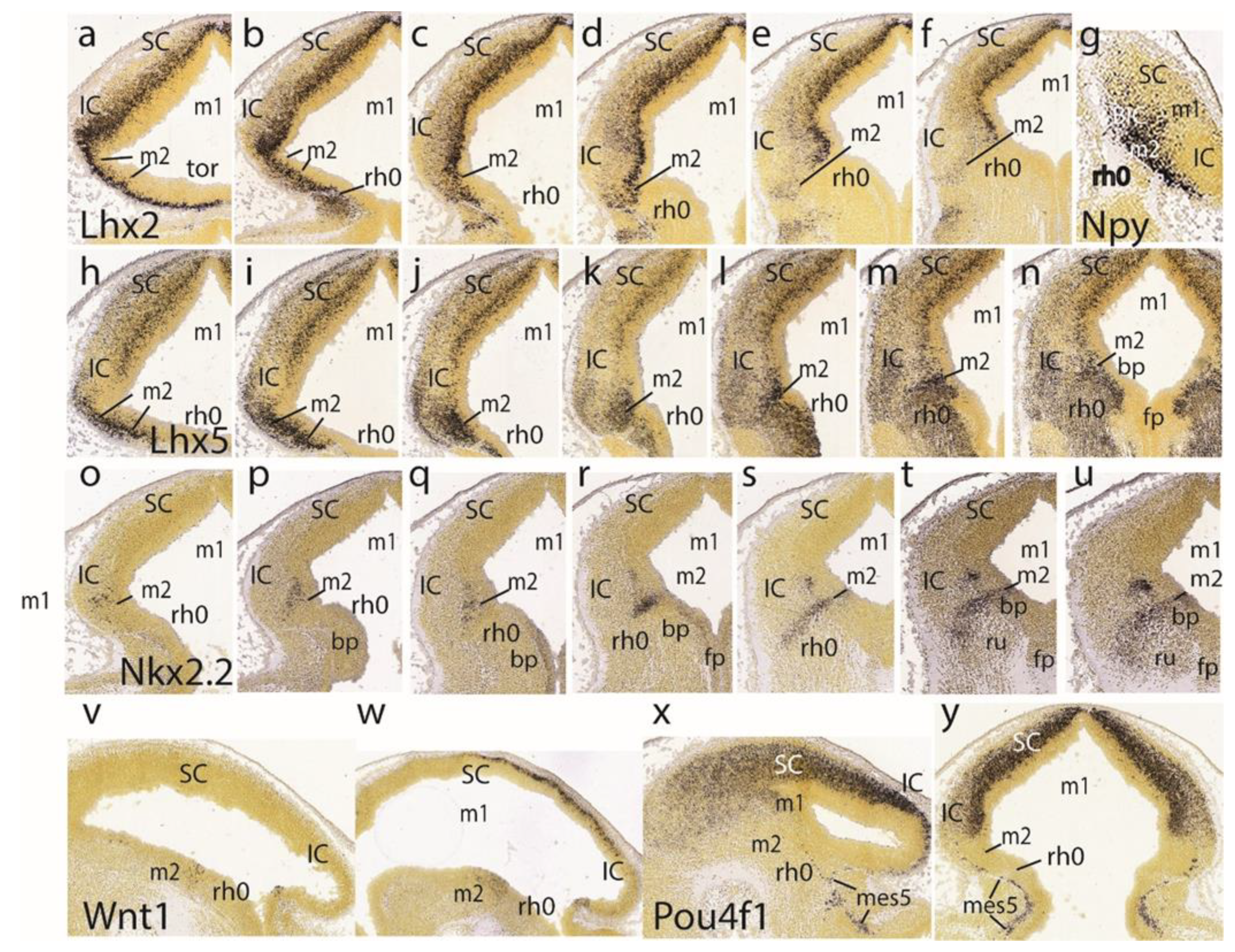
The
Lhx2 pattern is shown in a set of six dorsoventral horizontal sections. It consists of labeled deep mantle cells at the SC, IC, and preisthmus (not so with the isthmus), with a substantial superficial mass of IC that remains devoid of expression, particularly in the more ventral sections (
Figure 6a–f).
Figure 5g illustrates a very lateral parasagittal section reacted for
Npy. The signal is largely restricted to the preisthmus, though a number of positive cells seem to disperse tangentially across the BIC domain into the SC. The second row in
Figure 6 displays a series of dorsoventral horizontal sections illustrating
Lhx5 expression (
Figure 6h–n). We distinguish the positive SC and preisthmic domains (including the caudal sulcal area) and the intercalated negative IC domain. Note a subpial positive cell stream seems to invade the superficial IC mantle out of the preisthmus (
Figure 6i–k). There is also expression of this marker at the isthmus. The boundary across the IMB with the preisthmus is visible at the levels of
Figure 6h–l, partly thanks to the flattened posterior intraencephalic sulcus.
Nkx2.2-positive cell patches characterize the dorsal IC mass (
Figure 6o–q). More ventrally there is more compact signal associated with the caudal sulcal area along the dorsoventral dimension of the preisthmus (
Figure 6r,s). Finally, the last sections illustrate the confluence of the preisthmic transverse pattern with the longitudinal band coursing along the alar-basal boundary (
Figure 6t,u). These last sections are cut essentially transversal to the midbrain and typically show radial ventral distribution of
Nkx2.2-positive cells in the laterorubral tegmental area and dorsalward migration of a clump of periaqueductal positive neurons.
The
Wnt1 expression remains stable along the transverse IMB arch (two intersections are seen) and the midbrain roof plate (
Figure 6v,w).
Pou4f1, another marker of the migrated mes5 neurons, as well as a selective marker of the SC and IC mantle in m1 (not the preisthmus in m2), is shown in the parasagittal and horizontal sections (
Figure 6x,y). Some positive mes5 neurons remain fixed along the migration pathway targeting rh1 (the rh1/rh2 boundary is not violated).
