Transcriptomic and Metabolomic Analysis Unravels the Molecular Regulatory Mechanism of Fatty Acid Biosynthesis in Styrax tonkinensis Seeds under Methyl Jasmonate Treatment
Abstract
:1. Introduction
2. Results
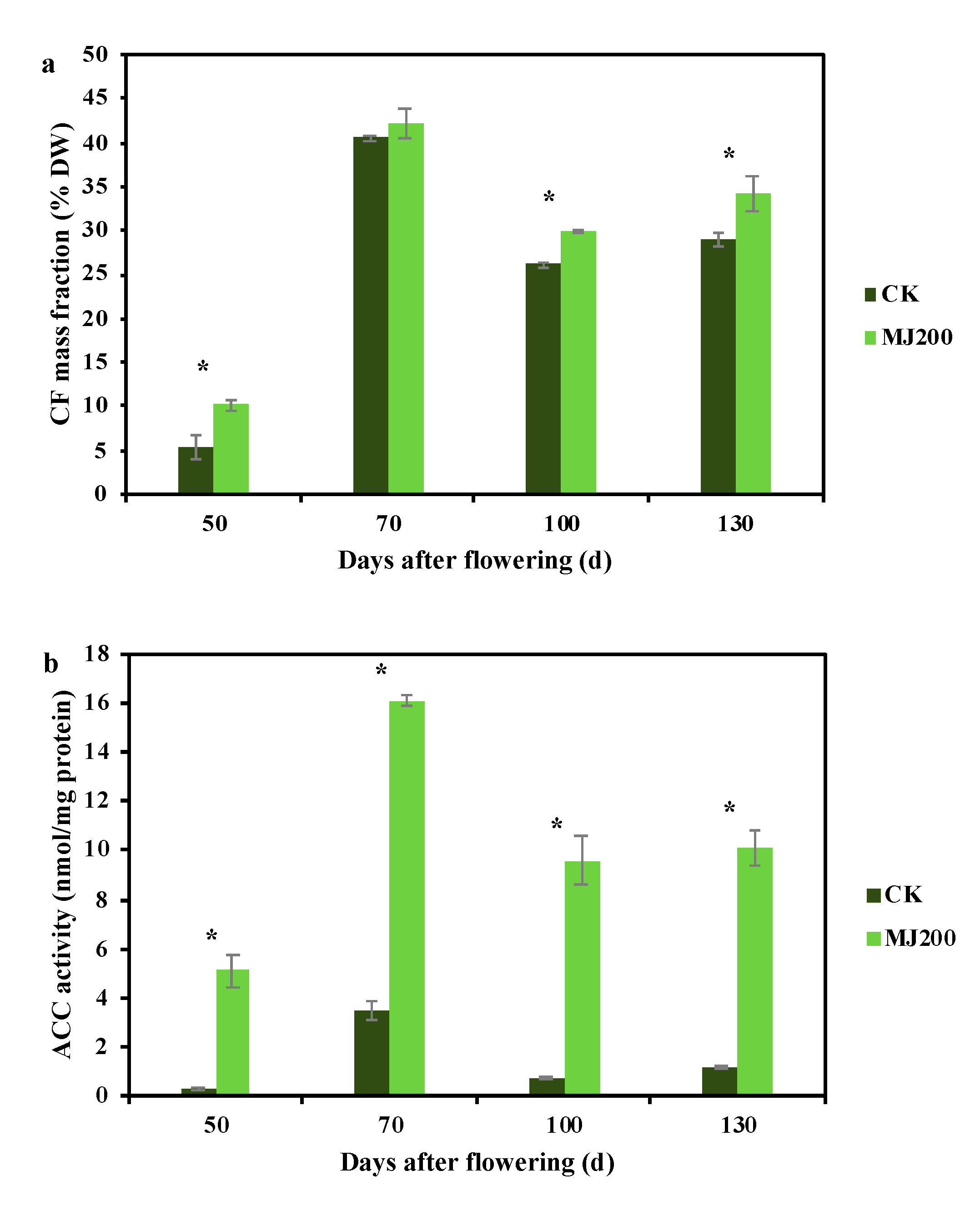
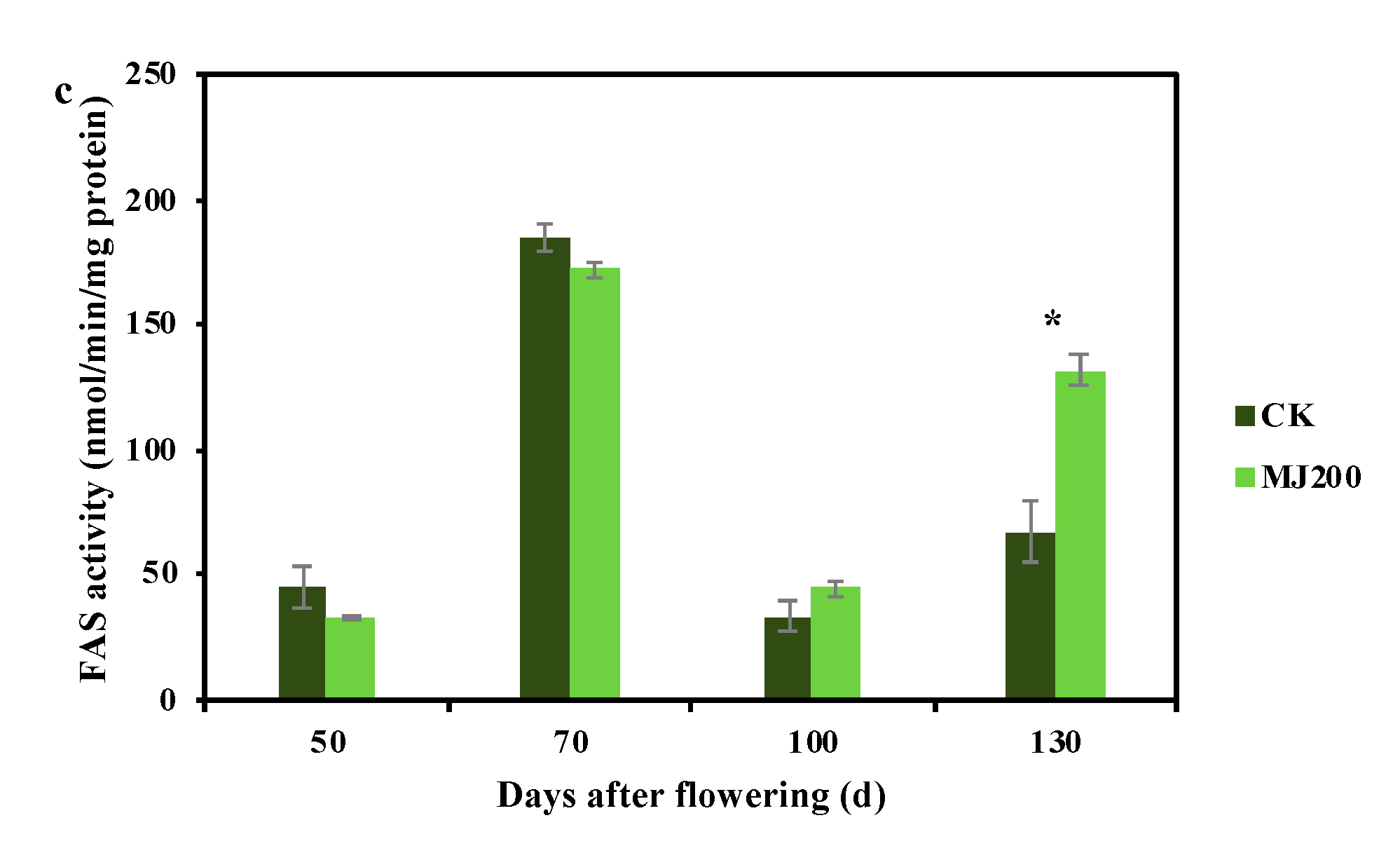
2.1. Dynamic Changing Trend of Crude Fat Mass Fraction and Fatty Acid Synthesis-Related Enzymes Activity under MJ200 Treatment
2.2. Dynamic Changing Trend of Fatty Acid Compositions under MJ200 Treatment
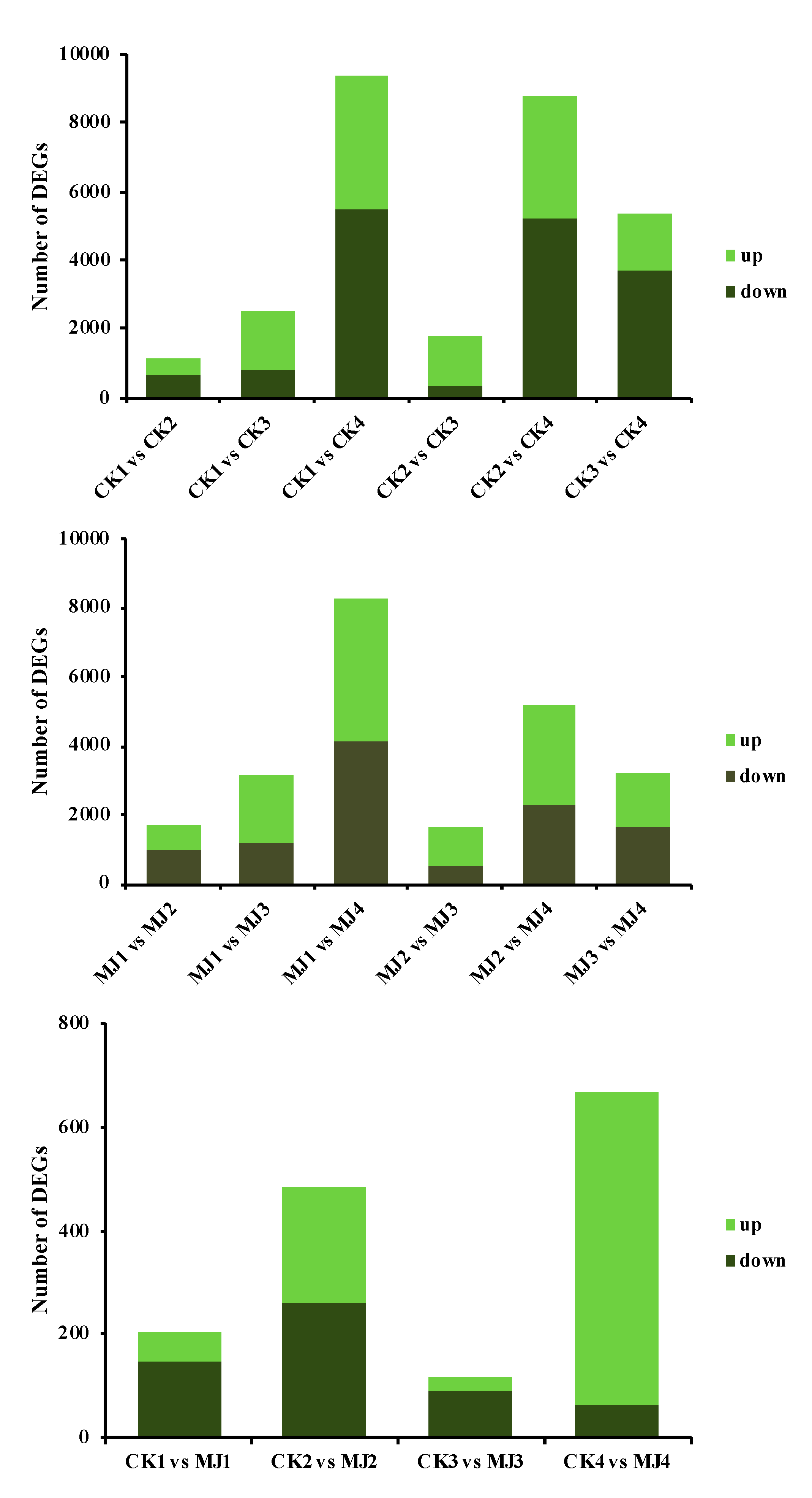
2.3. Analysis of Transcriptome Sequencing Data under MJ200 Treatment
2.3.1. Expression Level of Differentially Expressed Genes
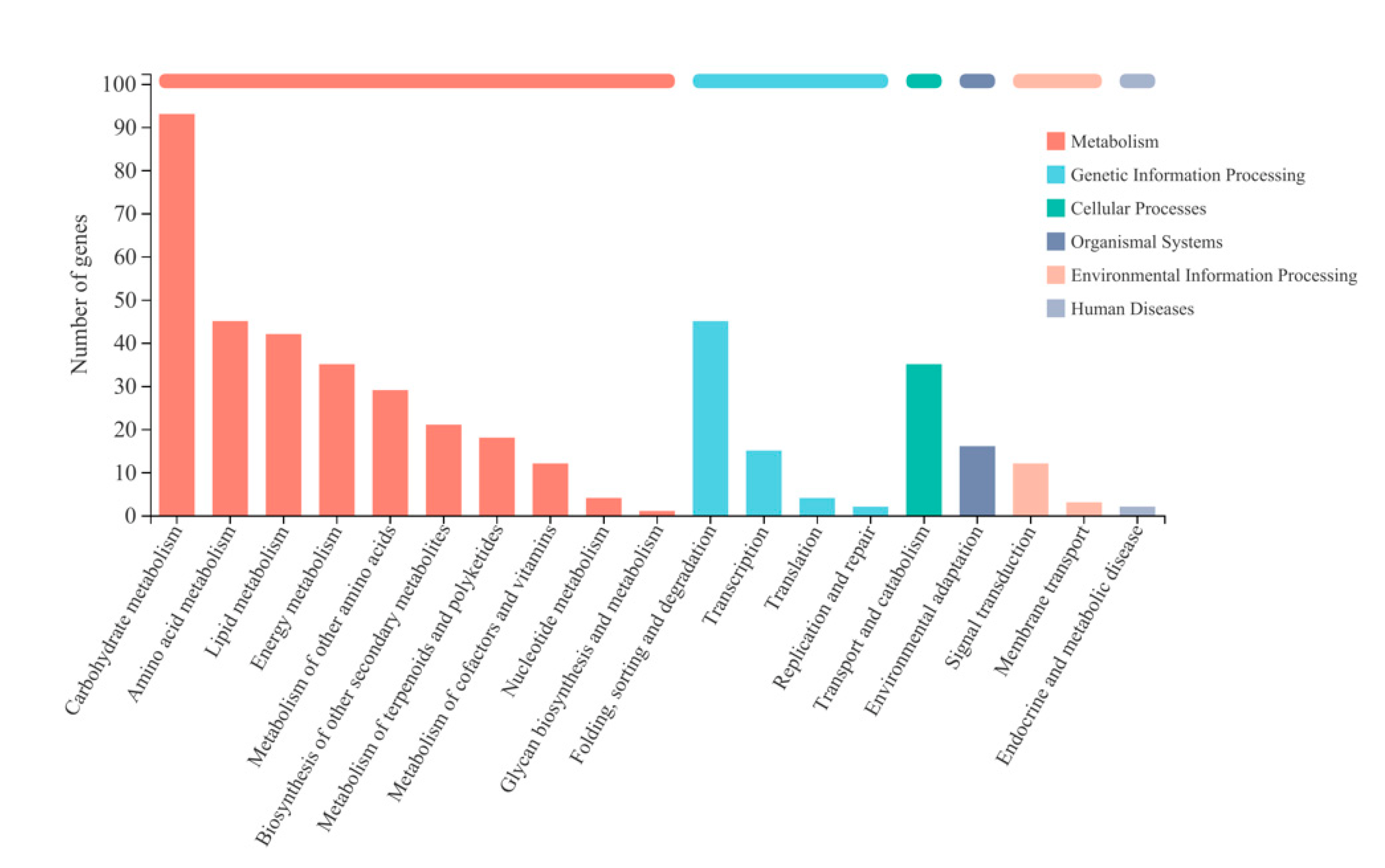
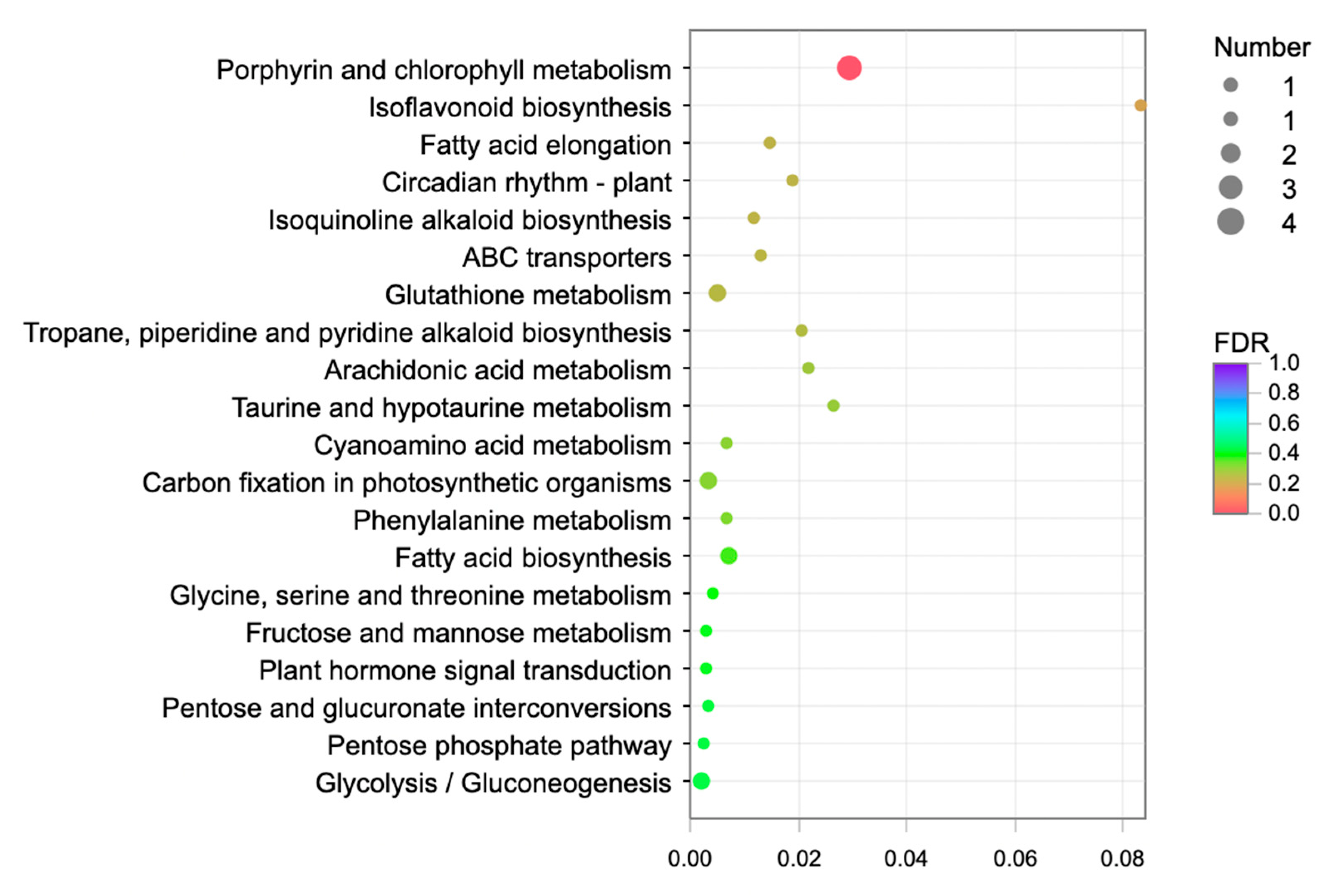
2.3.2. Enrichment Analysis of Differentially Expressed Genes
2.3.3. Identification and Expression Level of Genes Involved in Fatty Acid Biosynthesis
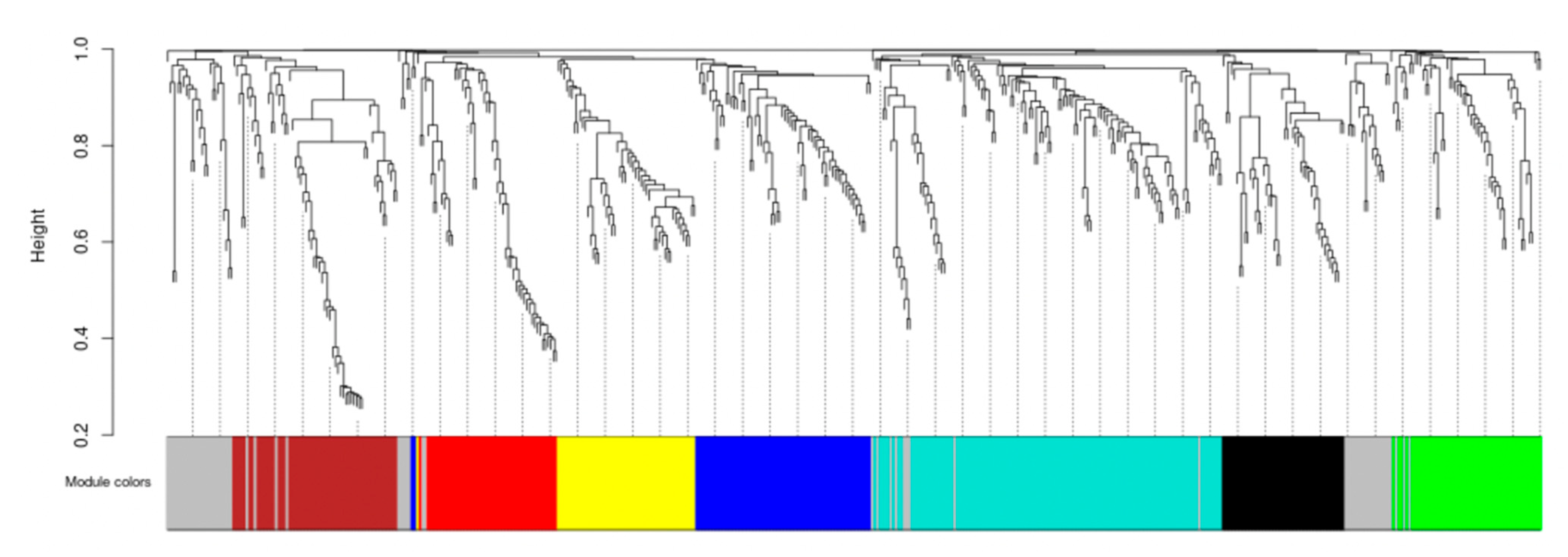
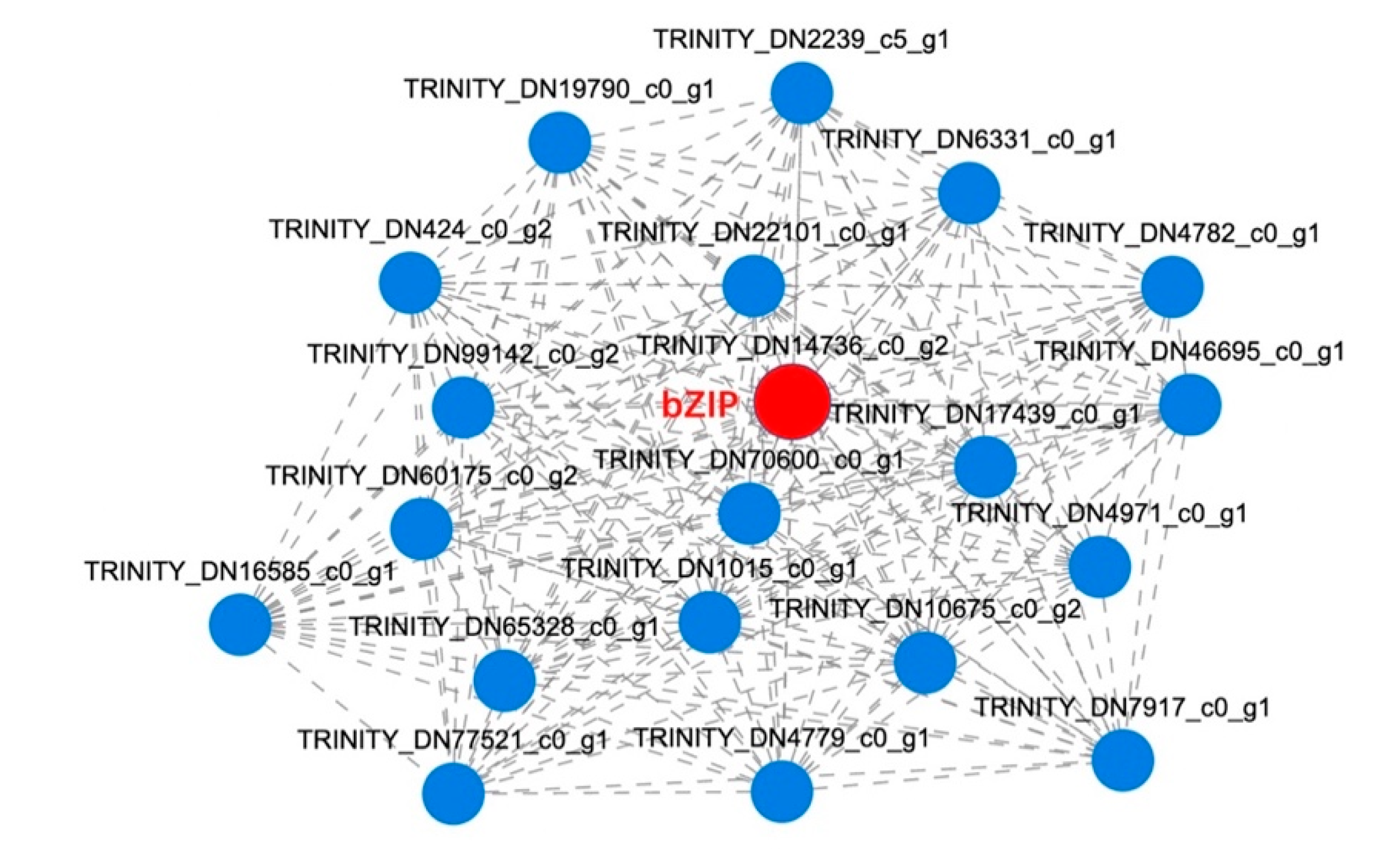
2.3.4. Co-Expression Network Analysis
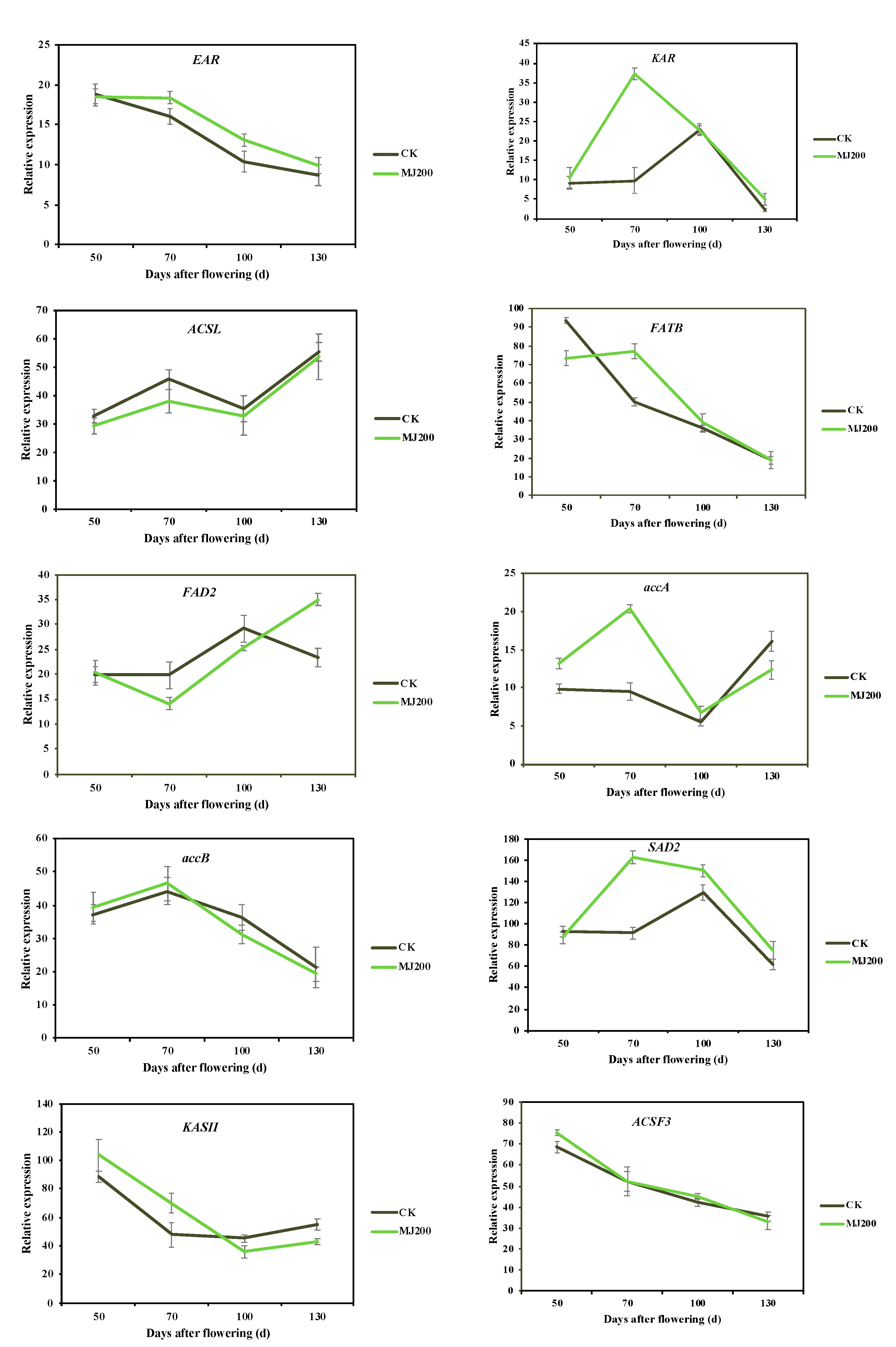
2.3.5. Analysis of qRT-PCR Validation
3. Discussion
3.1. MJ200 Improved Crude Fat Mass Fraction and Fatty Acid Synthesis-Related Enzyme Activity
3.2. MJ200 Altered Fatty Acid Composition and Concentration
3.3. MJ200 Conduced the Expression Level of Fatty Acid Biosynthesis-Related Genes
4. Materials and Methods
4.1. Plant Materials and Experimental Design
4.2. Sample Collection
- 50 DAF, the previous stage before the seeds’ dry matter rapidly increases
- 70 DAF, during the aforementioned steep rise in nutrient concentration
- 100 DAF, the stage that is marked by decreasing oil concentration and increasing starch concentration
- 130 DAF, the final maturation stage
4.3. Crude Fat Extraction
4.4. Targeted Metabolome Analysis
4.5. Enzyme Activity Assay
4.6. Transcriptome Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ohlrogge, J.; Browse, J. Lipid biosynthesis. Plant Cell 1995, 7, 957–970. [Google Scholar]
- Balla, T.; Sengupta, N.; Kim, Y.J. Lipid synthesis and transport are coupled to regulate membrane lipid dynamics in the endoplasmic reticulum. BBA-Mol. Cell Biol. Lipids 2020, 1865, 158461. [Google Scholar] [CrossRef]
- Goffman, F.D.; Alonso, A.P.; Schwender, J.; Shachar-Hill, Y.; Ohlrogge, J.B. Light enables a very high efficiency of carbon storage in developing embryos of rape seed. Plant Physiol. 2005, 138, 2269–2279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, D.K.; Ohlrogge, J.B.; Shachar, H.Y. The role of light in soybean seed filling metabolism. Plant 2009, 58, 220–234. [Google Scholar] [CrossRef]
