Sulforaphane Increase Mitochondrial Biogenesis-Related Gene Expression in the Hippocampus and Suppresses Age-Related Cognitive Decline in Mice
Abstract
:1. Introduction
2. Results
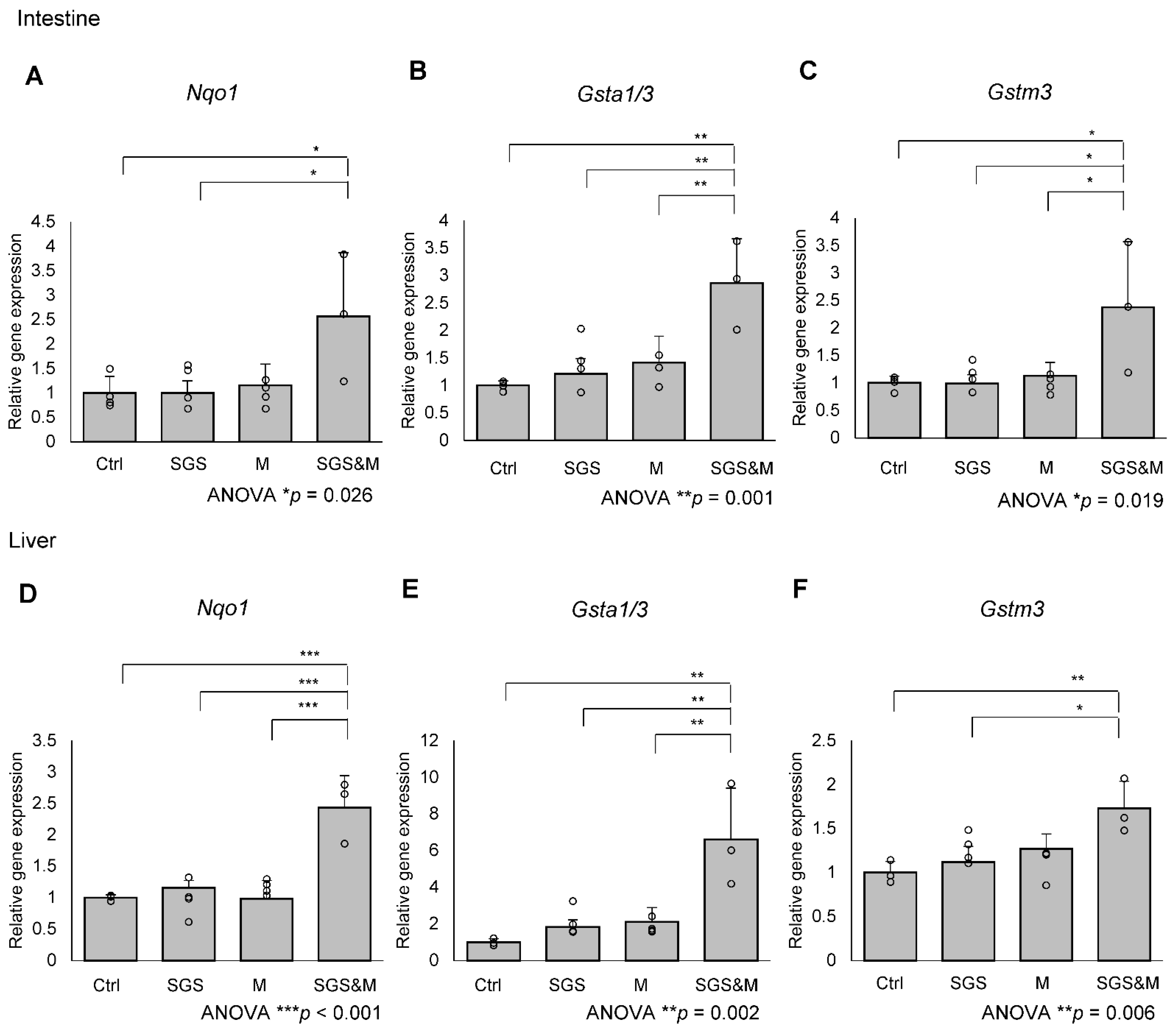
2.1. Co-Treatment with a Mixture of SGS and Mustard Extracts Efficiently Induces NRF2 Target Gene Expression in C57BL/6J Mice
