Nrf2 Transcriptional Activity Governs Intestine Development
Abstract
:1. Introduction
2. Results
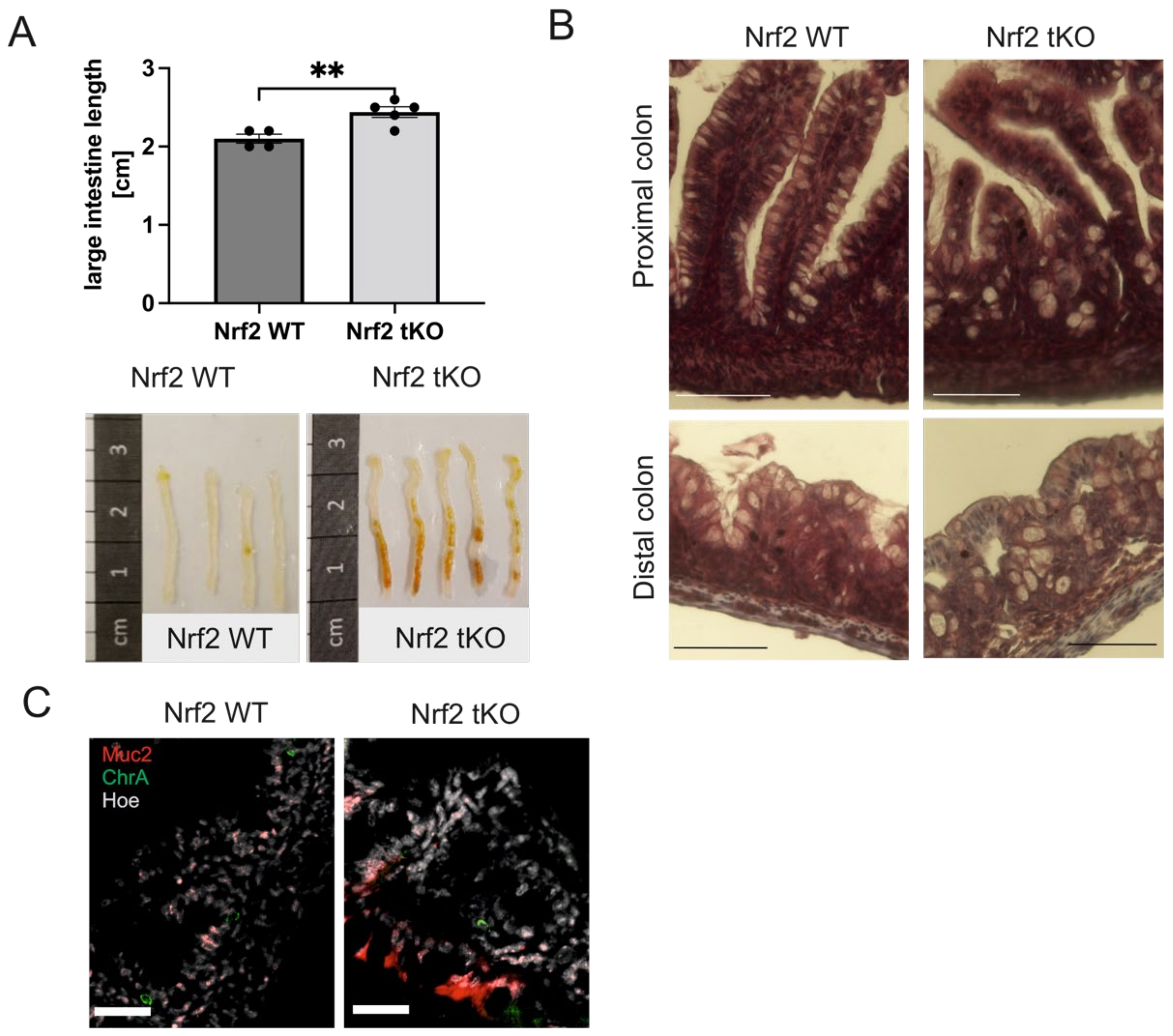
2.1. Nrf2 tKO Pups Have a Longer Colon with Coexistent Microscopic Alterations
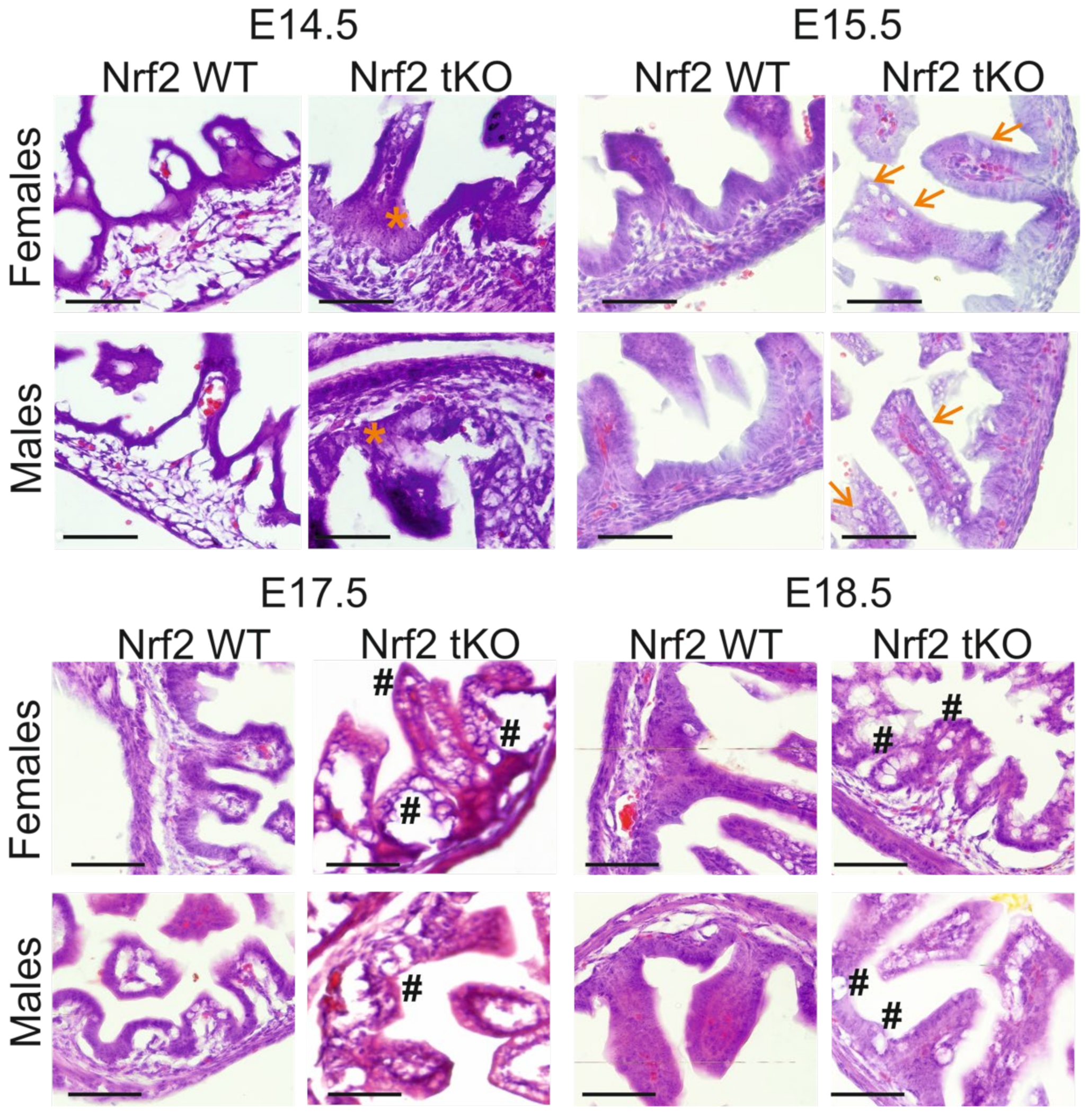
2.2. tKO Embryos Have Significantly Altered Crypt Distribution and Enlargement of Goblet Cells as Early as at Day 14.5 of Embryonic Development
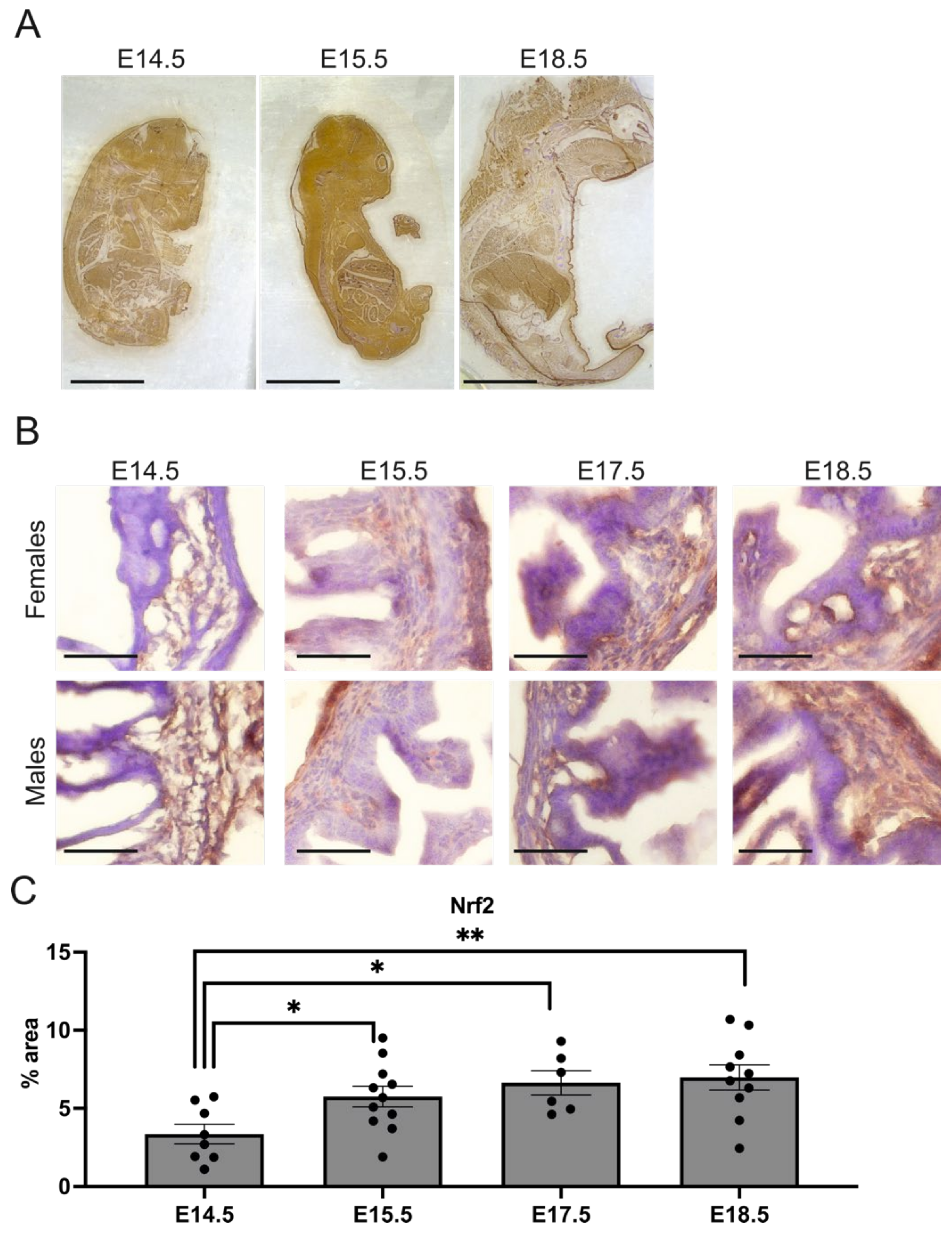
2.3. Nrf2 Level Is Increased in the Hindgut during Gestation
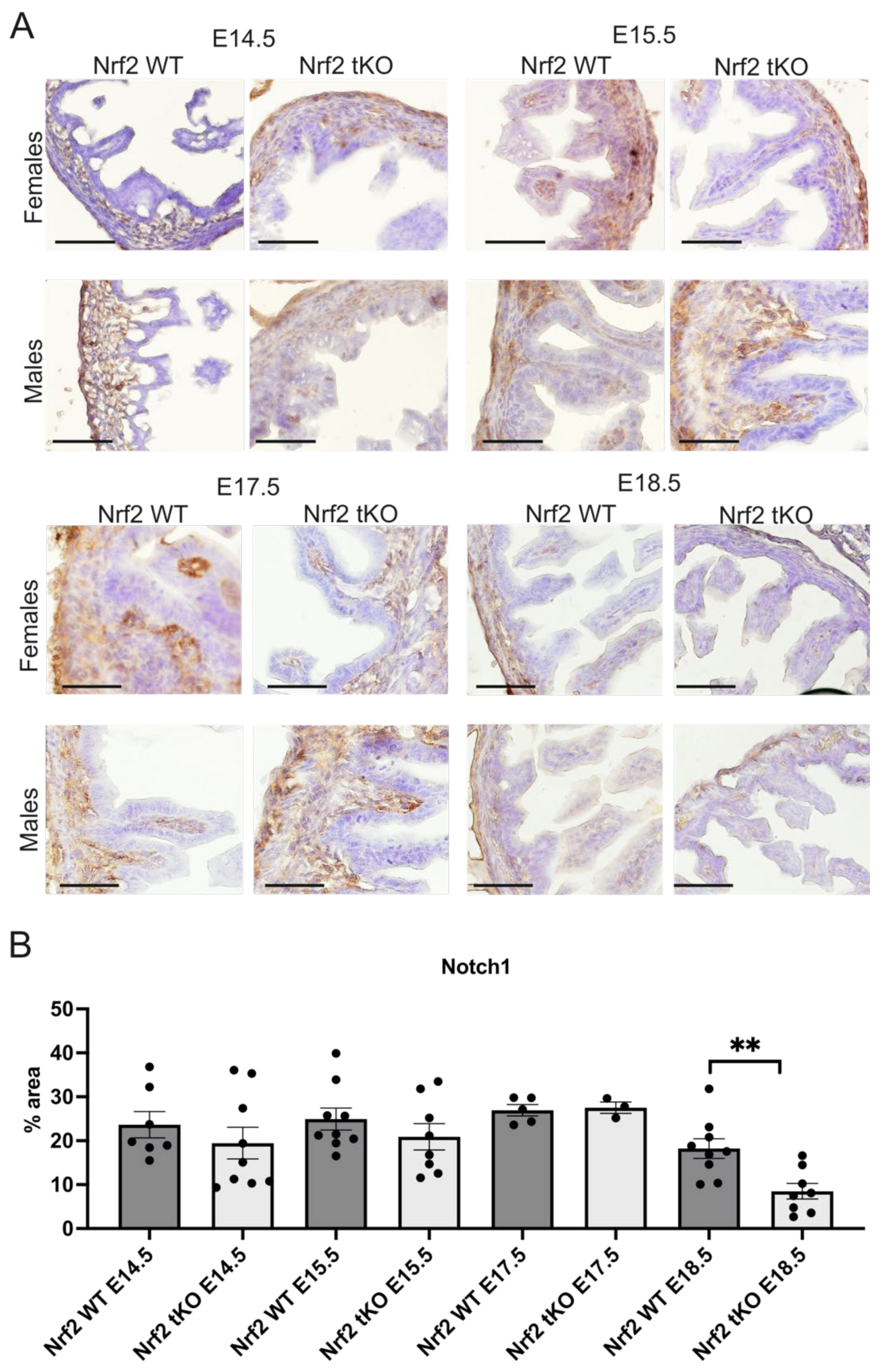
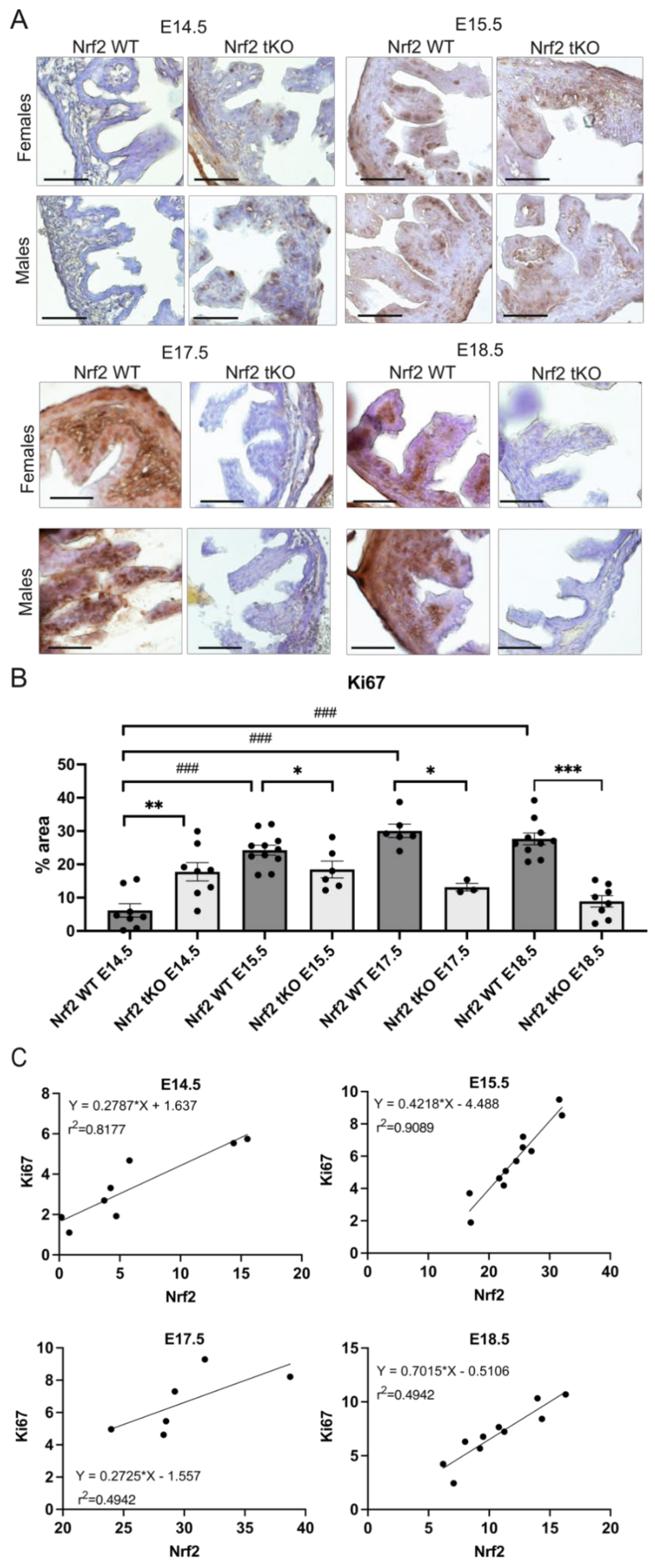
2.4. Nrf2 Transcriptional Activity May Influence the Proliferation and Differentiation at Different Stages of Embryonic Development
3. Discussion
4. Material and Methods
4.1. Animals
4.2. Embryo Preparation
4.3. Genotyping PCR
4.4. Histological and Immunofluorescent Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kikut, J.; Konecka, N.; Ziętek, M.; Szczuko, M. Inflammatory Bowel Disease Etiology: Current Knowledge. Pteridines 2018, 29, 206–214. [Google Scholar] [CrossRef]
- Tavakoli, P.; Vollmer-Conna, U.; Hadzi-Pavlovic, D.; Grimm, M.C. A Review of Inflammatory Bowel Disease: A Model of Microbial, Immune and Neuropsychological Integration. Public Health Rev. 2021, 42, 1603990. [Google Scholar] [CrossRef] [PubMed]
