Dual-Hit Model of Parkinson’s Disease: Impact of Dysbiosis on 6-Hydroxydopamine-Insulted Mice—Neuroprotective and Anti-Inflammatory Effects of Butyrate
Abstract
:1. Introduction
2. Results
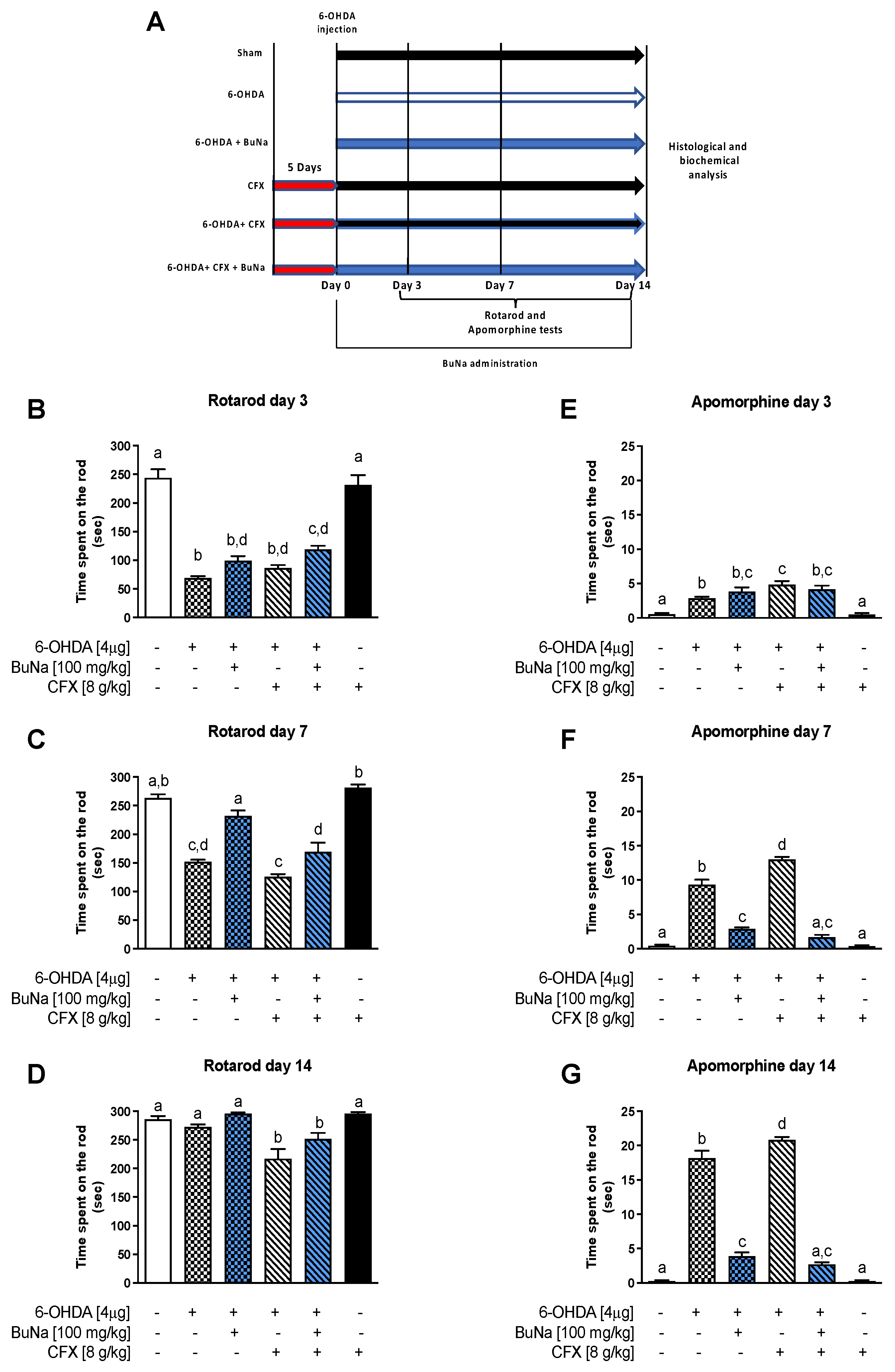
2.1. Motor Coordination and Apomorphine-Induced Rotational Behavior in CFX Dysbiotic PD Mice. BuNa Effect on Behavioral Traits
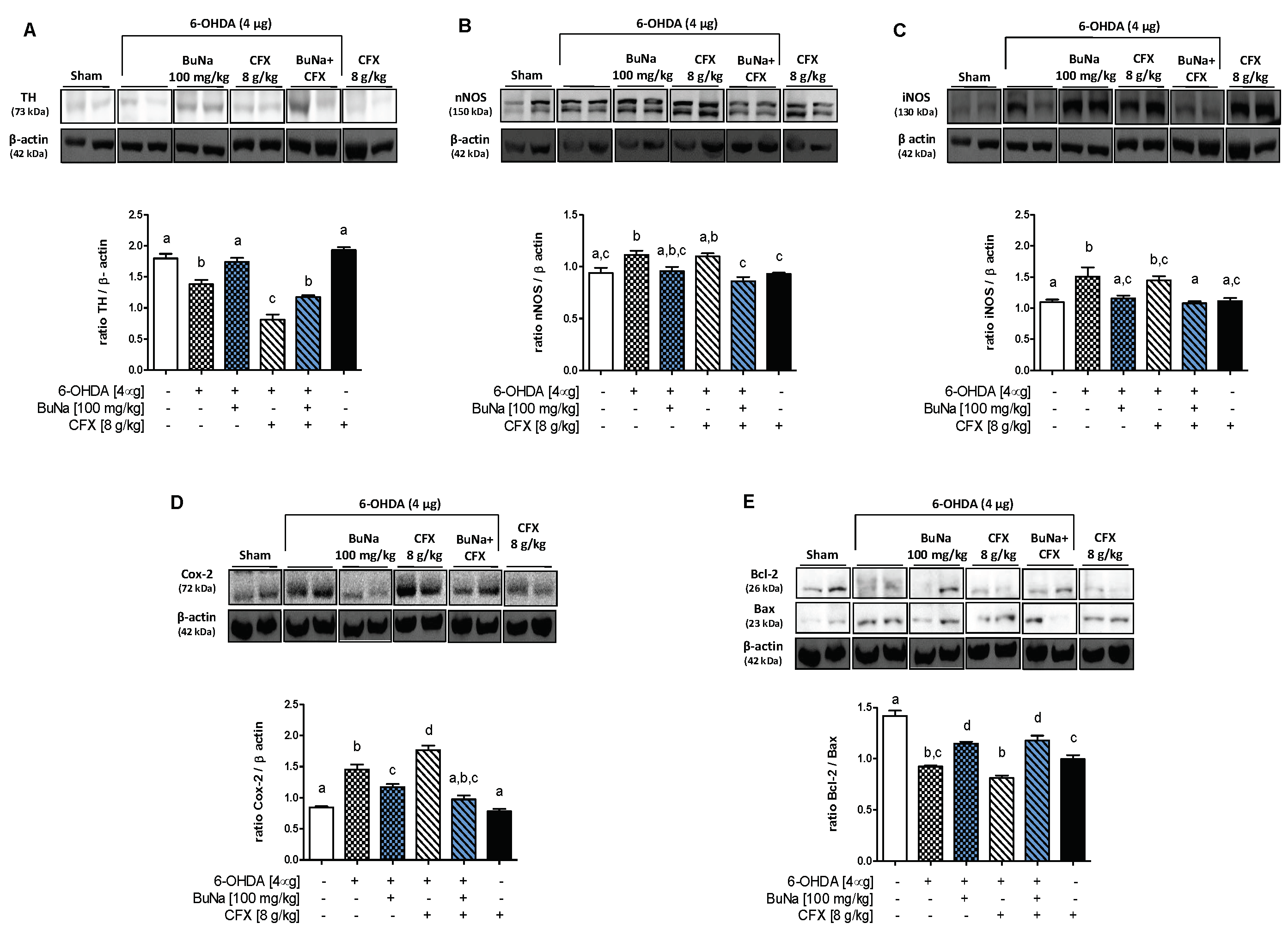
2.2. 6-OHDA-Induced Striatal Toxicity Is Worsened by CFX Pretreatment. Reversal Effect of BuNa
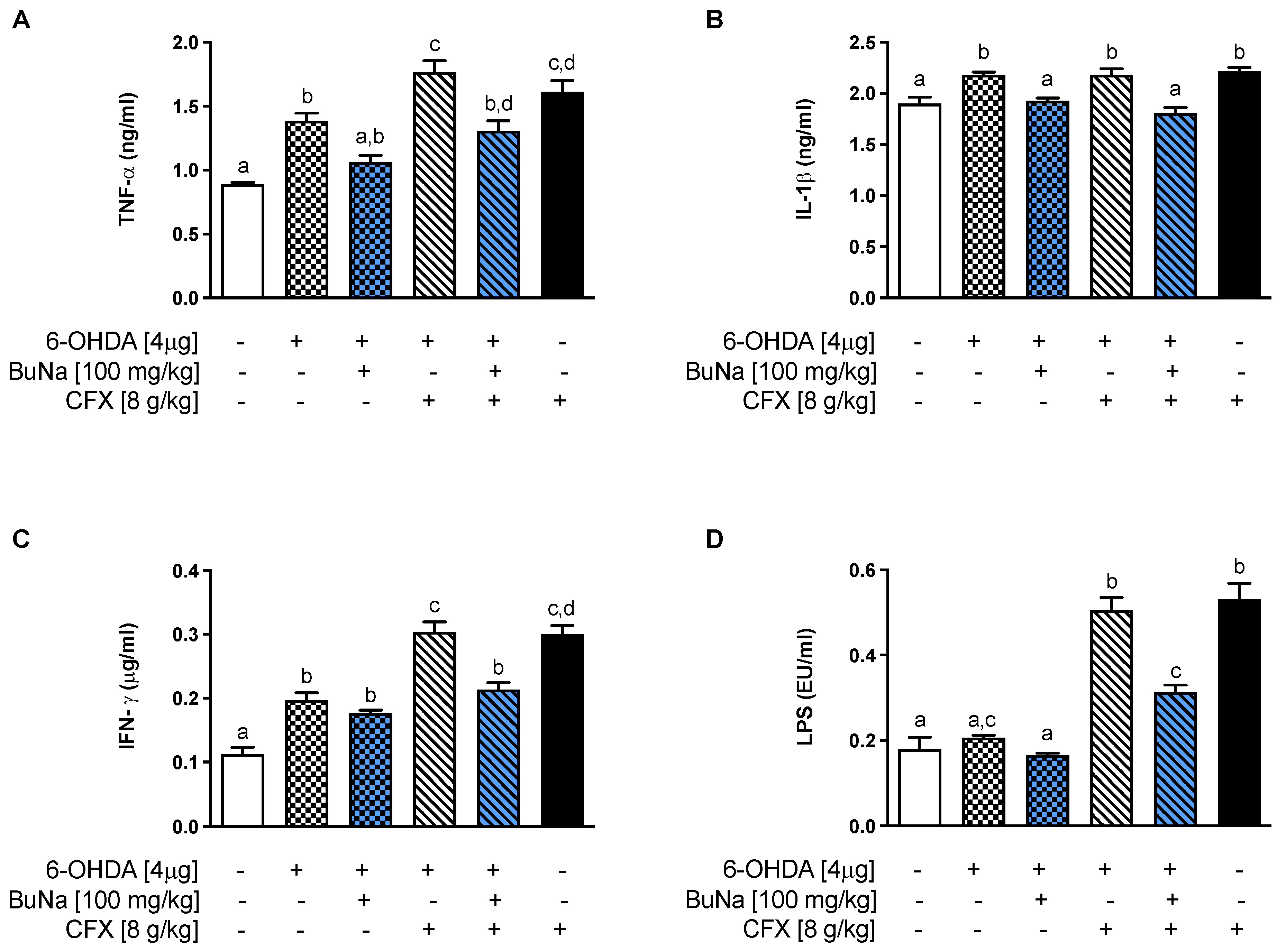
2.3. Alteration of Systemic Pro-Inflammatory Markers. Protective Effect of BuNa
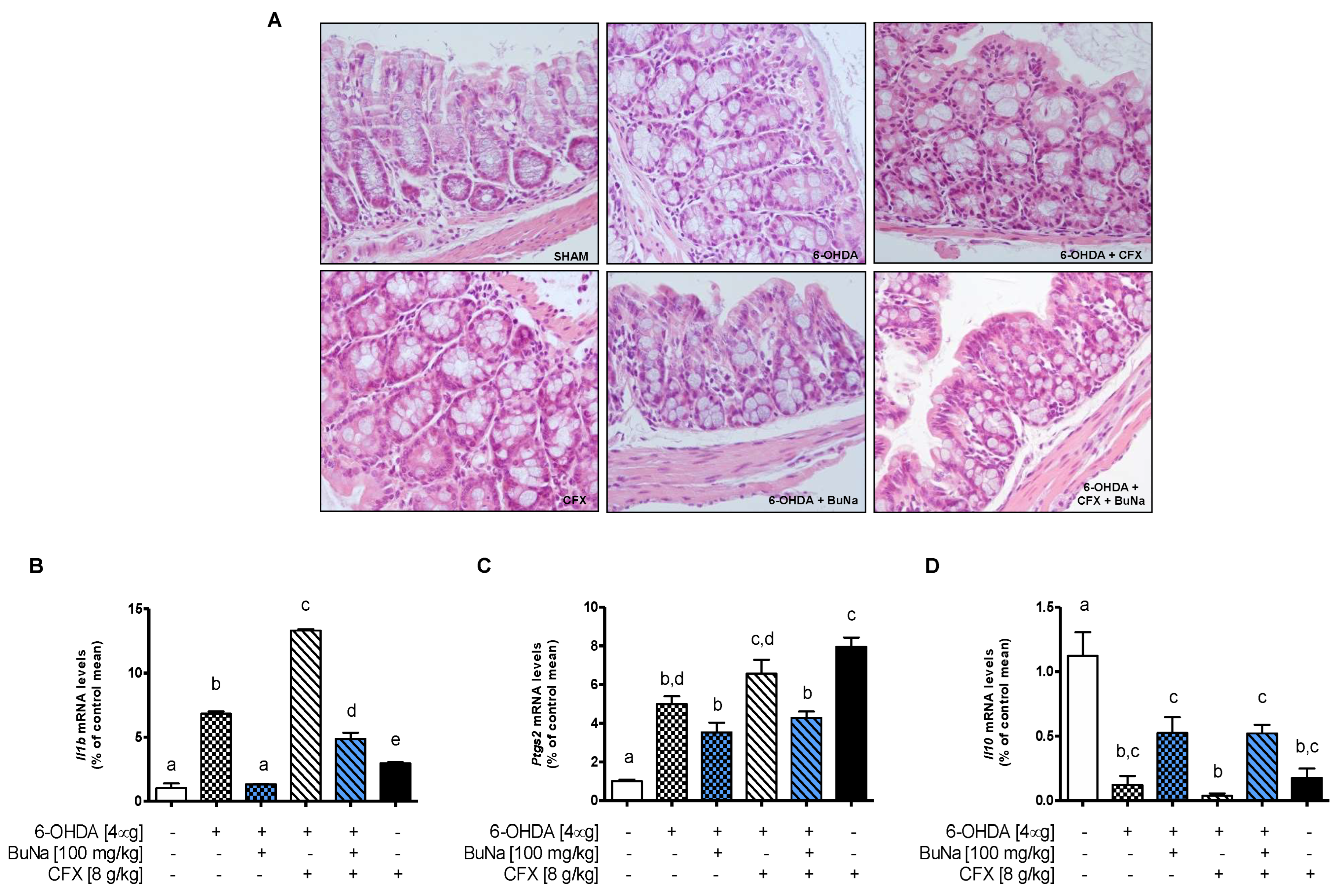
2.4. Colon Injury in Dysbiotic PD Mice and BuNa Effect on Gut Homeostasis
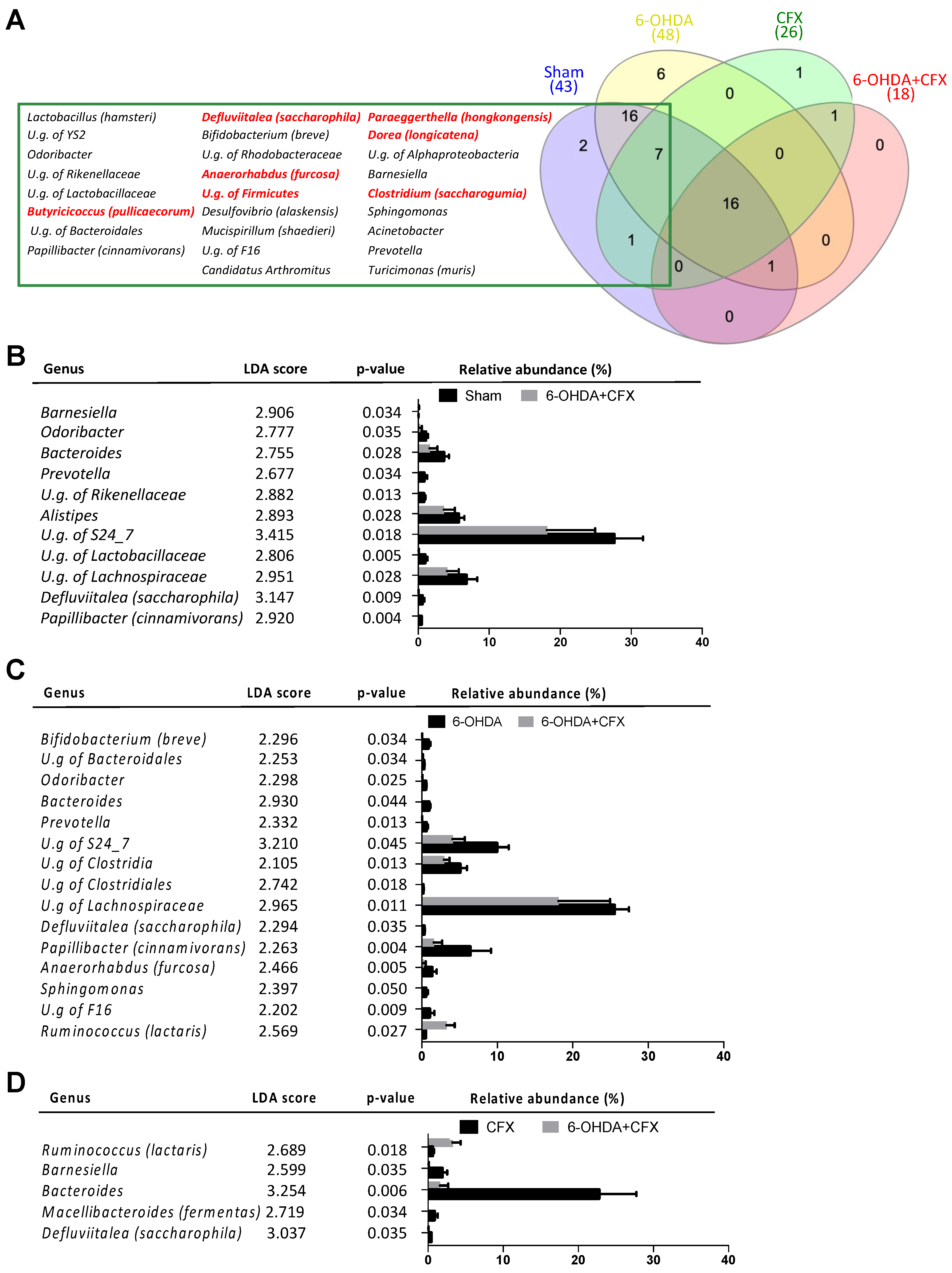
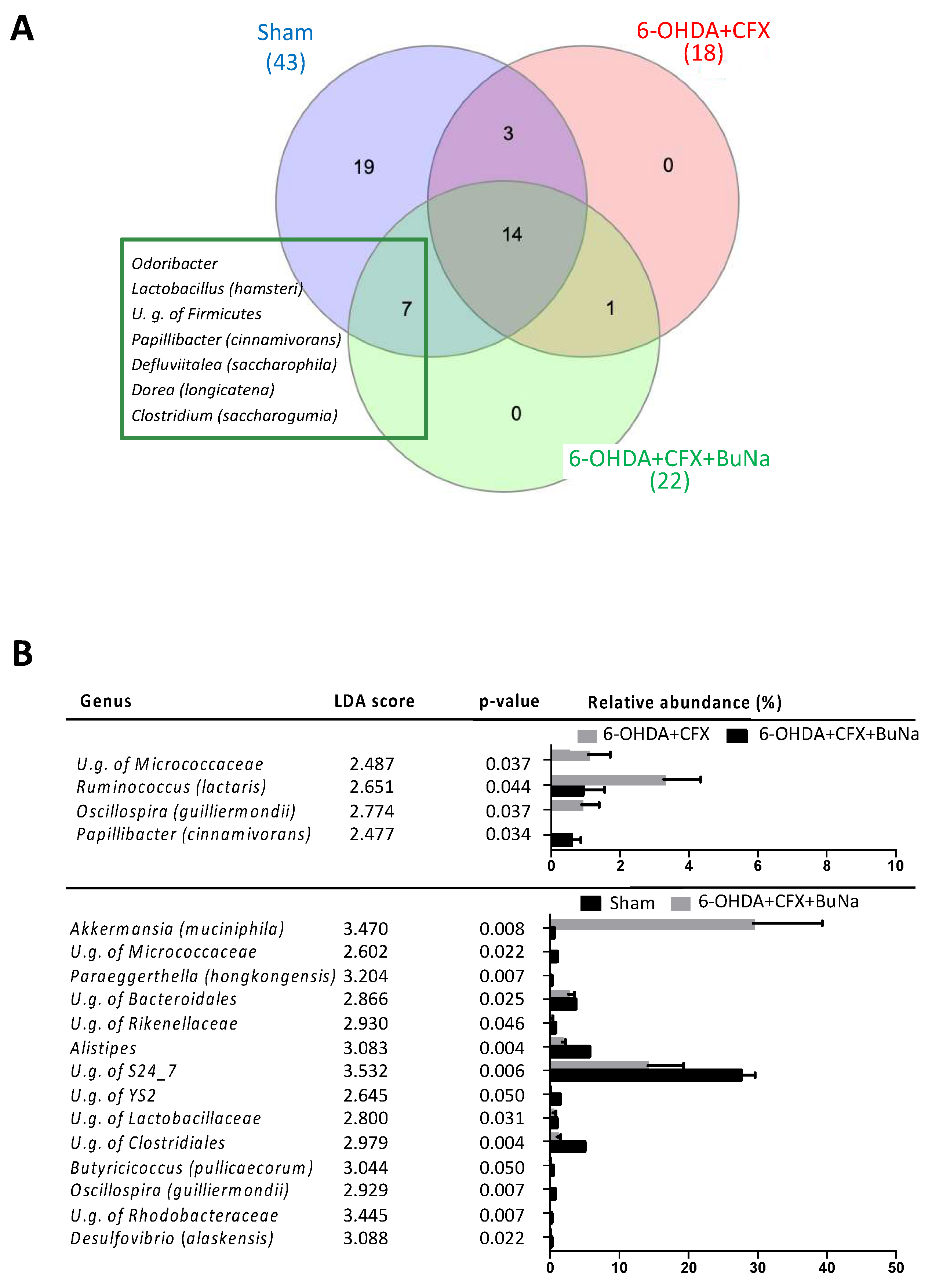
2.5. Antibiotic-Induced Dysbiosis in 6-OHDA-Lesioned Mice and BuNa Effect on Microbiota Composition
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Animals
4.3. Experimental Design and Drug Treatment
4.4. Rotarod Test
4.5. Apomorphine Test
4.6. Western Blot Analysis
4.7. Serum Parameters
4.8. Real-Time PCRA
4.9. Histopathological Analysis
4.10. Gut Microbiota-Based Studies
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef]
- Chen, H.; Burton, E.A.; Ross, G.W.; Huang, X.; Savica, R.; Abbott, R.D.; Ascherio, A.; Caviness, J.N.; Gao, X.; Gray, K.A.; et al. Research on the premotor symptoms of Parkinson’s disease: Clinical and etiological implications. Environ. Health Perspect. 2013, 121, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, C.H.; Del Tredici, K.; Braak, H. Parkinson’s disease: A dual-hit hypothesis. Neuropathol. Appl. Neurobiol. 2007, 33, 599–614. [Google Scholar] [CrossRef]
- Hawkes, C.H.; Del Tredici, K.; Braak, H. Parkinson’s disease: The dual hit theory revisited. Ann. N. Y. Acad. Sci. 2009, 1170, 615–622. [Google Scholar] [CrossRef]
- Braak, H.; Del Tredici, K.; Rub, U.; de Vos, R.A.; Jansen Steur, E.N.; Braak, E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 2003, 24, 197–211. [Google Scholar] [CrossRef]