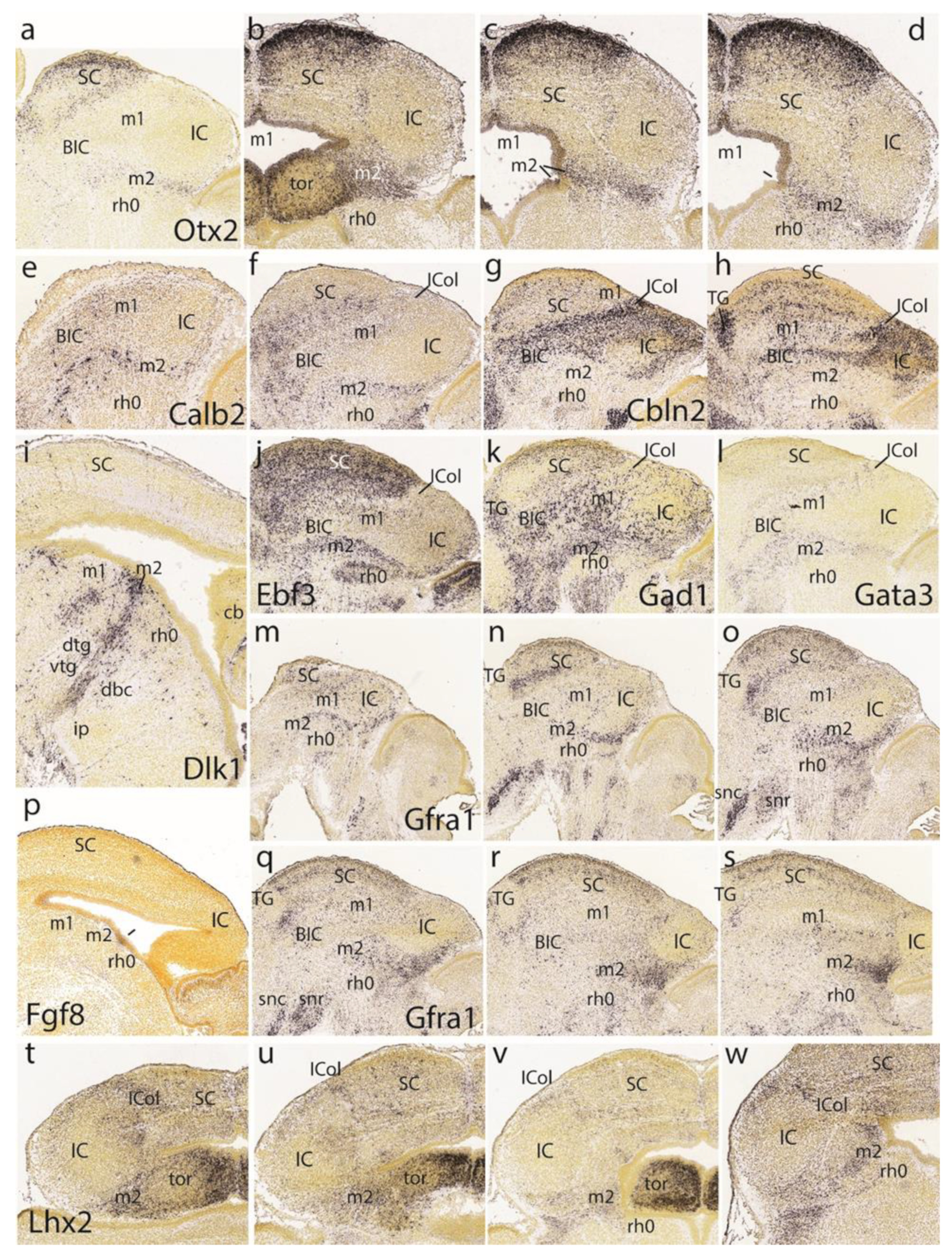
3.3. Midbrain and Isthmic Gene Patterns in E15.5 Mouse Embryos
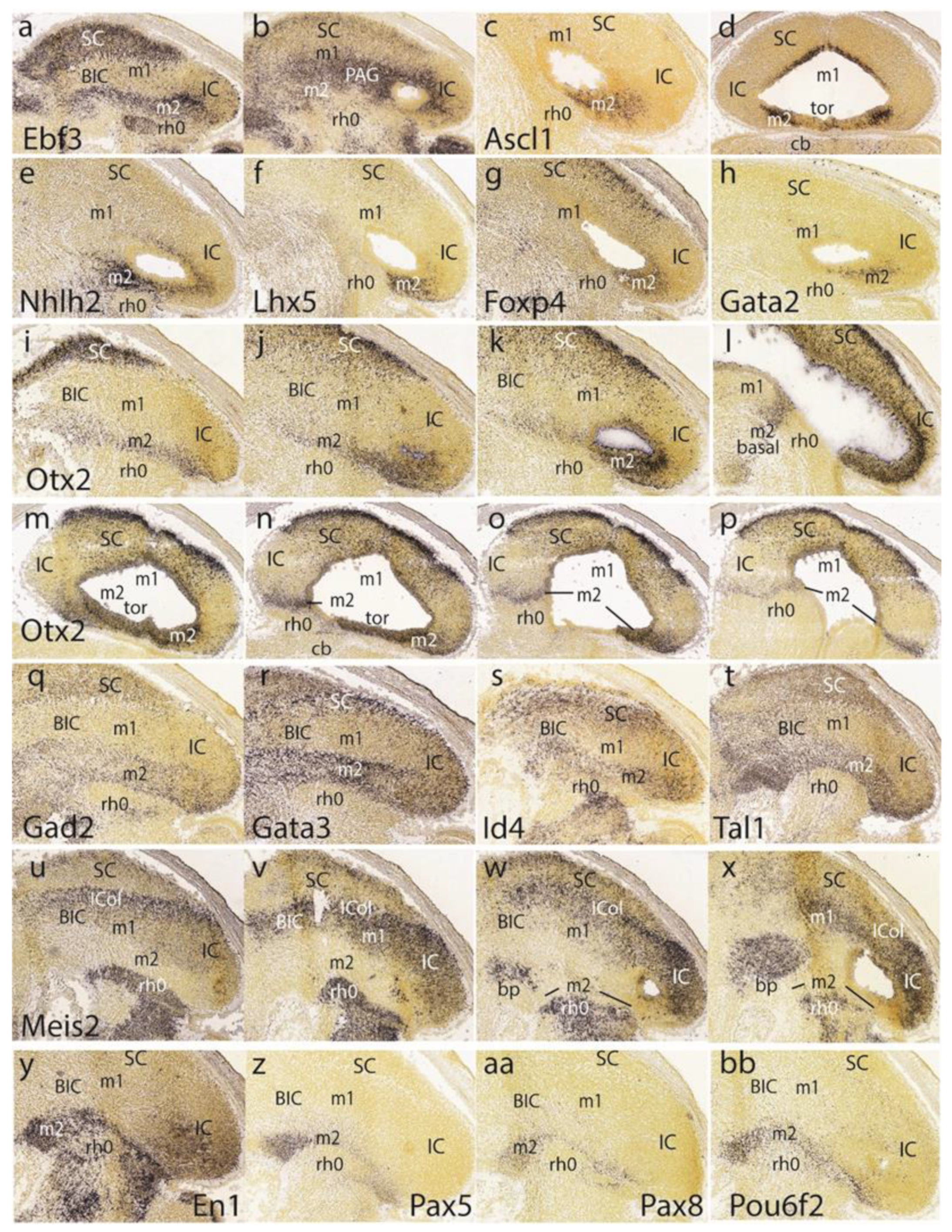
At E15.5,
Ebf3 is a marker of the preisthmus and SC/TG, leaving the BIC/IC domain essentially negative except for a few dispersed cells. There is also a negative boundary with the isthmus where a patch of signal appears in the mantle (
Figure 7a). In more medial parasagittal sections, extensive labeling of the midbrain periaqueductal gray formation is observed (
Figure 7b). The
Ascl1 signal appears only at the ventricular zone of the preisthmus and SC, leaving the IC and isthmus domains negative (
Figure 7c,d).