- Nan, S.Z.; Zhang, L.J.; Hu, X.W.; Miao, X.M.; Han, X.X.; Fu, H. Transcriptomic analysis reveals key genes involved in oil and linoleic acid biosynthesis during Artemisia sphaerocephala seed development. Int. J. Mol. Sci. 2021, 22, 8369. [Google Scholar] [CrossRef]
- Cardoso, A.L.; Neves, S.C.G.; da Silva, M.J. Kinetic study of alcoholysis of the fatty acids catalyzed by tin chloride (II): An alternative catalyst for biodiesel production. Energy Fuels 2009, 23, 1718–1722. [Google Scholar] [CrossRef]
- Wang, X.; Li, W.; Li, N.; Li, J. Omega-3 fatty acids-supplemented parenteral nutrition decreases hyperinflammatory response and attenuates systemic disease sequelae in severe acute pancreatitis: A randomized and controlled study. JPEN-J. Parenter Enter. Nutr. 2008, 32, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Bloedon, L.T.; Szapary, P.O. Flaxseed and cardiovascular risk. Nutr. Rev. 2004, 64, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.W.; Dai, Z.G.; Yang, Z.M.; Tang, Q.; Deng, C.; Xu, Y.; Wang, J.; Chen, J.; Zhao, D.; Zhang, S.; et al. Combined genome-wide association analysis and transcriptome sequencing to identify candidate genes for flax seed fatty acid metabolism. Plant Sci. 2019, 286, 98–107. [Google Scholar] [CrossRef]
- Baud, S.; Guyon, V.; Kronenberger, J.; Wuillème, S.; Miquel, M.; Caboche, M.; Lepiniec, L.; Rocha, C. Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J. 2003, 33, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Smooker, A.M.; Wells, R.; Morgan, C.; Beaudoin, F.; Cho, K.; Fraser, F.; Bancroft, I. The identification and mapping of candidate genes and QTL involved in the fatty acid desaturation pathway in Brassica napus. Theor. Appl. Genet. 2011, 122, 1075–1090. [Google Scholar] [CrossRef]
- Taylor, D.C.; Zhang, Y.; Kumar, A.; Francis, T.; Giblin, E.M.; Barton, D.; Ferrie, J.R.; Laroche, A.; Shah, S.; Zhu, W.; et al. Molecular modification of triacylglycerol accumulation by over-expression of DGAT1 to produce canola with increased seed oil content under field conditions. Botany 2009, 87, 533–543. [Google Scholar] [CrossRef] [Green Version]
- Jako, C.; Kumar, A.; Wei, Y.D.; Zou, J.T.; Barton, D.L.; Giblin, E.M.; Covello, P.S.; Taylor, D.C. Seed-specific overexpression of an Arabidopsis cDNA encoding a diacylglycerel acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001, 126, 861–874. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.W.; Zhang, B.; Hao, Y.J.; Huang, J.; Tian, A.G.; Liao, Y.; Zhang, J.S.; Chen, S.Y. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 2007, 52, 716–729. [Google Scholar] [CrossRef]
- Song, Q.X.; Li, Q.T.; Liu, Y.F.; Zhang, F.X.; Ma, B.; Zhang, W.K.; Man, W.Q.; Du, W.G.; Wang, G.D.; Chen, S.Y.; et al. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J. Exp. Bot. 2013, 64, 4329–4341. [Google Scholar] [CrossRef]