- Kaur, G.; Behl, T.; Bungau, S.; Kumar, A.; Uddin, M.S.; Mehta, V.; Zengin, G.; Mathew, B.; Shah, M.A.; Arora, S. Dysregulation of the Gut-Brain Axis, Dysbiosis and Influence of Numerous Factors on Gut Microbiota Associated Parkinson’s Disease. Curr. Neuropharmacol. 2021, 19, 233–247. [Google Scholar] [CrossRef]
- Elfil, M.; Kamel, S.; Kandil, M.; Koo, B.B.; Schaefer, S.M. Implications of the Gut Microbiome in Parkinson’s Disease. Mov. Disord. 2020, 35, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.F.; Shan, C.; Zhuang, S.Y.; Zhuang, Q.Q.; Ghosh, A.; Zhu, K.C.; Kong, X.K.; Wang, S.M.; Gong, Y.L.; Yang, Y.Y.; et al. Gut microbiota-derived propionate mediates the neuroprotective effect of osteocalcin in a mouse model of Parkinson’s disease. Microbiome 2021, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Dodiya, H.B.; Forsyth, C.B.; Voigt, R.M.; Engen, P.A.; Patel, J.; Shaikh, M.; Green, S.J.; Naqib, A.; Roy, A.; Kordower, J.H.; et al. Chronic stress-induced gut dysfunction exacerbates Parkinson’s disease phenotype and pathology in a rotenone-induced mouse model of Parkinson’s disease. Neurobiol. Dis. 2020, 135, 104352. [Google Scholar] [CrossRef]
- Sun, M.F.; Zhu, Y.L.; Zhou, Z.L.; Jia, X.B.; Xu, Y.D.; Yang, Q.; Cui, C.; Shen, Y.Q. Neuroprotective effects of fecal microbiota transplantation on MPTP-induced Parkinson’s disease mice: Gut microbiota, glial reaction and TLR4/TNF-alpha signaling pathway. Brain Behav. Immun. 2018, 70, 48–60. [Google Scholar] [CrossRef]
- Gouda, N.A.; Elkamhawy, A.; Cho, J. Emerging Therapeutic Strategies for Parkinson’s Disease and Future Prospects: A 2021 Update. Biomedicines 2022, 10, 371. [Google Scholar] [CrossRef] [PubMed]
- Lama, A.; Pirozzi, C.; Avagliano, C.; Annunziata, C.; Mollica, M.P.; Calignano, A.; Meli, R.; Raso, G.M. Nutraceuticals: An integrative approach to starve Parkinson’s disease. Brain Behav. Immun. Health 2020, 2, 100037. [Google Scholar] [CrossRef] [PubMed]
- Raso, G.M.; Avagliano, C.; Calignano, A. Response to comment by Juan Segura-Aguilar: New preclinical model are required to discover neuroprotective compound in Parkinson’s disease. Pharmacol. Res. 2017, 119, 491–492. [Google Scholar] [CrossRef]
- Mou, Y.; Du, Y.; Zhou, L.; Yue, J.; Hu, X.; Liu, Y.; Chen, S.; Lin, X.; Zhang, G.; Xiao, H.; et al. Gut Microbiota Interact With the Brain Through Systemic Chronic Inflammation: Implications on Neuroinflammation, Neurodegeneration, and Aging. Front. Immunol. 2022, 13, 796288. [Google Scholar] [CrossRef]