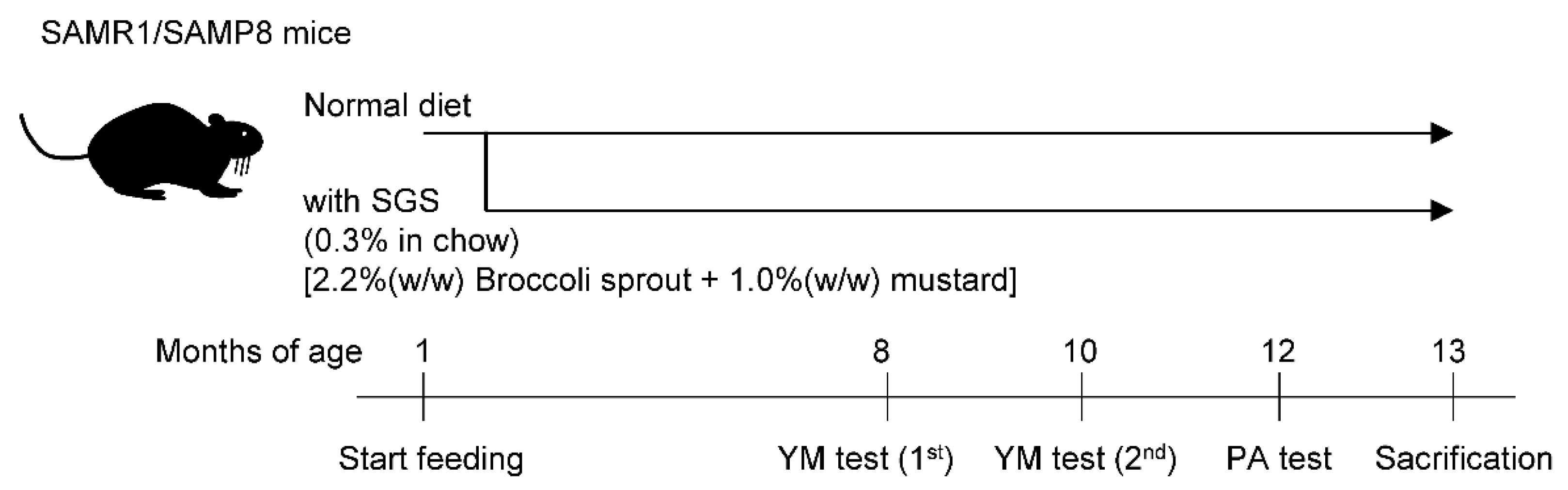
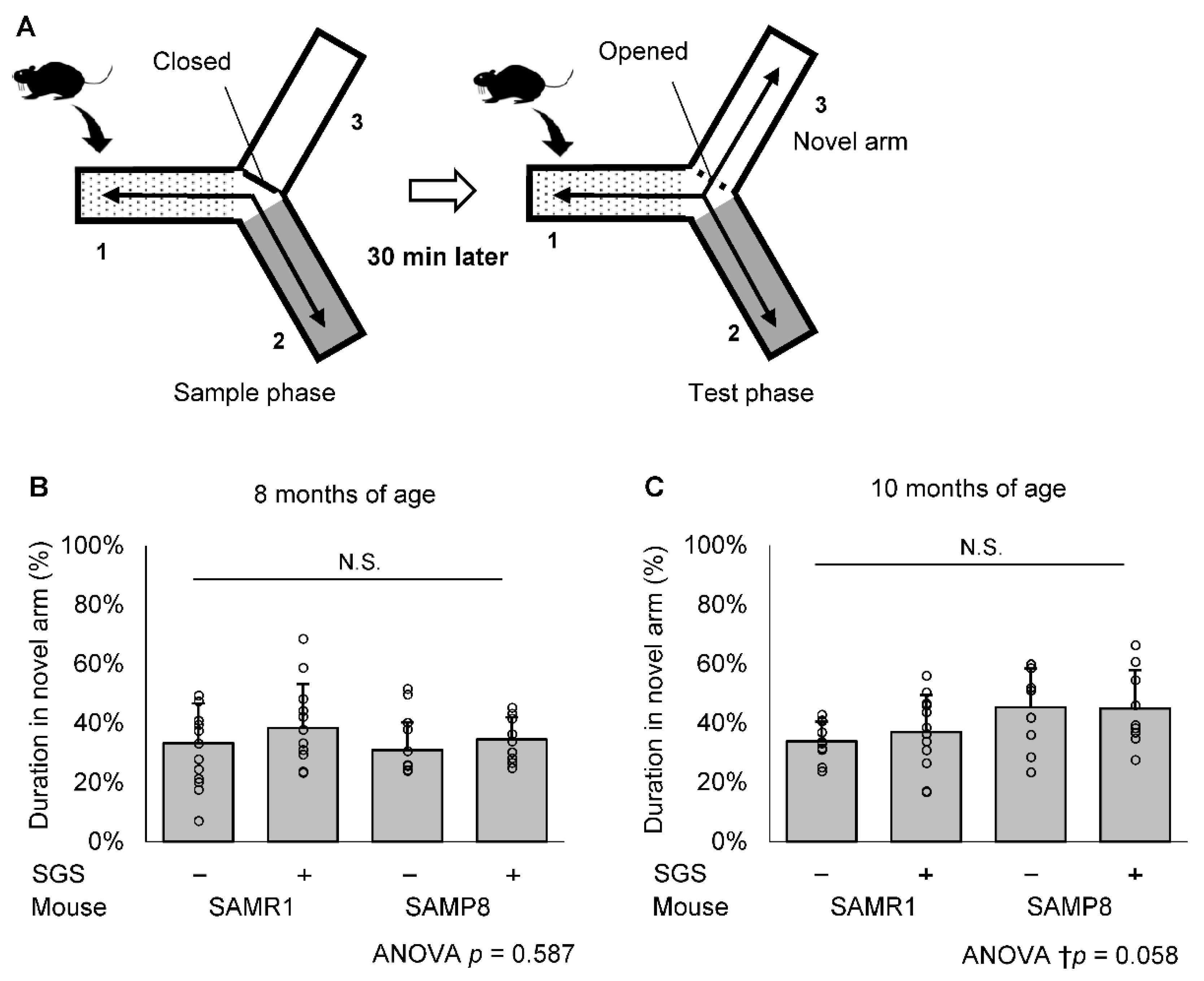
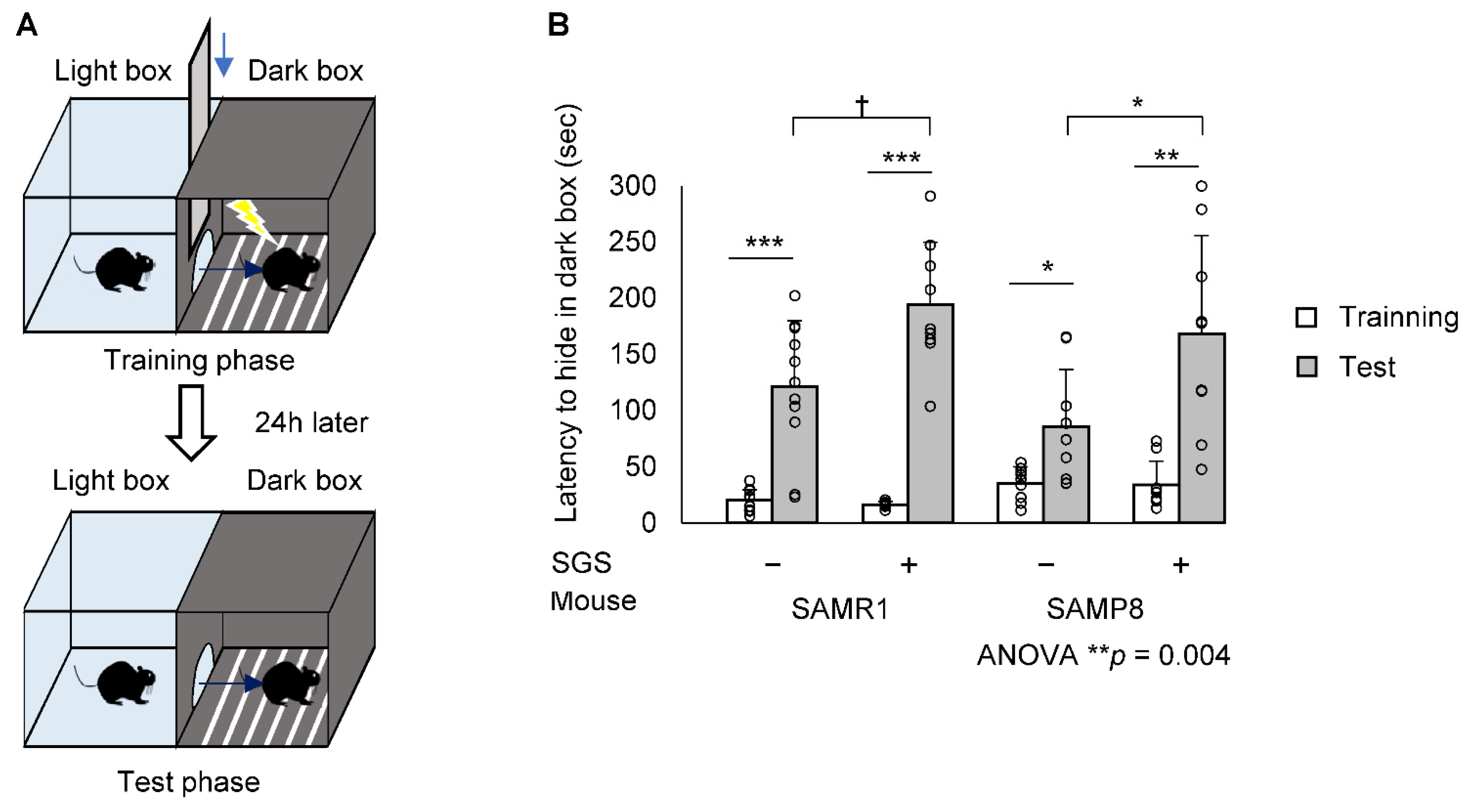
2.2. SGS Intake Prevents Age-Related Cognitive Decline in SAMP8