- Liu, J.; Hua, W.; Zhan, G.M.; Wei, F.; Wang, X.F.; Liu, G.H.; Wang, H.Z. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef]
- Pouvreau, B.; Baud, S.; Vernoud, V.; Morin, V.; Cyrille, P.; Gendrot, G.; Pichon, J.P.; Rouster, J.; Paul, W.; Rogowsky, P.M. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011, 156, 674–686. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Allen, W.B.; Zheng, P.Z.; Li, C.J.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.G.; Tang, M.; Sun, D.J.; Zhu, H.G.; Wei, J.; Chen, F.; Tang, L. Influence of climatic factors and soil types on seed weight and oil content of Jatropha curcas in Guangxi, China. Proc. Environ. Sci. 2012, 12, 439–444. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Z.; Chen, Y.; Shockey, J.; Han, X.J.; Wang, Y.D. Proteomic analysis of tung tree (Vernicia fordii) oilseeds during the developmental stages. Molecules 2016, 21, 1486. [Google Scholar] [CrossRef]
- Fu, X.N.; Su, J.L.; Hou, L.; Zhang, K.; Li, H.W.; Liu, X.Y.; Jia, C.X.Z.; Xu, J. Physicochemical and thermal characteristics of Moringa oleifera seed oil. Adv. Compos. Hybrid Mater. 2021, 4, 685–695. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Luo, Y.; Wang, X.J.; Yu, F.Y. Fruit spray of 24-epibrassinolide and fruit shade alter pericarp photosynthesis activity and seed lipid accumulation in Styrax tonkinensis. J. Plant Growth Regul. 2018, 37, 1066–1084. [Google Scholar] [CrossRef]
- Chen, C.; Wang, X.J.; Cao, Y.Y.; Wu, Q.K.; Yu, F.Y. Time course morphological and histological changes in differentiating floral buds of Styrax tonkinensis. Int. J. Agric. Biol. 2019, 22, 1491–1496. [Google Scholar]
- Xu, L.P.; Yu, F.Y. Corolla structure and fragrance components in Styrax tonkinensis. Trees 2015, 29, 1127–1134. [Google Scholar] [CrossRef]
- Chen, C.; Cao, Y.Y.; Wang, X.J.; Wu, Q.K.; Yu, F.Y. Do stored reserves and endogenous hormones in overwintering twigs determine flower bud differentiation of summer blooming plant-Styrax tonkinensis. Int. J. Agric. Biol. 2019, 22, 815–820. [Google Scholar]
- Xie, Q.; Ma, R.; Guo, X.Q.; Chen, H.; Wang, J. Benzoinum from Styrax tonkinensis (Pierre) Craib ex Hart exerts a NVU protective effect by inhibiting cell apoptosis in cerebral ischaemia rats. J. Ethnopharmacol. 2021, 265, 113355. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Wang, X.J.; Luo, Y.; Yu, F.Y. Carbon competition between fatty acids and starch during benzoin seeds maturation slows oil accumulation speed. Trees 2017, 31, 1025–1039. [Google Scholar] [CrossRef]