The preisthmus (with or without the SC) appears selectively labeled by
Nhlh2,
Lhx5,
Foxp4, and
Gata2 (
Figure 7e–h).
Gata2 shows a linear pattern of expression at the boundary of the preisthmus with the IC that recalls the pattern of
Fgf15 at E13.5. These sites can be easily ascribed to the preisthmus (and SC) by comparison with
Otx2 expression in three lateromedial parasagittal sections (
Figure 7i–k). We confirmed that the IC keeps
Otx2 expression in its ventricular zone (
Figure 7k,l). A paramedian parasagittal section also illustrates the
Otx2 signal in the basal preisthmus, representing a quite narrow transverse and radial interstice placed in a retrorubral position (
Figure 7l). Four dorsoventral horizontal sections (note there is left–right obliquity of these sections; both sides are of interest) labeled likewise for
Otx2 illustrate the wide dorsal extent of the positive alar preisthmic domain (next to the negative IC) at the level where it makes contact with the local roof plate (
Figure 7m). More ventrally, the alar preisthmus gets reduced to a narrower
Otx2-positive band that underlines the caudal aspect of the IC and limits caudally with the negative isthmus domain (
Figure 7n–p). It is appreciated in these images (particularly on the left side) that at E15.5 the isthmus has penetrated significantly topographically into the midbrain vesicle (intussusception by morphogenetic deformation of the IMB). The four underlying patterns of the markers
Gad2, Gata3, Id4, and
Tal1, identify specifically the preisthmus and show a curved boundary with the isthmus, with development of a marked convexity towards the midbrain (
Figure 7q–t).
A set of four parasagittal sections next illustrate the
Meis2 pattern, which is negative in the preisthmus and most of the SC but positive in the BIC/IC complex (
Figure 7u–x). Note also that the dorsal rim of the latter domain shows a stronger signal than its ventral rim, as is observed particularly in
Figure 7w. This corresponds to the incipient differentiation along this more strongly labeled locus of the separate
intercollicular domain (ICol), intercalated between the SC and IC. There is also a separate patch of
Meis2 expression in the basal m1 area. The isthmus and other hindbrain prepontine areas are also strongly positive.
The superficial preisthmic mantle stratum begins to differentiate into what we call the ‘subbrachial nucleus’ (SubB; [
21]). This primordium adopts a characteristic curved shape above the isthmic superficial elements (due to the curving of the IMB). It shows a
ventral head-like thickening (oriented topographically rostral) and a longer
dorsal tail portion (oriented topographically caudal). We show this characteristic image displayed by four different gene markers (
En1,
Pax5,
Pax8,
Pou6f2) in
Figure 7y,z,aa,bb.
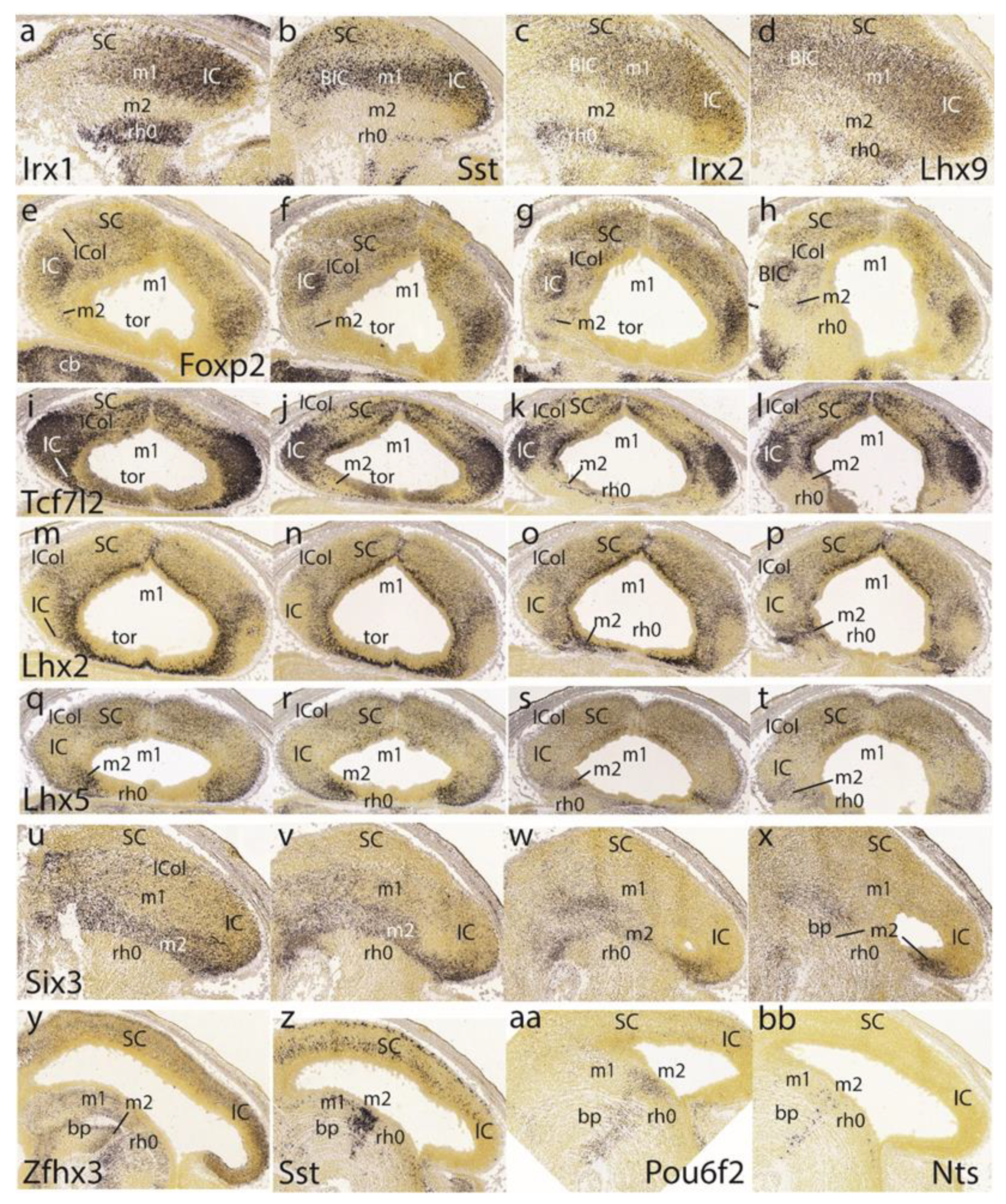