- Fazilaty, H.; Brügger, M.D.; Valenta, T.; Szczerba, B.M.; Berkova, L.; Doumpas, N.; Hausmann, G.; Scharl, M.; Basler, K. Tracing Colonic Embryonic Transcriptional Profiles and Their Reactivation upon Intestinal Damage. Cell Rep. 2021, 36, 109484. [Google Scholar] [CrossRef] [PubMed]
- Elmentaite, R.; Ross, A.D.B.; Roberts, K.; James, K.R.; Ortmann, D.; Gomes, T.; Nayak, K.; Tuck, L.; Pritchard, S.; Bayraktar, O.A.; et al. Single-Cell Sequencing of Developing Human Gut Reveals Transcriptional Links to Childhood Crohn’s Disease. Dev. Cell 2020, 55, 771–783.e5. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.M.; Hill, D.R.; Aurora, M.; Spence, J.R. Morphogenesis and Maturation of the Embryonic and Postnatal Intestine. Semin. Cell Dev. Biol. 2017, 66, 81–93. [Google Scholar] [CrossRef]
- Spence, J.R.; Lauf, R.; Shroyer, N.F. Vertebrate Intestinal Endoderm Development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2011, 240, 501–520. [Google Scholar] [CrossRef] [Green Version]
- Kopacz, A.; Werner, E.; Kloska, D.; Hajduk, K.; Fichna, J.; Jozkowicz, A.; Piechota-Polanczyk, A. Nrf2 Transcriptional Activity in the Mouse Affects the Physiological Response to Tribromoethanol. Biomed. Pharmacother. 2020, 128, 110317. [Google Scholar] [CrossRef]