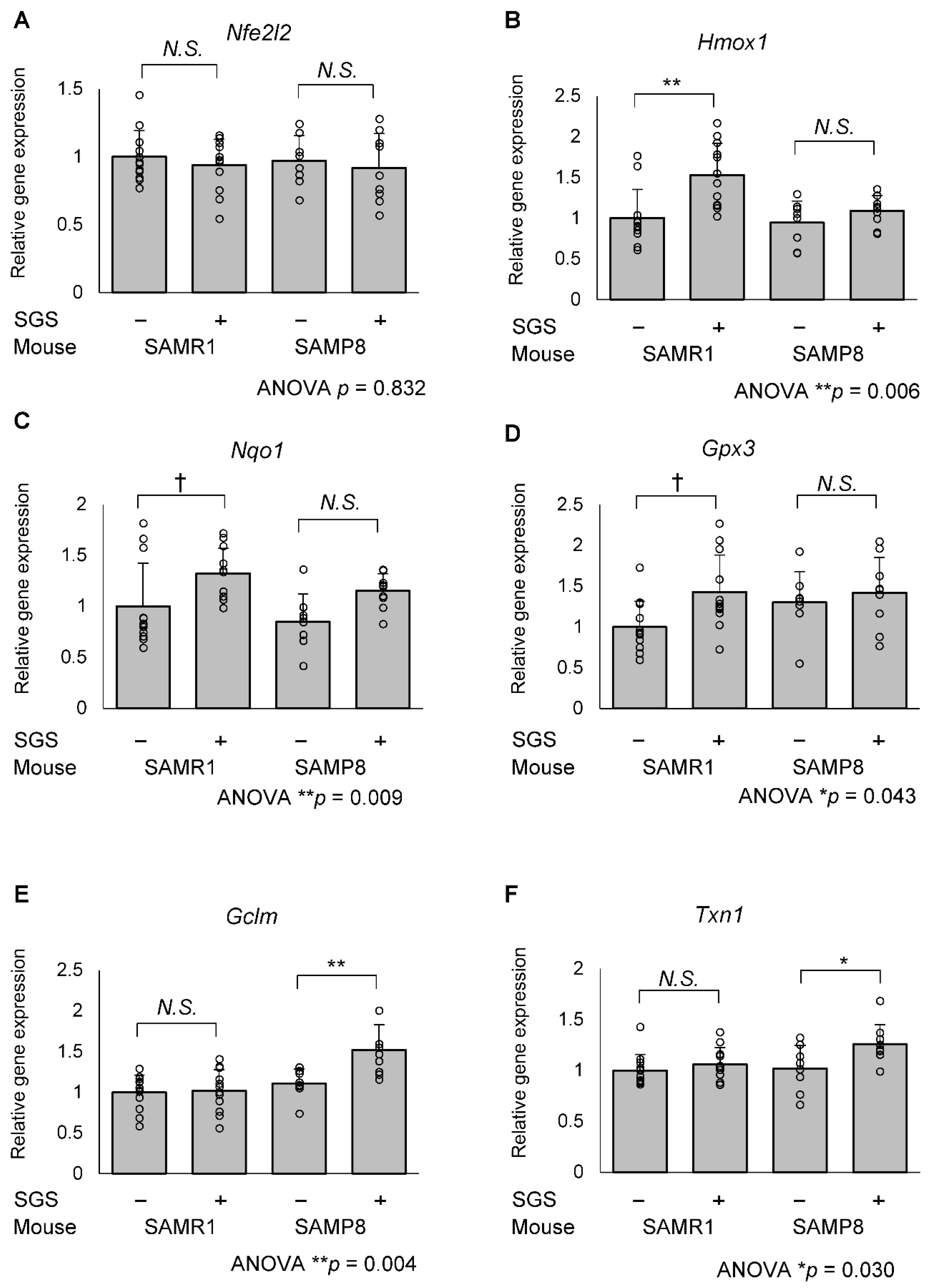
2.3. SGS Intake Differentially Modulates the Expression of NRF2/ARE Pathway Genes in the Hippocampi of SAMR1 and SAMP8 Mice