The preisthmus is delineated perfectly in negative by the markers
Irx1,
Sst,
Irx2, and
Lhx9. All of them label distinctly parts of the isthmic mantle at rh0 (well delimited from rh1), as well as the BIC/IC complex (
Figure 8a–d). In horizontal sections,
Foxp2 shows deep to superficial gradiental labeling of the IC primordium plus a weaker SC signal. There is also some separate weak signal at the intermediate stratum of the preisthmic mantle, caudal to the IC (
Figure 8e–h). The positive patch at the left bottom corner of
Figure 8h is an unidentified superficial isthmic mass. Note that at E15.5, the midbrain caudal sulcal area is hard to identify due to its progressive flattening.
Another good marker for the IC is
Tcf7l2 (
Figure 8i–l). The collicular labeling occupies the whole mantle, while more rostrally, possibly at levels through the BIC nucleus, this mass appears separated by a negative periventricular stratum from the corresponding positive ventricular zone. The SC shows a wholly different pattern and the preisthmus is essentially negative except for a few periventricular cells. Similar sections labeled for
Lhx2 illustrate in contrast a negative IC core nucleus with an underlying positive periventricular stratum, which is continuous with the preisthmic mantle (
Figure 8m–p; here the preisthmus reaches the roof plate in
Figure 8m,n) as well as with the SC counterpart. The superficial mantle intercalated between the negative IC and SC domains shows an increased signal of
Lhx2 (ICol;
Figure 8m–p); this transitional area corresponds to the incipient intercollicular area introduced above with
Meis2. The IC domain appears fully and selectively negative with
Lhx5, whereas the SC is weakly labeled in a diffuse pattern (
Figure 8q–t). In contrast, the preisthmus selectively shows its characteristic mantle signal (the isthmus being wholly negative). The preisthmic domain dwindles in extent ventralwards.
The gene
Six3 shows positive cells distributed through lateral parts of the BIC/IC complex, but is otherwise more strongly represented along the preisthmus and is absent from the isthmus and SC regions (
Figure 8u–x).
The last row in
Figure 8 is dedicated to examples of markers selectively labeling the basal or tegmental preisthmus, namely
Zfhx3,
Sst,
Pou6f2, and
Nts (
Figure 8y,z,aa,bb). The differential distributions of the labeled elements suggest that various neuronal types are more or less mixed within the narrow preisthmic tegmentum. We do not show the local dopaminergic or serotonergic neurons reported by our group [
21,
27].
3.4. Midbrain and Isthmic Gene Patterns in E18.5 Mouse Embryos
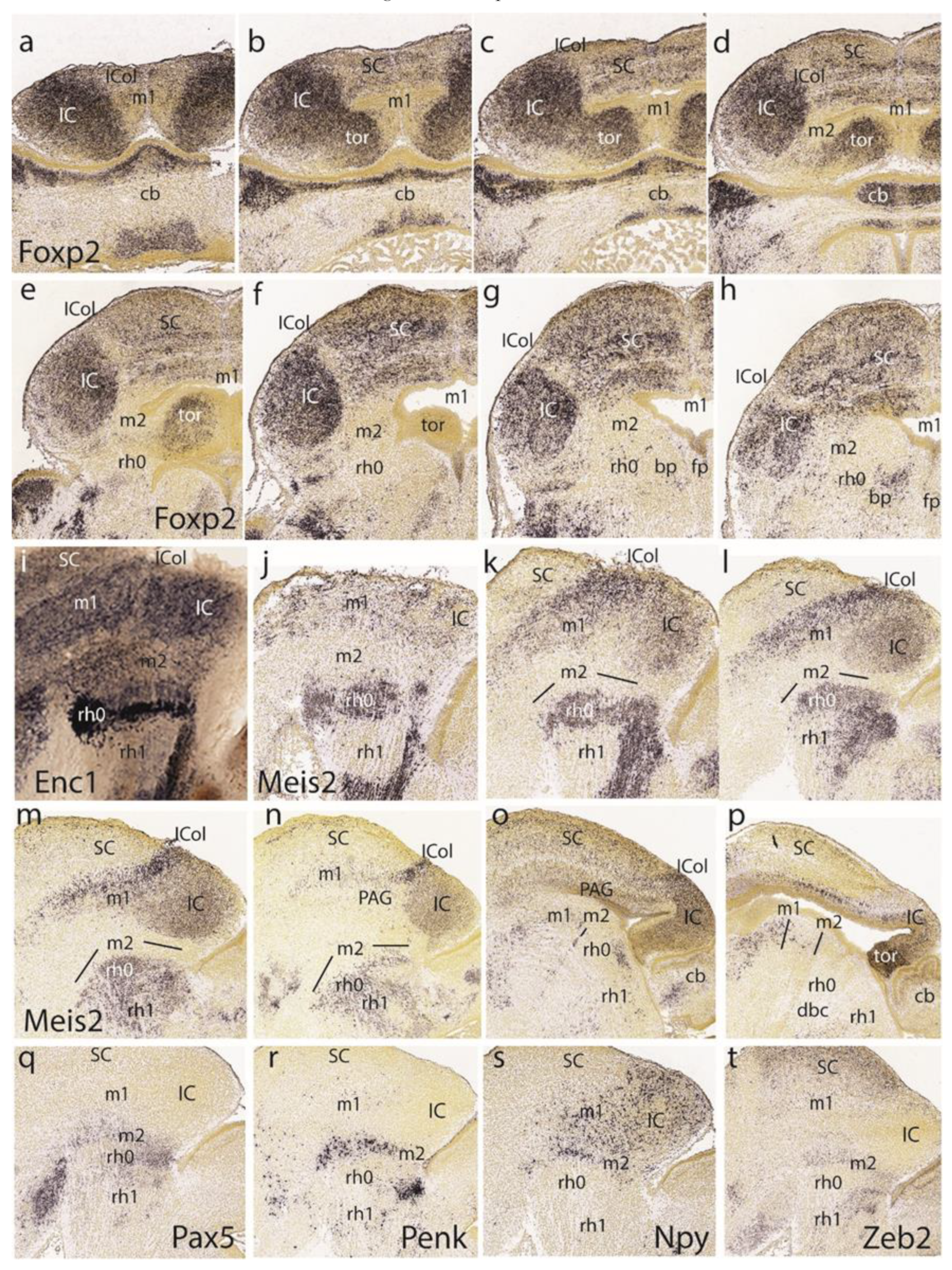
The first row in
Figure 9 presents the E18.5
Otx2 pattern. As before, the SC and preisthmus domains stand out, while the largely negative BIC/IC complex only appears labeled at its ventricular zone (
Figure 9a–d).