- Gonzalez-Correa, C.A.; Mulett-Vasquez, E.; Miranda, D.A.; Gonzalez-Correa, C.H.; Gomez-Buitrago, P.A. The colon revisited or the key to wellness, health and disease. Med. Hypotheses 2017, 108, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Antonini, M.; Lo Conte, M.; Sorini, C.; Falcone, M. How the Interplay Between the Commensal Microbiota, Gut Barrier Integrity, and Mucosal Immunity Regulates Brain Autoimmunity. Front. Immunol. 2019, 10, 1937. [Google Scholar] [CrossRef]
- Liao, J.F.; Cheng, Y.F.; You, S.T.; Kuo, W.C.; Huang, C.W.; Chiou, J.J.; Hsu, C.C.; Hsieh-Li, H.M.; Wang, S.; Tsai, Y.C. Lactobacillus plantarum PS128 alleviates neurodegenerative progression in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced mouse models of Parkinson’s disease. Brain Behav. Immun. 2020, 90, 26–46. [Google Scholar] [CrossRef]
- Liu, X.; Du, Z.R.; Wang, X.; Sun, X.R.; Zhao, Q.; Zhao, F.; Wong, W.T.; Wong, K.H.; Dong, X.L. Polymannuronic acid prebiotic plus Lacticaseibacillus rhamnosus GG probiotic as a novel synbiotic promoted their separate neuroprotection against Parkinson’s disease. Food Res. Int. 2022, 155, 111067. [Google Scholar] [CrossRef] [PubMed]
- Unger, M.M.; Spiegel, J.; Dillmann, K.U.; Grundmann, D.; Philippeit, H.; Burmann, J.; Fassbender, K.; Schwiertz, A.; Schafer, K.H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Nuzum, N.D.; Loughman, A.; Szymlek-Gay, E.A.; Hendy, A.; Teo, W.P.; Macpherson, H. Gut microbiota differences between healthy older adults and individuals with Parkinson’s disease: A systematic review. Neurosci. Biobehav. Rev. 2020, 112, 227–241. [Google Scholar] [CrossRef]
- Shin, C.; Lim, Y.; Lim, H.; Ahn, T.B. Plasma Short-Chain Fatty Acids in Patients With Parkinson’s Disease. Mov. Disord. 2020, 35, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Bullich, C.; Keshavarzian, A.; Garssen, J.; Kraneveld, A.; Perez-Pardo, P. Gut Vibes in Parkinson’s Disease: The Microbiota-Gut-Brain Axis. Mov. Disord. Clin. Pract. 