- Wu, Q.K.; Cao, Y.Y.; Chen, C.; Gao, Z.Z.; Yu, F.Y.; Guy, R.D. Transcriptome analysis of metabolic pathways associated with oil accumulation in developing seed kernels of Styrax tonkinensis, a woody biodiesel species. BMC Plant Biol. 2020, 20, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zafari, M.; Ebadi, A.; Sedghi, M.; Jahanbakhsh, S. Alleviating effect of 24-epibrassinolide on seed oil content and fatty acid composition under drought stress in safflower. J. Food Compos. Anal. 2020, 92, 103544. [Google Scholar] [CrossRef]
- Ghassemi-Golezani, K.; Farhangi-Abriz, S. Changes in oil accumulation and fatty acid composition of soybean seeds under salt stress in response to salicylic acid and jasmonic acid. Russ. J. Plant Physiol. 2018, 65, 229–236. [Google Scholar] [CrossRef]
- Demole, E.; Lederer, E.; Mercier, D. Isolement et determination de la structure du jasmonate de methyle, constituant odorant caracteristique de l’essence de jasmin. Helv. Chim. Acta 1962, 45, 675–685. [Google Scholar] [CrossRef]
- D’Onofrio, C.; Matarese, F.; Cuzzola, A. Effect of methyl jasmonate on the aroma of Sangiovese grapes and wines. Food Chem. 2018, 242, 352–361. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Ni, M.; Yu, F.Y. Methyl jasmonate application and flowering stage affect scent emission of Styrax japonicus. Flavour Frag. J. 2021, 36, 497–504. [Google Scholar] [CrossRef]
- Lei, G.J.; Sun, L.; Sun, Y.; Zhu, X.F.; Li, X.; Zheng, S.J. Jasmonic acid alleviates cadmium toxicity in Arabidopsis via suppression of cadmium uptake and translocation. J. Integr. Plant Biol. 2020, 62, 218–227. [Google Scholar] [CrossRef]
- Liu, H.R.; Meng, F.L.; Miao, H.Y.; Chen, S.S.; Yin, T.T.; Hu, S.S.; Shao, Z.Y.; Liu, Y.Y.; Gao, L.X.; Zhu, C.Q.; et al. Effects of postharvest methyl jasmonate treatment on main health promoting components and volatile organic compounds in cherry tomato fruits. Food Chem. 2018, 263, 194–200. [Google Scholar] [CrossRef]
- Mohamed, H.I.; Latif, H.H. Improvement of drought tolerance of soybean plants by using methyl jasmonate. Physiol. Mol. Biol. Plants 2017, 23, 545–556. [Google Scholar] [CrossRef]
- Zalewski, K.; Czaplicki, S.; Rafalowski, R.; Stryiński, R.; Okorski, A.; Nitkiewicz, B. The effect of exogenous methyl jasmonate on the fatty acid composition of germinating triticale kernels (×Triticosecale Wittmack, cv. Ugo). Curr. Plant Biol. 2021, 28, 100225. [Google Scholar] [CrossRef]
- Xiong, B.; Zhang, Z.; Dong, S. Biodiesel from Lindera glauca oil, a potential non-food feedstock in Southern China. Ind. Crop. Prod. 2018, 122, 107–113. [Google Scholar] [CrossRef]
- Dai, G.; Yang, J.; Lu, S.; Huang, C.H.; Jin, J.; Jiang, P.; Yan, P.B. The potential impact of invasive woody oil plants on protected areas in china under future climate conditions. Sci. Rep. 2018, 8, 1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q. Effects of Exogenous Gibberellin and Methyl Jasmonate on Fatty Acid Metabolism in Camelina sativa. Master’s Thesis, Tianjin University of Science and Technology, Tianjin, China, 2018. (In Chinese). [Google Scholar]