The lateral parasagittal section in
Figure 9a illustrates the characteristic headed worm shape of the subbrachial nucleus (the superficial stratum of the preisthmus), as described at E15.5. The three horizontal sections in
Figure 9b–d show a sequence through the positive preisthmus, passing from its enlarged dorsal part to its much narrower ventral portion. Note the posterior intraencephalic sulcus is recognizable at levels of
Figure 9c,d and only part of its ependymal ventricular zone expresses the
Otx2 marker. This indicates that the bottom of the sulcus may represent the IMB, so that its rostral lip belongs to m2, while the caudal lip is isthmic (representing the organizer). A number of positive preisthmic cells seem to violate this boundary and disperse in the neighboring isthmic mantle (
Figure 9c,d). The IC core is negative and appears surrounded deeply and superficially by a dispersed shell of positive cells. The negative deep stratum lying under the deep IC shell corresponds to the local periaqueductal gray (PAG). Labeling in the SC is restricted to intermediate and superficial cell layers, plus the ventricular zone. Its own part of the PAG is also negative.
Interestingly, the dorsalmost
Otx2 section shown (
Figure 9b) illustrates dorsomedial continuity of the positive preisthmic mantle with symmetric bulges at the dorsocaudal midbrain midline. We interpret these bulges as derived from the dorsomedial toral undifferentiated domain distinguished at the same position at the preceding stages. These bulges emerge between E15.5 and E18.5, indicating late growth. Their significance will be considered in the Discussion. We will explore their genoarchitectonic profile and histogenetic evolution here and in the next section.
The second row in
Figure 9 shows the
Calb2 (calretinin) and
Cbln2 patterns in parasagittal sections (
Figure 9e–h, respectively).
Calb2 appears selectively in the subbrachial preisthmic nucleus (a subpial formation), showing its typical head-tail configuration (
Figure 9e). A deeper section interests the intermediate preisthmic stratum, forming the incipient cuneiform nucleus (
Figure 9f). The isthmus and BIC/IC domains are negative. In contrast,
Cbln2 leaves unlabeled the preisthmus and selectively marked the BIC/IC/ICol complex. Note the ICol area appears more intensely labeled than the underlying BIC/IC. The SC appears labeled in several layers and can be perfectly distinguished in
Figure 9h from the massively labeled rostral tectal gray (TG) formation (another midbrain retinorecipient locus; see [
26]).
The basal or tegmental preisthmus appears distinctly labeled with
Dlk1 (
Figure 9i). The underlying
Figure 9p shows a remnant of
Fgf8 activity at a ventricular locus supposed to be just caudally adjacent to the ventricular projection of the basal preisthmus in
Figure 9i.
We examine again the
Ebf3 pattern at E18.5 (centered on the preisthmus and the SC, plus isolated isthmic cell masses;
Figure 9j), as well as the
Gad1 pattern (
Figure 9k), which illustrates the presence of GABAergic neurons in the preisthmus BIC nucleus and IC shell, and some layers of the SC. The isthmus proper remains unlabeled.
Gata 3 (
Figure 9l) weakly labels the preisthmus and faintly labels the ICol area.
The pattern of
Gfra1 is shown in six lateromedial parasagittal section levels (
Figure 9m–o,q–s). The major labeled domain is the preisthmus, always found under the negative BIC/IC complex. The three more lateral sections interest the subbrachial and cuneiform nuclei, whereas the medial sections illustrate the preisthmic PAG area and end at the dorsomedial toral region. The superficial and intermediate strata of the SC are positive, and it can be observed that the rostral tectal gray visual formation is wholly negative with
Gfra1 (TG;
Figure 9m–o,q–s).
The last row in
Figure 9 displays four dorsoventral horizontal sections labeled with
Lhx2 (
Figure 9t–w). These sections illustrate the dorsomedial continuity of the positive preisthmic mantle with the symmetric toral bulges that have emerged out of the paramedian roof area, just in front of the isthmus.
Lhx2 distinctly labels the ventricular zone and part of the mantle stratum of the toral bulges. Their protrusion into the midbrain ventricle causes the tip of the toral bulges to appear tangentially sectioned in
Figure 9v. The last section in
Figure 8w shows a standard view of a more ventral part of the IC and the caudally adjacent labeled preisthmic territory.
The described toral phenomenon was further studied by examining the
Foxp2 marker, which does not label the preisthmus (and the ICol area) and strongly labels the IC core and the BIC nucleus, apart from multiple layers in the SC. A series of eight dorsoventral horizontal sections (
Figure 10a–h) show that the IC core extends in a hook pattern, passing above the preisthmus and into the dorsomedial toral area next to the roof plate. Moreover,
Foxp2 also labels the mantle of the toral bulges, in continuity with the IC core (
Figure 10a–d). There is, though, a sand-clock-like shape with an interposed, less-labeled constriction corresponding to the IC-preisthmic boundary (
Figure 10b–d). The toral mantle disappears progressively into
Figure 10e,f, where mainly the ventricular lining is cut tangentially. The last section shows the positive BIC nucleus, which continues rostralward toward the IC core, limited dorsally by the negative ICol area. The last three sections show some positive preisthmic patches caudal to the IC occupying a superficial stratum (subbrachial nucleus), as well as the caudally adjoining positive parabigeminal isthmic nucleus (SubB; PBG;
Figure 10f–h).