2019, 6, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Aho, V.T.E.; Pereira, P.A.B.; Voutilainen, S.; Paulin, L.; Pekkonen, E.; Auvinen, P.; Scheperjans, F. Gut microbiota in Parkinson’s disease: Temporal stability and relations to disease progression. EBioMedicine 2019, 44, 691–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrov, V.A.; Saltykova, I.V.; Zhukova, I.A.; Alifirova, V.M.; Zhukova, N.G.; Dorofeeva, Y.B.; Tyakht, A.V.; Kovarsky, B.A.; Alekseev, D.G.; Kostryukova, E.S.; et al. Analysis of Gut Microbiota in Patients with Parkinson’s Disease. Bull. Exp. Biol. Med. 2017, 162, 734–737. [Google Scholar] [CrossRef]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480.e12. [Google Scholar] [CrossRef] [Green Version]
- Davie, J.R. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003, 133 (Suppl. 7), 2485S–2493S. [Google Scholar] [CrossRef]
- Luhrs, H.; Gerke, T.; Muller, J.G.; Melcher, R.; Schauber, J.; Boxberge, F.; Scheppach, W.; Menzel, T. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand. J. Gastroenterol. 2002, 37, 458–466. [Google Scholar] [CrossRef]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef] [Green Version]
- Avagliano, C.; Russo, R.; De Caro, C.; Cristiano, C.; La Rana, G.; Piegari, G.; Paciello, O.; Citraro, R.; Russo, E.; De Sarro, G.; et al. Palmitoylethanolamide protects mice against 6-OHDA-induced neurotoxicity and endoplasmic reticulum stress: In vivo and in vitro evidence. Pharmacol. Res. 2016, 113, 276–289. [Google Scholar] [CrossRef]
- Lama, A.; Annunziata, C.; Coretti, L.; Pirozzi, C.; Di Guida, F.; Izzo, A.N.; Cristiano, C.; Mollica, M.P.; Chiariotti, L.; Pelagalli, A.; et al. N-(1-carbamoyl-2-phenylethyl) butyramide reduces antibiotic-induced intestinal injury, innate immune activation and modulates microbiota composition. Sci. Rep. 2019, 9, 4832. [Google Scholar] [CrossRef]
- McQuade, R.M.; Singleton, L.M.; Wu, H.; Lee, S.; Constable, R.; Di Natale, M.; Ringuet, M.T.; Berger, J.P.; Kauhausen, J.; Parish, C.L.; et al. The association of enteric neuropathy with gut phenotypes in acute and progressive models of Parkinson’s disease. Sci. Rep. 2021, 11, 7934. [Google Scholar] [CrossRef] [PubMed]