- Flores, G.; Luisa, R.D.C.M. Enhancement of nutritionally significant constituents of black currant seeds by chemical elicitor application. Food Chem. 2016, 194, 1260–1265. [Google Scholar] [CrossRef]
- Ramos, M.J.; Fernández, C.M.; Casas, A.; Rodrguez, L.; Prez, N. Influence of fatty acid composition of raw materials on biodiesel properties. Bioresour. Technol. 2009, 100, 261–268. [Google Scholar] [CrossRef]
- Reddy, A.N.R.; Saleh, A.A.; Islam, M.S.; Hamdan, S.; Rahman, M.R.; Masjuki, H.H. Experimental evaluation of fatty acid composition influence on Jatropha biodiesel physicochemical properties. J. Renew. Sustain. Energy 2018, 10, 013103. [Google Scholar] [CrossRef]
- Knothe, G.; Razon, L.F.; Castro, M.E.G. Fatty acids, triterpenes and cycloalkanes in ficus seed oils. Plant Physiol. Biochem. 2019, 135, 127–131. [Google Scholar] [CrossRef]
- Lwin, M.M.; Jin, Y.; Yang, X.P.; Shen, H.; Qi, C.; Wang, Y.Z.; Zin, P.P.; Tuang, Z.K.; Lu, S.Y.; Yang, W.N. Nutritional evaluation, fatty acid analysis and anticancer properties investigation of seeds from kan-zaw, a potential diet oil and medicinal plant. Oil Crop Sci. 2021, 6, 122–127. [Google Scholar] [CrossRef]
- Zhou, X.J.; Wang, Y.Y.; Xu, Y.N.; Yan, R.S.; Zhao, P.; Liu, W.Z. De novo characterization of flower bud transcriptomes and the development of EST-SSR markers for the endangered tree Tapiscia sinensis. Int. J. Mol. Sci. 2015, 16, 12855–12870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norton, G.J.; Lou-Hing, D.E.; Meharg, A.A.; Price, A.H. Rice-arsenate interactions in hydroponics: Whole genome transcriptional analysis. J. Exp. Bot. 2008, 59, 2267–2276. [Google Scholar] [CrossRef] [Green Version]
- Li, C.F.; Zhu, Y.; Yu, Y.; Zhao, Q.Y.; Wang, S.J.; Wang, X.C.; Yao, M.Z.; Luo, D.; Li, X.; Chen, L.; et al. Global transcriptome and gene regulation network for secondary metabolism biosynthesis of tea plant (Camellia sinensis). BMC Genom. 2015, 16, 560. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Yin, C.M.; Xiang, L.; Jiang, W.T.; Xu, S.Z.; Mao, Z.Q. Transcription strategies related to photosynthesis and nitrogen metabolism of wheat in response to nitrogen deficiency. BMC Plant Biol. 2020, 20, 448. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.Z.; Wang, L.; Fu, J.M.; Wu, Y.; Du, H.Y.; Tan, X.F.; Zou, F.; Li, F.D. Transcriptome sequencing discovers genes related to fatty acid biosynthesis in the seeds of Eucommia ulmoides. Genes Genom. 2016, 38, 275–283. [Google Scholar] [CrossRef]