- Kopacz, A.; Kloska, D.; Forman, H.J.; Jozkowicz, A.; Grochot-Przeczek, A. Beyond Repression of Nrf2: An Update on Keap1. Free Radic. Biol. Med. 2020, 157, 63–74. [Google Scholar] [CrossRef]
- Piechota-Polanczyk, A.; Fichna, J. Review Article: The Role of Oxidative Stress in Pathogenesis and Treatment of Inflammatory Bowel Diseases. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2014, 387, 605–620. [Google Scholar] [CrossRef] [Green Version]
- Piotrowska, M.; Swierczynski, M.; Fichna, J.; Piechota-Polanczyk, A. The Nrf2 in the Pathophysiology of the Intestine: Molecular Mechanisms and Therapeutic Implications for Inflammatory Bowel Diseases. Pharmacol. Res. 2021, 163, 105243. [Google Scholar] [CrossRef]
- Cuadrado, A.; Manda, G.; Hassan, A.; Alcaraz, M.J.; Barbas, C.; Daiber, A.; Ghezzi, P.; León, R.; López, M.G.; Oliva, B.; et al. Transcription Factor NRF2 as a Therapeutic Target for Chronic Diseases: A Systems Medicine Approach. Pharmacol. Rev. 2018, 70, 348–383. [Google Scholar] [CrossRef] [Green Version]