- Blandini, F.; Balestra, B.; Levandis, G.; Cervio, M.; Greco, R.; Tassorelli, C.; Colucci, M.; Faniglione, M.; Bazzini, E.; Nappi, G.; et al. Functional and neurochemical changes of the gastrointestinal tract in a rodent model of Parkinson’s disease. Neurosci. Lett. 2009, 467, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Colucci, M.; Cervio, M.; Faniglione, M.; De Angelis, S.; Pajoro, M.; Levandis, G.; Tassorelli, C.; Blandini, F.; Feletti, F.; De Giorgio, R.; et al. Intestinal dysmotility and enteric neurochemical changes in a Parkinson’s disease rat model. Auton. Neurosci. 2012, 169, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.Y.; Yang, J.; Zhang, X.; Zhu, J. Gastrointestinal non-motor dysfunction in Parkinson’s disease model rats with 6-hydroxydopamine. Physiol. Res. 2019, 68, 295–303. [Google Scholar] [CrossRef]
- Chai, X.Y.; Diwakarla, S.; Pustovit, R.V.; McQuade, R.M.; Di Natale, M.; Ermine, C.M.; Parish, C.L.; Finkelstein, D.I.; Furness, J.B. Investigation of nerve pathways mediating colorectal dysfunction in Parkinson’s disease model produced by lesion of nigrostriatal dopaminergic neurons. Neurogastroenterol. Motil. 2020, 32, e13893. [Google Scholar] [CrossRef] [PubMed]
- Leclair-Visonneau, L.; Clairembault, T.; Volteau, C.; Chapelet, G.; Le Dily, S.; Vavasseur, F.; Coron, E.; Preterre, C.; Neunlist, M.; Pereon, Y.; et al. Colonic neuropathology is not associated with autonomic dysfunction in Parkinson’s disease. Parkinsonism Relat. Disord. 2019, 61, 224–227. [Google Scholar] [CrossRef]
- Devriese, S.; Eeckhaut, V.; Geirnaert, A.; Van den Bossche, L.; Hindryckx, P.; Van de Wiele, T.; Van Immerseel, F.; Ducatelle, R.; De Vos, M.; Laukens, D. Reduced Mucosa-associated Butyricicoccus Activity in Patients with Ulcerative Colitis Correlates with Aberrant Claudin-1 Expression. J. Crohn’s Colitis 2017, 11, 229–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.M.; Van de Wiele, T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef] [Green Version]
- Qin, Y.; Havulinna, A.S.; Liu, Y.; Jousilahti, P.; Ritchie, S.C.; Tokolyi, A.; Sanders, J.G.; Valsta, L.; Brozynska, M.; Zhu, Q.; et al. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat. Genet. 2022, 54, 134–142. [Google Scholar] [CrossRef]