Indeed, the isthmic subpial PGB nucleus can be now recognized as a group of superficial patches. These patches are labeled distinctly with
Enc1(
Figure 9i) or with
Meis2 (
Figure 9j), apart from other, deeper neurons (
Figure 10k–n). This pattern clearly distinguishes the isthmic mantle from both the preisthmus and rh1 (
Figure 10i–l). Note the better developed, higher
Meis2 signal of the ICol contrasting with the BIC/IC complex (
Figure 10k–o). The toral bulge seen in
Figure 10p also strongly expresses
Meis2.
In the last row of
Figure 10, we show four similarly lateral parasagittal sections illustrating the superficial preisthmic signals of
Pax5,
Penk,
Npy, and
Zeb2 (
Figure 10q–t). The
Npy signal obviously is also present in the IC, though this may represent migrated cells.
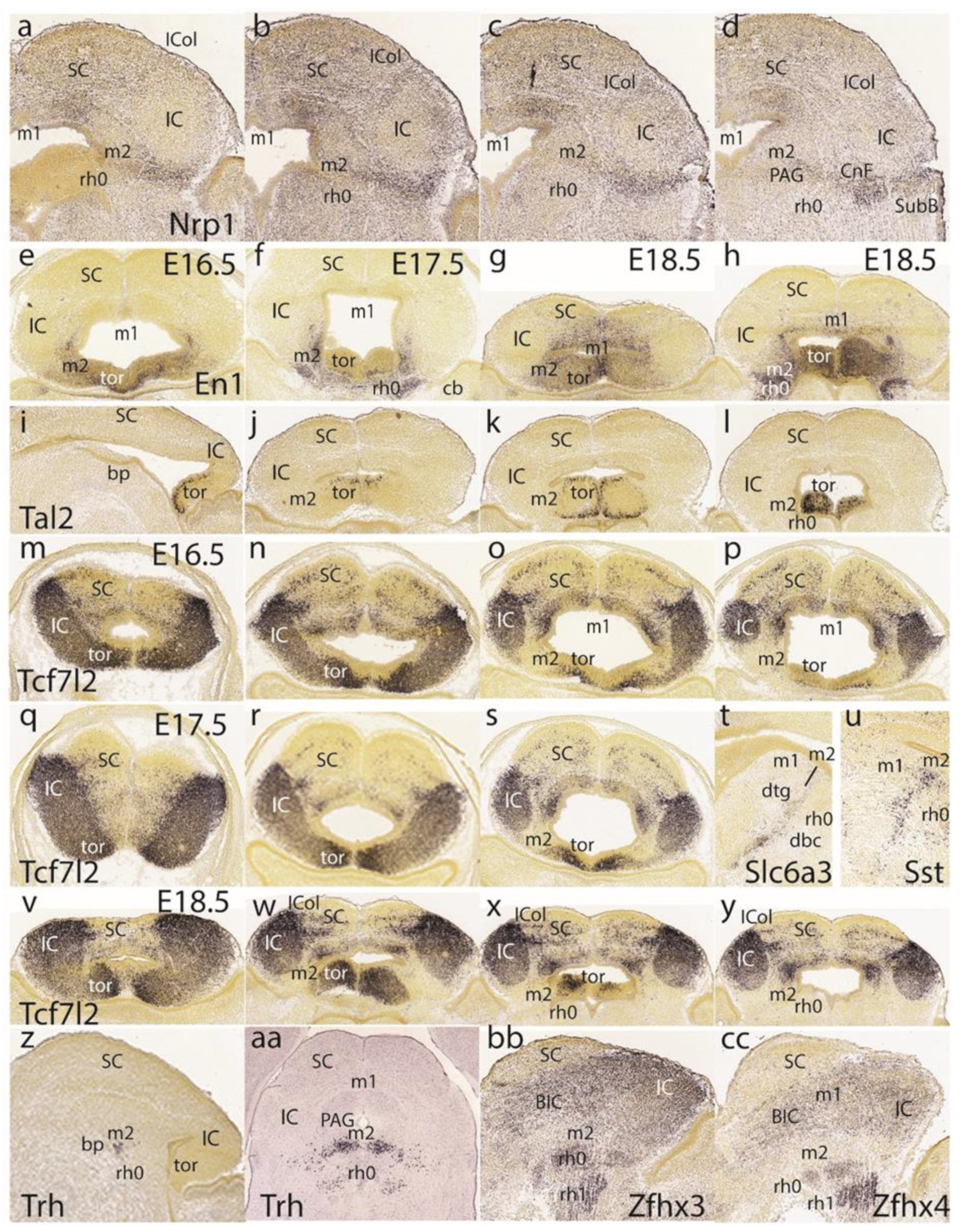
Dorsoventral sections displaying
Nrp1 expression reveal labeling of intermediate and superficial strata of the preisthmus, with less signal at the corresponding PAG portion (
Figure 11a–d). On the other hand, the PAG under the SC seems specifically labeled, similarly to the IC shell.
We next present images of the
En1 signal to clarify sequential changes at the dorsomedial toral area. At E16.5 (
Figure 11e), the toral area is positive jointly with the preisthmus (not so the IC), but remains undifferentiated, hardly starting to bulge into the ventricular space. At 17.5 (
Figure 11f), the toral bulges are more evident and show the
En1 signal at their mantle layer. At E18.5 (
Figure 11g,h), the dorsomedial toral bulges are fully formed, protruding strongly in the ventricle and maintaining
En1 expression. Further evidence on these formations was found in the
Tal2 expression pattern at E18.5, illustrated in one sagittal section (
Figure 11i) and three horizontal sections (
Figure 11j–l). This marker mainly labels the ventricular zone of the toral bulges, being absent from both the IC and the interposed preisthmic territory.