- Arisawa, T.; Tahara, T.; Shibata, T.; Nagasaka, M.; Nakamura, M.; Kamiya, Y.; Fujita, H.; Yoshioka, D.; Okubo, M.; Sakata, M.; et al. Nrf2 Gene Promoter Polymorphism Is Associated with Ulcerative Colitis in a Japanese Population. Hepato-gastroenterology 2008, 55, 394–397. [Google Scholar]
- Chan, K.; Lu, R.; Chang, J.C.; Kan, Y.W. NRF2, a Member of the NFE2 Family of Transcription Factors, Is Not Essential for Murine Erythropoiesis, Growth, and Development. Proc. Natl. Acad. Sci. USA 1996, 93, 13943–13948. [Google Scholar] [CrossRef] [Green Version]
- Wakabayashi, N.; Chartoumpekis, D.V.; Kensler, T.W. Crosstalk between Nrf2 and Notch Signaling. Free Radic. Biol. Med. 2015, 88, 158–167. [Google Scholar] [CrossRef] [Green Version]
- Yagishita, Y.; McCallum, M.L.; Kensler, T.W.; Wakabayashi, N. Constitutive Activation of Nrf2 in Mice Expands Enterogenesis in Small Intestine Through Negative Regulation of Math1. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 503–524. [Google Scholar] [CrossRef]
- Kloska, D.; Kopacz, A.; Cysewski, D.; Aepfelbacher, M.; Dulak, J.; Jozkowicz, A.; Grochot-Przeczek, A. Nrf2 Sequesters Keap1 Preventing Podosome Disassembly: A Quintessential Duet Moonlights in Endothelium. Antioxid. Redox Signal. 2019, 30, 1709–1730. [Google Scholar] [CrossRef]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/Small Maf Heterodimer Mediates the Induction of Phase II Detoxifying Enzyme Genes through Antioxidant Response Elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef]