- Coleman, R.; Lewin, T.M.; Muoio, D.M. Physiological and nutritional regulation of enzymes of triacylglycerol synthesis. Annu. Rev. Nutr. 2000, 20, 77–103. [Google Scholar] [CrossRef]
- Wang, Y.F.; Zhao, J.; Chen, H.; Zhang, Q.; Zheng, Y.S.; Li, D.D. Molecular cloning and characterization of long-chain acyl-CoA synthetase 9 from the mesocarp of African oil palm (Elaeis guineensis Jacq.). Sci. Hortic. 2021, 276, 109751. [Google Scholar] [CrossRef]
- Zhang, C.L.; Zhang, Y.L.; Hu, X.; Xiao, X.; Wang, G.L.; You, C.X.; Li, Y.Y.; Hao, Y.J. An apple long-chain acyl-CoA synthetase, MdLACS4, induced early flowering and enhances abiotic stress resistance in Arabidopsis. Plant Sci. 2020, 297, 110529. [Google Scholar] [CrossRef]
- Dörmann, P.; Kridl, J.C.; Ohlrogge, J.B. Cloning and expression in Escherichia coli of a cDNA coding for the oleoyl-acyl carrier protein thioesterase from corianderer (Coriandrum sativum L.). BBA-Lipids Lipid Metab. 1994, 1212, 134–136. [Google Scholar] [CrossRef]
- Mekhedov, S.; Martínez, O.; Ohlrogge, J. Toward a functional catalog of the plant genome. A survey of gene for lipid biosynthesis. Plant Physiol. 2000, 122, 389–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Thuillier, I.; Venegas-Calerón, M.; Moreno-Pérez, A.J.; Salas, J.J.; Garcés, R.; Wettstein-Knowles, P.; Martínez-Force, E. Sunflower (Helianthus annuus) fatty acid synthase complex: β-Ketoacyl-[acyl carrier protein] reductase genes. Plant Physiol. Biochem. 2021, 166, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, Y.; Takaiwa, F.; Shojik, K. Structure and expression of two seed-specific cDNA encoding stearoyl-acyl carrier protein desaturase from sesame, Sesamum indicum L. Plant Cell Physiol. 1996, 37, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.J.; Xu, G.; Du, H.W.; Hu, J.Y.; Liu, Z.J.; Li, H.; Li, J.S.; Yang, X.H. Natural variations in stearoyl-acp desaturase genes affect the conversion of stearic to oleic acid in maize kernel. Theor. Appl. Genet. 2017, 130, 151–161. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Mendes, A.; Kelly, A.A.; Van, E.H.; Shaw, E.; Powers, S.J.; Kurup, S.; Eastmond, P.J. bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase 3. Plant Cell 2013, 25, 3104–3116. [Google Scholar] [CrossRef] [Green Version]
- Peng, S.F.; Lu, J.; Zhang, Z.; Ma, L.; Liu, C.X.; Chen, Y.Z. Global transcriptome and correlation analysis reveal cultivar specific molecular signatures associated with fruit development and fatty acid determination in Camellia oleifera Abel. Int. J. Genom. 2020, 2020, 6162802. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, X. Comparative transcriptomic analysis identifies genes responsible for fruit count and oil yield in the oil tea plant Camellia chekiangoleosa. Sci. Rep. 2018, 8, 6637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aznar-Moreno, J.A.; Durrett, T.P. Simultaneous targeting of multiple gene homeologs to alter seed oil production in Camelina sativa. Plant Cell Physiol. 2017, 58, 1260–1267. [Google Scholar] [CrossRef]