- Mondot, S.; Lepage, P.; Seksik, P.; Allez, M.; Treton, X.; Bouhnik, Y.; Colombel, J.F.; Leclerc, M.; Pochart, P.; Dore, J.; et al. Structural robustness of the gut mucosal microbiota is associated with Crohn’s disease remission after surgery. Gut 2016, 65, 954–962. [Google Scholar] [CrossRef]
- Dan, Z.; Mao, X.; Liu, Q.; Guo, M.; Zhuang, Y.; Liu, Z.; Chen, K.; Chen, J.; Xu, R.; Tang, J.; et al. Altered gut microbial profile is associated with abnormal metabolism activity of Autism Spectrum Disorder. Gut Microbes 2020, 11, 1246–1267. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Gao, X.; Zhao, H.; Luo, X.; Li, J.; Tan, X.; Wang, L.; Zhao, J.B.; Wang, J.; Yang, G.; et al. Depicting the composition of gut microbiota in children with tic disorders: An exploratory study. J. Child. Psychol. Psychiatry 2021, 62, 1246–1254. [Google Scholar] [CrossRef] [PubMed]
- Cantu-Jungles, T.M.; Rasmussen, H.E.; Hamaker, B.R. Potential of Prebiotic Butyrogenic Fibers in Parkinson’s Disease. Front. Neurol. 2019, 10, 663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St Laurent, R.; O’Brien, L.M.; Ahmad, S.T. Sodium butyrate improves locomotor impairment and early mortality in a rotenone-induced Drosophila model of Parkinson’s disease. Neuroscience 2013, 246, 382–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.X.; Du, D.; Zheng, T.; Fang, Y.; Chen, Y.S.; Yi, H.L.; He, Q.Y.; Gao, D.W.; Shi, Q.L. Detecting dopaminergic neuronal degeneration using diffusion tensor imaging in a rotenone-induced rat model of Parkinson’s disease: Fractional anisotropy and mean diffusivity values. Neural. Regen. Res. 2017, 12, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Funakohi-Tago, M.; Sakata, T.; Fujiwara, S.; Sakakura, A.; Sugai, T.; Tago, K.; Tamura, H. Hydroxytyrosol butyrate inhibits 6-OHDA-induced apoptosis through activation of the Nrf2/HO-1 axis in SH-SY5Y cells. Eur. J. Pharmacol. 2018, 834, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.; Cristiano, C.; Avagliano, C.; De Caro, C.; La Rana, G.; Raso, G.M.; Canani, R.B.; Meli, R.; Calignano, A. Gut-brain Axis: Role of Lipids in the Regulation of Inflammation, Pain and CNS Diseases. Curr. Med. Chem. 2018, 25, 3930–3952. [Google Scholar] [CrossRef]