- Kweider, N.; Huppertz, B.; Rath, W.; Lambertz, J.; Caspers, R.; ElMoursi, M.; Pecks, U.; Kadyrov, M.; Fragoulis, A.; Pufe, T.; et al. The Effects of Nrf2 Deletion on Placental Morphology and Exchange Capacity in the Mouse. J. Matern.-Fetal Neonatal Med. 2017, 30, 2068–2073. [Google Scholar] [CrossRef]
- Wakabayashi, N.; Itoh, K.; Wakabayashi, J.; Motohashi, H.; Noda, S.; Takahashi, S.; Imakado, S.; Kotsuji, T.; Otsuka, F.; Roop, D.R.; et al. Keap1-Null Mutation Leads to Postnatal Lethality Due to Constitutive Nrf2 Activation. Nat. Genet. 2003, 35, 238–245. [Google Scholar] [CrossRef]
- Yang, S.; Yu, M. Role of Goblet Cells in Intestinal Barrier and Mucosal Immunity. J. Inflamm. Res. 2021, 14, 3171–3183. [Google Scholar] [CrossRef]
- Johansson, M.E.V.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The Inner of the Two Muc2 Mucin-Dependent Mucus Layers in Colon Is Devoid of Bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuckin, M.A.; Lindén, S.K.; Sutton, P.; Florin, T.H. Mucin Dynamics and Enteric Pathogens. Nat. Rev. Microbiol. 