- Yao, Q.Y.; Huang, H.; Tong, Y.; Xia, E.H.; Gao, L.Z. Transcriptome analysis identifies candidate genes related to triacylglycerol and pigment biosynthesis and photoperiodic flowering in the ornamental and oil-producing plant, Camellia reticulata (Theaceae). Front. Plant Sci. 2016, 7, 163. [Google Scholar] [CrossRef] [Green Version]
- Brusselmans, K.; Vrolix, R.; Verhoeven, G.; Swinnen, J.V. Induction of cancer cell apoptosis by flavonoids is associated with their ability to inhibit fatty acid synthase activity. J. Biol. Chem. 2005, 280, 5636–5645. [Google Scholar] [CrossRef] [Green Version]
- Bays, N.; Hill, A.D.; Kariv, I. A simplified scintillation proximity assay for fatty acid synthase activity: Development and comparison with other FAS activity assays. J. Biomol. Screen. 2009, 14, 636–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Mao, X.Z.; Huang, J.J.; Ding, Y.; Wu, J.M.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L.P. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| FA Composition (%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| 50 DAF | 70 DAF | 100 DAF | 130 DAF | |||||
| CK | MJ200 | CK | MJ200 | CK | MJ200 | CK | MJ200 | |
| C14:0 | 0.17 ± 0.06 | 0.14 ± 0.05 | 0.08 ± 0.04 | 0.07 ± 0.02 | 0.08 ± 0.05 | 0.09 ± 0.04 | 0.08 ± 0.06 | 0.08 ± 0.03 |
| C15:0 | 0.10 ± 0.02 | 0.05 ± 0.01 | 0.04 ± 0.02 | 0.04 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.02 | 0.02 ± 0.01 | 0.02 ± 0.01 |
| C16:0 | 25.53 ± 2.02 | 22.55 ± 3.11 | 17.29 ± 3.15 | 16.18 ± 1.33 | 11.73 ± 0.94 | 11.93 ± 2.08 | 10.41 ± 2.26 | 10.26 ± 1.77 |
| C16:1 | 0.24 ± 0.02 | 0.24 ± 0.02 | 0.14 ± 0.04 | 0.13 ± 0.02 | 0.18 ± 0.01 | 0.15 ± 0.06 | 0.13 ± 0.08 | 0.14 ± 0.05 |
| C17:0 | 0.64 ± 0.03 | 0.67 ± 0.04 | 0.79 ± 0.32 | 0.73 ± 0.07 | 0.43 ± 0.16 | 0.45 ± 0.12 | 0.37 ± 0.11 | 0.35 ± 0.03 |
| C17:1 | 0.41 ± 0.03 | 0.33 ± 0.01 | 0.37 ± 0.18 | 0.40 ± 0.03 | 0.31 ± 0.10 | 0.28 ± 0.02 | 0.24 ± 0.06 | 0.22 ± 0.04 |
| C18:0 | 5.95 ± 0.78 | 6.39 ± 0.48 | 3.17 ± 0.74 | 2.80 ± 0.21 | 2.58 ± 1.27 | 3.28 ± 1.93 | 3.79 ± 1.57 | 3.71 ± 0.55 |
| C18:1 | 13.80 ± 1.30 | 13.39 ± 1.12 | 29.36 ± 2.00 | 29.93 ± 3.29 | 23.33 ± 2.65 | 22.28 ± 2.44 | 11.65 ± 2.06 | 11.71 ± 1.33 |
| C18:2 | 24.86 ± 1.70 | 24.74 ± 1.60 | 53.86 ± 5.04 | 55.09 ± 5.21 | 53.43 ± 6.00 | 52.14 ± 5.60 | 44.25 ± 7.00 | 44.43 ± 4.98 |