- Srivastav, S.; Neupane, S.; Bhurtel, S.; Katila, N.; Maharjan, S.; Choi, H.; Hong, J.T.; Choi, D.Y. Probiotics mixture increases butyrate, and subsequently rescues the nigral dopaminergic neurons from MPTP and rotenone-induced neurotoxicity. J. Nutr. Biochem. 2019, 69, 73–86. [Google Scholar] [CrossRef] [Green Version]
- Canani, R.B.; Costanzo, M.D.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.; De Caro, C.; Avagliano, C.; Cristiano, C.; La Rana, G.; Raso, G.M.; Canani, R.B.; Meli, R.; Calignano, A. Sodium butyrate and its synthetic amide derivative modulate nociceptive behaviors in mice. Pharmacol. Res. 2016, 103, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Simeoli, R.; Raso, G.M.; Pirozzi, C.; Lama, A.; Santoro, A.; Russo, R.; Montero-Melendez, T.; Canani, R.B.; Calignano, A.; Perretti, M.; et al. An orally administered butyrate-releasing derivative reduces neutrophil recruitment and inflammation in dextran sulphate sodium-induced murine colitis. Br. J. Pharmacol. 2017, 174, 1484–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, T.H.; Park, J.H.; Jeon, W.M.; Han, K.S. Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr. Res. Pract. 2015, 9, 343–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paxinos, G.; Franklin, K.B. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Lama, A.; Pirozzi, C.; Annunziata, C.; Morgese, M.G.; Senzacqua, M.; Severi, I.; Calignano, A.; Trabace, L.; Giordano, A.; Meli, R.; et al. Palmitoylethanolamide counteracts brain fog improving depressive-like behaviour in obese mice: Possible role of synaptic plasticity and neurogenesis. Br. J. Pharmacol. 2021, 178, 845–859. [Google Scholar] [CrossRef]
- Lama, A.; Pirozzi, C.; Severi, I.; Morgese, M.G.; Senzacqua, M.; Annunziata, C.; Comella, F.; Del Piano, F.; Schiavone, S.; Petrosino, S.; et al. Palmitoylethanolamide dampens neuroinflammation and anxiety-like behavior in obese mice. Brain Behav. Immun. 2022, 102, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Fan, H.Y.; Yang, M.Y.; Zhang, Z.K.; Liu, K. Therapeutic effect of a hydroxynaphthoquinone fraction on dextran sulfate sodium-induced ulcerative colitis. World J. Gastroenterol. 2014, 20, 15310–15318. [Google Scholar] [CrossRef]
- Wang, R.; Shen, L.; Li, H.; Peng, H. Eriodictyol attenuates dextran sodium sulphate-induced colitis in mice by regulating the sonic hedgehog signalling pathway. Pharm. Biol. 2021, 59, 974–985. [Google Scholar] [CrossRef]