2011, 9, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.; Min, M.; Yang, C.; Tian, C.; Gookin, S.; Carter, D.; Spencer, S.L. Ki67 Is a Graded Rather than a Binary Marker of Proliferation versus Quiescence. Cell Rep. 2018, 24, 1105–1112.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Zhang, Y.; Wei, Y.; Liu, G.; Liu, Y.; Gao, Q.; Zou, L.; Zeng, W.; Zhang, N. Activation of AKT Pathway by Nrf2/PDGFA Feedback Loop Contributes to HCC Progression. Oncotarget 2016, 7, 65389–65402. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Huang, G.; Cheng, H. Transcription Factor Nrf2 Induces the Up-Regulation of LncRNA TUG1 to Promote Progression and Adriamycin Resistance in Urothelial Carcinoma of the Bladder. Cancer Manag. Res. 2019, 11, 6079–6090. [Google Scholar] [CrossRef] [Green Version]
- Bellanti, F.; di Bello, G.; Iannelli, G.; Pannone, G.; Pedicillo, M.C.; Boulter, L.; Lu, W.-Y.; Tamborra, R.; Villani, R.; Vendemiale, G.; et al. Inhibition of Nuclear Factor (Erythroid-Derived 2)-like 2 Promotes Hepatic Progenitor Cell Activation and Differentiation. NPJ Regen. Med. 2021, 6, 28. [Google Scholar] [CrossRef]
- Shirasaki, K.; Taguchi, K.; Unno, M.; Motohashi, H.; Yamamoto, M. NF-E2-Related Factor 2 Promotes Compensatory Liver Hypertrophy after Portal Vein Branch Ligation in Mice. Hepatology 2014, 59, 2371–2382. [Google Scholar] [CrossRef]