| C18:3 | 18.63 ± 1.85 | 20.16 ± 1.31 | 8.52 ± 1.50 | 7.81 ± 0.61 | 6.89 ± 1.17 | 7.81 ± 1.88 | 9.99 ± 2.62 | 9.65 ± 1.57 |
| C20:0 | 2.30 ± 0.22 | 2.67 ± 0.20 | 0.85 ± 0.16 | 0.90 ± 0.05 | 0.24 ± 0.14 | 0.41 ± 0.15 | 0.38 ± 0.13 | 0.38 ± 0.11 |
| C20:1 | 1.45 ± 0.25 | 2.14 ± 0.28 | 0.61 ± 0.24 | 0.63 ± 0.03 | 0.24 ± 0.08 | 0.30 ± 0.05 | 0.37 ± 0.18 | 0.33 ± 0.10 |
| C20:2 | 0.41 ± 0.01 | 0.71 ± 0.03 | 0.22 ± 0.01 | 0.50 ± 0.01 | 0.06 ± 0.03 | 0.09 ± 0.07 | 0.05 ± 0.01 | 0.05 ± 0.02 |
| C20:3 | 0.37 ± 0.01 | 0.48 ± 0.01 | 0.20 ± 0.01 | 0.30 ± 0.01 | 0.03 ± 0.01 | 0.04 ± 0.01 | 0.02 ± 0.02 | 0.02 ± 0.01 |
| C21:0 | 0.61 ± 0.02 | 0.67 ± 0.03 | 0.30 ± 0.04 | 0.33 ± 0.01 | 0.04 ± 0.02 | 0.06 ± 0.03 | 0.04 ± 0.01 | 0.03 ± 0.01 |
| C22:0 | 1.72 ± 0.22 | 1.57 ± 0.08 | 0.65 ± 0.11 | 0.70 ± 0.02 | 0.16 ± 0.01 | 0.25 ± 0.13 | 0.20 ± 0.03 | 0.18 ± 0.03 |
| C22:1 | 0.17 ± 0.06 | 0.10 ± 0.04 | 0.06 ± 0.04 | 0.07 ± 0.04 | 0.05 ± 0.03 | 0.04 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 |
| C23:0 | 0.54 ± 0.02 | 0.67 ± 0.02 | 0.28 ± 0.03 | 0.40 ± 0.01 | 0.06 ± 0.03 | 0.08 ± 0.06 | 0.06 ± 0.01 | 0.05 ± 0.01 |
| C24:0 | 1.62 ± 0.16 | 1.67 ± 0.07 | 0.71 ± 0.11 | 0.87 ± 0.04 | 0.14 ± 0.10 | 0.25 ± 0.15 | 0.17 ± 0.08 | 0.14 ± 0.10 |
| C24:1 | 0.47 ± 0.01 | 0.67 ± 0.01 | 0.28 ± 0.02 | 0.43 ± 0.01 | 0.06 ± 0.04 | 0.06 ± 0.01 | 0.04 ± 0.02 | 0.02 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Chen, H.; Han, C.; Liu, Z.; Ni, M.; Wu, Q.; Yu, F. Transcriptomic and Metabolomic Analysis Unravels the Molecular Regulatory Mechanism of Fatty Acid Biosynthesis in Styrax tonkinensis Seeds under Methyl Jasmonate Treatment. Int. J. Mol. Sci. 2022, 23, 6190. https://doi.org/10.3390/ijms23116190
Chen C, Chen H, Han C, Liu Z, Ni M, Wu Q, Yu F. Transcriptomic and Metabolomic Analysis Unravels the Molecular Regulatory Mechanism of Fatty Acid Biosynthesis in Styrax tonkinensis Seeds under Methyl Jasmonate Treatment. International Journal of Molecular Sciences. 2022; 23(11):6190. https://doi.org/10.3390/ijms23116190
Chicago/Turabian StyleChen, Chen, Hong Chen, Chao Han, Zemao Liu, Ming Ni, Qikui Wu, and Fangyuan Yu. 2022. "Transcriptomic and Metabolomic Analysis Unravels the Molecular Regulatory Mechanism of Fatty Acid Biosynthesis in Styrax tonkinensis Seeds under Methyl Jasmonate Treatment" International Journal of Molecular Sciences 23, no. 11: 6190. https://doi.org/10.3390/ijms23116190
APA StyleChen, C., Chen, H., Han, C., Liu, Z., Ni, M., Wu, Q., & Yu, F. (2022). Transcriptomic and Metabolomic Analysis Unravels the Molecular Regulatory Mechanism of Fatty Acid Biosynthesis in Styrax tonkinensis Seeds under Methyl Jasmonate Treatment. International Journal of Molecular Sciences, 23(11), 6190. https://doi.org/10.3390/ijms23116190