We also examined the
Tcf7l2 signal at E16.5 (
Figure 11m–p), E17.5 (
Figure 11q–s), and E18.5 (
Figure 11v–y). At E16.5 and at section levels above the preisthmus, this marker is already continuously expressed from the IC core into the immature dorsomedial toral area, leaving the roof plate negative (
Figure 11m,n). Once the series intersects the preisthmus more ventrally, this appears negative, though a remnant of the positive toral area is still present (
Figure 11o,p). A similar pattern was observed at E17.5 (
Figure 11q–s) as well as at E18.5 (
Figure 11v–y). This pattern suggests that the toral area is a dorsocaudal prolongation of the IC domain, which ends near the dorsal part of the preisthmus. However, the selective labeling of the toral area by markers such as
En1 and
Tal2 suggest that there are aspects of its molecular profile comparable to the preisthmus that are absent at the IC, irrespective of shared
Tcf7l2 expression.
The free space at the end of row five in
Figure 11 was used to insert two views of selective labeling at the basal tegmental preisthmus with
Slc6a3 and
Sst (
Figure 11t,u). A singular basal preisthmic dot-like area of expression seen periventricularly in sagittal sections labeled with
Trh led to an examination of the coronal sections and the finding that there is a
Trh-positive distinct cell group in the ventral PAG associated with the preisthmus, out of which less numerous similar cells spread out laterally and ventrally (
Figure 11z,aa). Finally, both
Zfhx3 and
Zfhx4 were found to selectively label isthmic superficial masses and other rh1 cell masses, as well as the BIC/IC complex, leaving the preisthmus domain devoid of significant expression (similar to
Enc1 and
Meis2 described above).
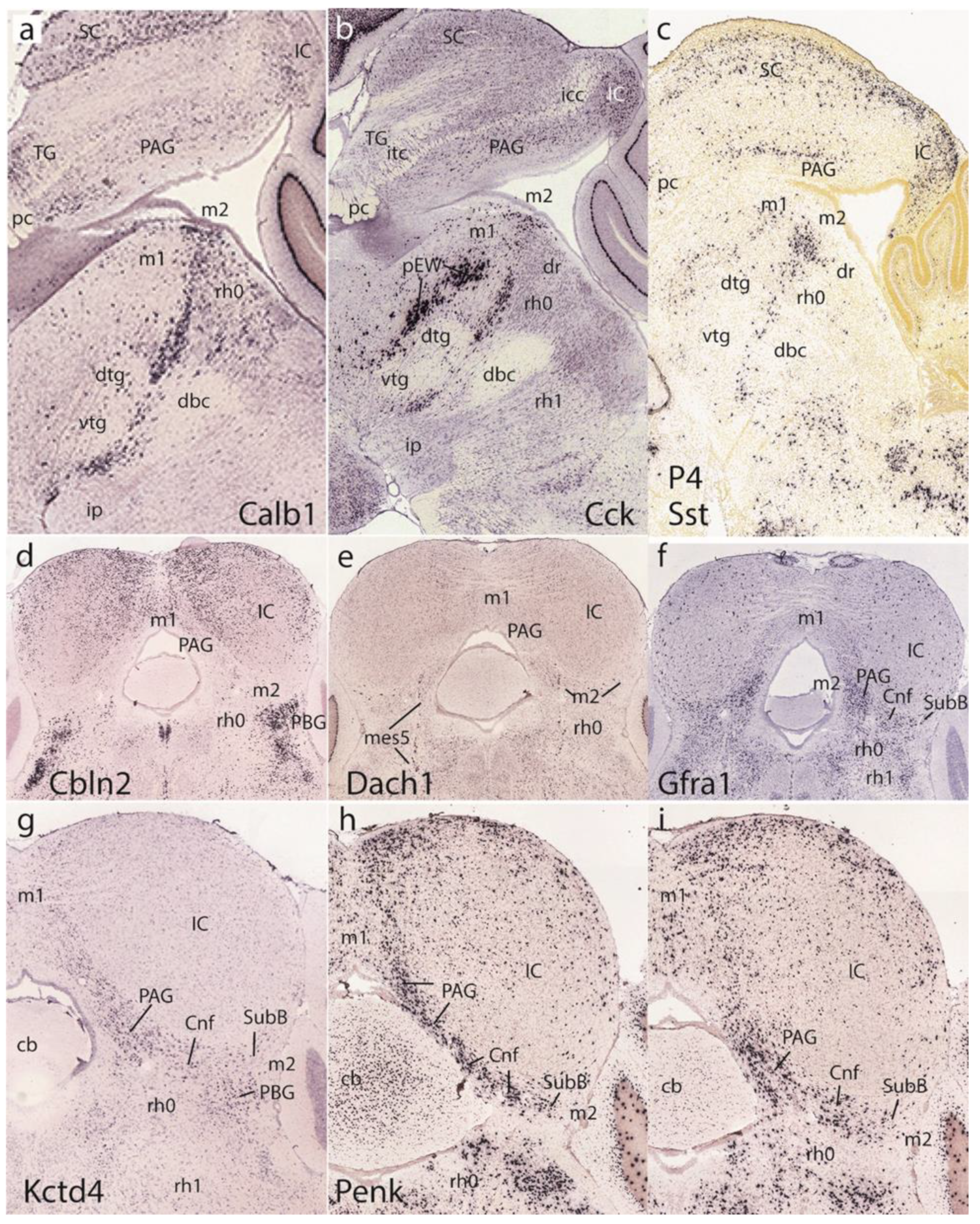
3.5. Midbrain and Isthmic Gene Patterns in Postnatal Mice
Figure 12, corresponding to postnatal results, shows uniformly adult brain material. The first row of images is dedicated to the basal preisthmus, represented by an elongated and narrow tissue band at the caudal end of the basal midbrain where cells expressing selective markers such as
Calb1,
Cck, and
Sst are found, among other markers such as dopamine and serotonin (
Figure 12a–c). The two first images clearly show the dorsal and ventral tegmental decussations, which characterize the m1 ventral midline or floor, placed just in front of the m2 floor. Behind m2 we see the IMB limit and at some distance the decussation of the brachium conjunctivum, known to decussate across the isthmic floor ([
8]; this is irrespective of the fact that some outdated sources place it in the midbrain).
Figure 12a,b also distinctly show the prepontine interpeduncular nucleus complex lying just caudal to the superficial end of the basal m2 at the apex of the cephalic flexure (see [
29]), as well as the major mass of the dorsal raphe nucleus periventricularly (dr; see [
27]).