- Al-Sawaf, O.; Fragoulis, A.; Rosen, C.; Keimes, N.; Liehn, E.A.; Hölzle, F.; Kan, Y.W.; Pufe, T.; Sönmez, T.T.; Wruck, C.J. Nrf2 Augments Skeletal Muscle Regeneration after Ischaemia–Reperfusion Injury. J. Pathol. 2014, 234, 538–547. [Google Scholar] [CrossRef]
- Kärkkäinen, V.; Pomeshchik, Y.; Savchenko, E.; Dhungana, H.; Kurronen, A.; Lehtonen, S.; Naumenko, N.; Tavi, P.; Levonen, A.-L.; Yamamoto, M.; et al. Nrf2 Regulates Neurogenesis and Protects Neural Progenitor Cells Against Aβ Toxicity. Stem Cells 2014, 32, 1904–1916. [Google Scholar] [CrossRef]
- Yang, W.; Sun, Z.; Yang, B.; Wang, Q. Nrf2-Knockout Protects from Intestinal Injuries in C57BL/6J Mice Following Abdominal Irradiation with γ Rays. Int. J. Mol. Sci. 2017, 18, 1656. [Google Scholar] [CrossRef]
- Goeden, N.; Bonnin, A. Ex Vivo Perfusion of Mid-to-Late-Gestation Mouse Placenta for Maternal-Fetal Interaction Studies during Pregnancy. Nat. Protoc. 2013, 8, 66–74. [Google Scholar] [CrossRef] [Green Version]
- Tunster, S.J. Genetic Sex Determination of Mice by Simplex PCR. Biol. Sex Differ. 