- Coretti, L.; Cristiano, C.; Florio, E.; Scala, G.; Lama, A.; Keller, S.; Cuomo, M.; Russo, R.; Pero, R.; Paciello, O.; et al. Sex-related alterations of gut microbiota composition in the BTBR mouse model of autism spectrum disorder. Sci. Rep. 2017, 7, 45356. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faust, K.; Raes, J. CoNet app: Inference of biological association networks using Cytoscape. F1000Research 2016, 5, 1519. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avagliano, C.; Coretti, L.; Lama, A.; Pirozzi, C.; De Caro, C.; De Biase, D.; Turco, L.; Mollica, M.P.; Paciello, O.; Calignano, A.; et al. Dual-Hit Model of Parkinson’s Disease: Impact of Dysbiosis on 6-Hydroxydopamine-Insulted Mice—Neuroprotective and Anti-Inflammatory Effects of Butyrate. Int. J. Mol. Sci. 2022, 23, 6367. https://doi.org/10.3390/ijms23126367
Avagliano C, Coretti L, Lama A, Pirozzi C, De Caro C, De Biase D, Turco L, Mollica MP, Paciello O, Calignano A, et al. Dual-Hit Model of Parkinson’s Disease: Impact of Dysbiosis on 6-Hydroxydopamine-Insulted Mice—Neuroprotective and Anti-Inflammatory Effects of Butyrate. International Journal of Molecular Sciences. 2022; 23(12):6367. https://doi.org/10.3390/ijms23126367
Chicago/Turabian StyleAvagliano, Carmen, Lorena Coretti, Adriano Lama, Claudio Pirozzi, Carmen De Caro, Davide De Biase, Luigia Turco, Maria Pina Mollica, Orlando Paciello, Antonio Calignano, and et al. 2022. "Dual-Hit Model of Parkinson’s Disease: Impact of Dysbiosis on 6-Hydroxydopamine-Insulted Mice—Neuroprotective and Anti-Inflammatory Effects of Butyrate" International Journal of Molecular Sciences 23, no. 12: 6367. https://doi.org/10.3390/ijms23126367
APA StyleAvagliano, C., Coretti, L., Lama, A., Pirozzi, C., De Caro, C., De Biase, D., Turco, L., Mollica, M. P., Paciello, O., Calignano, A., Meli, R., Lembo, F., & Mattace Raso, G. (2022). Dual-Hit Model of Parkinson’s Disease: Impact of Dysbiosis on 6-Hydroxydopamine-Insulted Mice—Neuroprotective and Anti-Inflammatory Effects of Butyrate. International Journal of Molecular Sciences, 23(12), 6367. https://doi.org/10.3390/ijms23126367