The second and third rows show coronal sections representing actually horizontal sections across the IMB and the preisthmus, labeled respectively with
Cbln2,
Dach1,
Gfra1, Kctd4, and
Penk (
Figure 12d–i).
Cbln2 is a marker of the superficial isthmic and rh1 nuclei, which labels as well parts of the IC, but leaves the intercalated preisthmic domain wholly negative. This lack of signal can be followed from the PAG across the intermediate cuneiform area into the superficial derivative, the subbrachial nucleus (PAG, Cnf, SubB;
Figure 12d).
Dach1 weakly labels a few neurons distributed across the three strata of the preisthmic domain and leaves unlabeled the IC and the isthmus (
Figure 12e).
Gfra1 is a preisthmus marker and the corresponding sector of the PAG appears densely labeled (note the boundary with the IC PAG sector), as well as the adjacent Cnf nucleus (
Figure 12f). There are also some weakly positive isthmic cells.
Kctd4 selectively labels preisthmic elements and illustrates here mainly the preisthmic PAG and the adjacent Cnf nucleus (
Figure 12g). The last two sections in
Figure 12h,i show the
Penk signal at the preisthmic domain (again PAG and Cnf) as well as at the underlying superficial isthmus (the PBG nucleus).
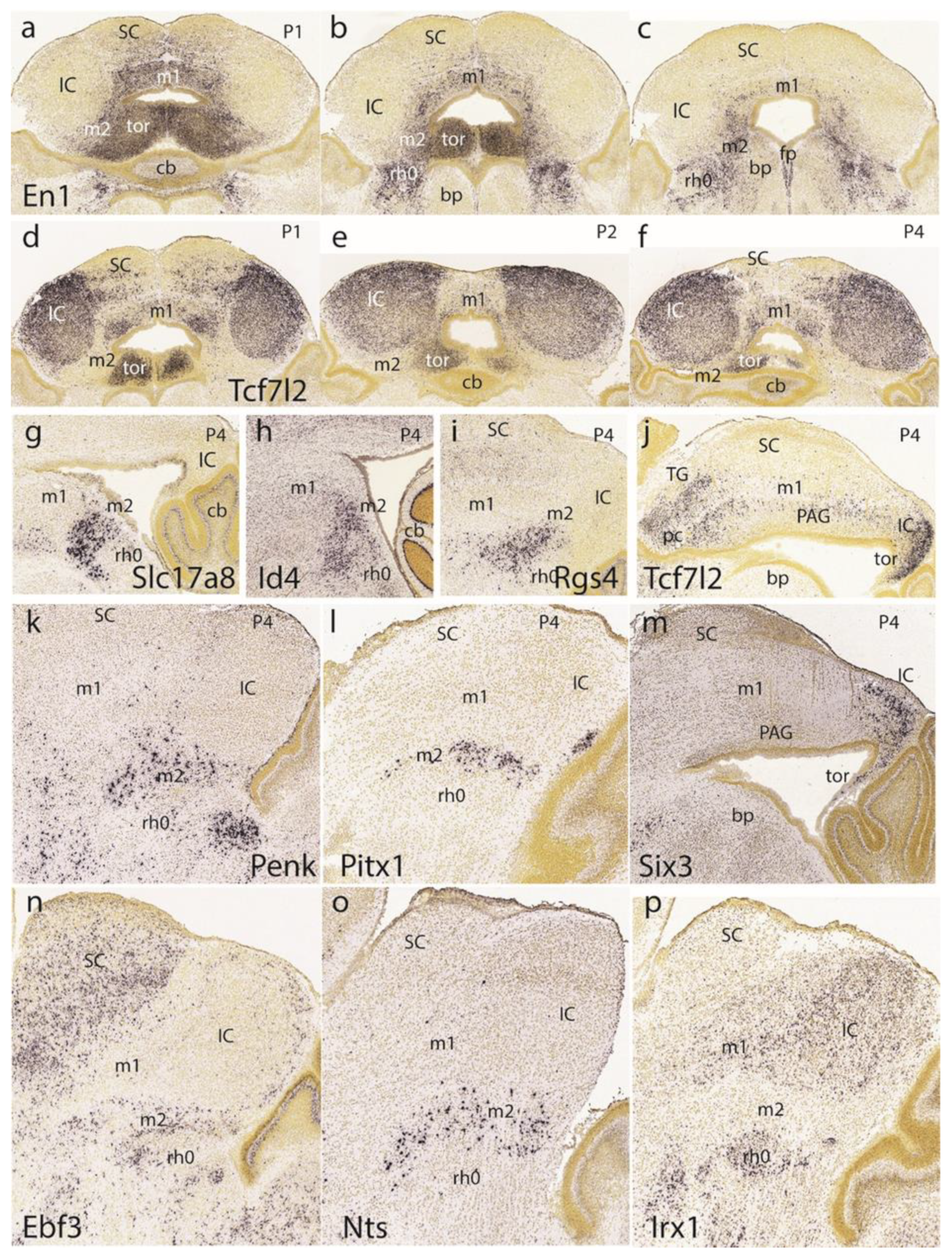
Returning to the dorsomedial toral area, we first show it at P1 labeled selectively in connection with the preisthmus with
En1 (
Figure 13a–c). Labeling is massive at the toral region and decreases somewhat into the adjacent preisthmus. There is no signal in the IC core. Complex additional details characterize the isthmic derivatives that express
En1, a marker that is also present at the diverse sectors of the PAG (
Figure 13a–c). Next, we examined the
Tcf7l2 signal at the IC and the toral region at P1 (
Figure 13d), P2 (
Figure 13e), and P4 (
Figure 13f). We observed over these stages that the toral bulges gradually become flattened. Sagittal sections at P4 labeled with
Tcf7l2 or
Six3 show the marked flattening of the former toral bulges (tor), practically changing into the thin roof-related membrane that separates the rostral cerebellar lobules from the midbrain ventricular cavity (
Figure 13j,m).
The sagittal sections shown in
Figure 13g–i correspond to the selective labeling of preisthmic patches of PAG with the markers
Slc17a8,
Id4, and
Rgs4, respectively. The sagittal images in
Figure 13k,l illustrate superficial labeling of the subbrachial preisthmic nucleus (SubB) with
Penk and
Pitx1, respectively. Other markers also selectively labeling the SubB nucleus are
Ebf3 and
Nts (
Figure 13n,o), whereas
Irx1 labels the complementary isthmus and BIC/IC domains (
Figure 13p).