2017, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Arganda-Carreras, I.; Kaynig, V.; Rueden, C.; Eliceiri, K.W.; Schindelin, J.; Cardona, A.; Sebastian Seung, H. Trainable Weka Segmentation: A Machine Learning Tool for Microscopy Pixel Classification. Bioinformatics 2017, 33, 2424–2426. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopacz, A.; Kloska, D.; Klimczyk, D.; Kopec, M.; Jozkowicz, A.; Piechota-Polanczyk, A. Nrf2 Transcriptional Activity Governs Intestine Development. Int. J. Mol. Sci. 2022, 23, 6175. https://doi.org/10.3390/ijms23116175
Kopacz A, Kloska D, Klimczyk D, Kopec M, Jozkowicz A, Piechota-Polanczyk A. Nrf2 Transcriptional Activity Governs Intestine Development. International Journal of Molecular Sciences. 2022; 23(11):6175. https://doi.org/10.3390/ijms23116175
Chicago/Turabian StyleKopacz, Aleksandra, Damian Kloska, Dominika Klimczyk, Magdalena Kopec, Alicja Jozkowicz, and Aleksandra Piechota-Polanczyk. 2022. "Nrf2 Transcriptional Activity Governs Intestine Development" International Journal of Molecular Sciences 23, no. 11: 6175. https://doi.org/10.3390/ijms23116175
APA StyleKopacz, A., Kloska, D., Klimczyk, D., Kopec, M., Jozkowicz, A., & Piechota-Polanczyk, A. (2022). Nrf2 Transcriptional Activity Governs Intestine Development. International Journal of Molecular Sciences, 23(11), 6175. https://doi.org/10.3390/ijms23116175