2.4. SGS Intake Transcriptionally Increases Mitochondrial Master Regulators in the Hippocampi of SAMP8 Mice
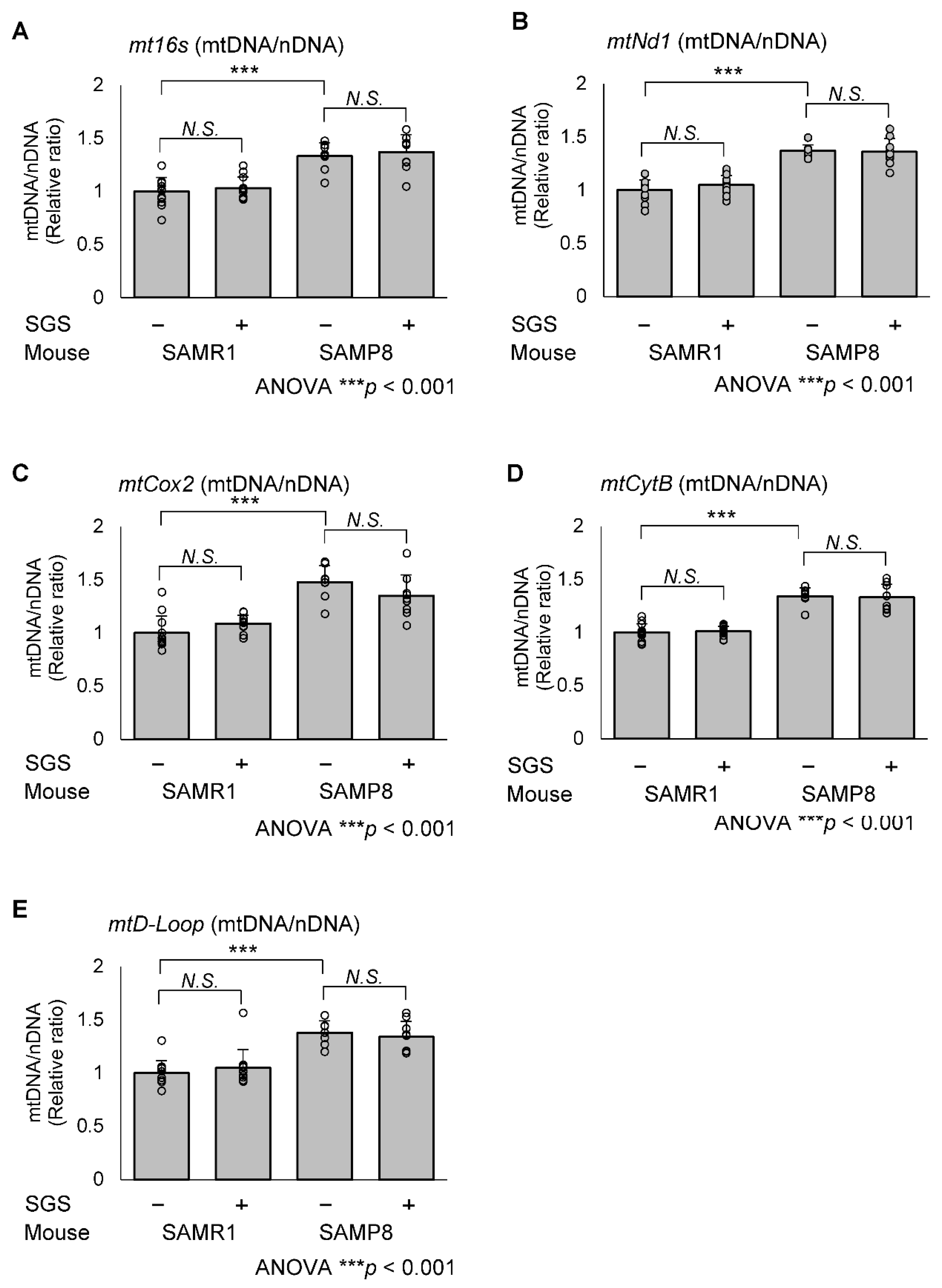
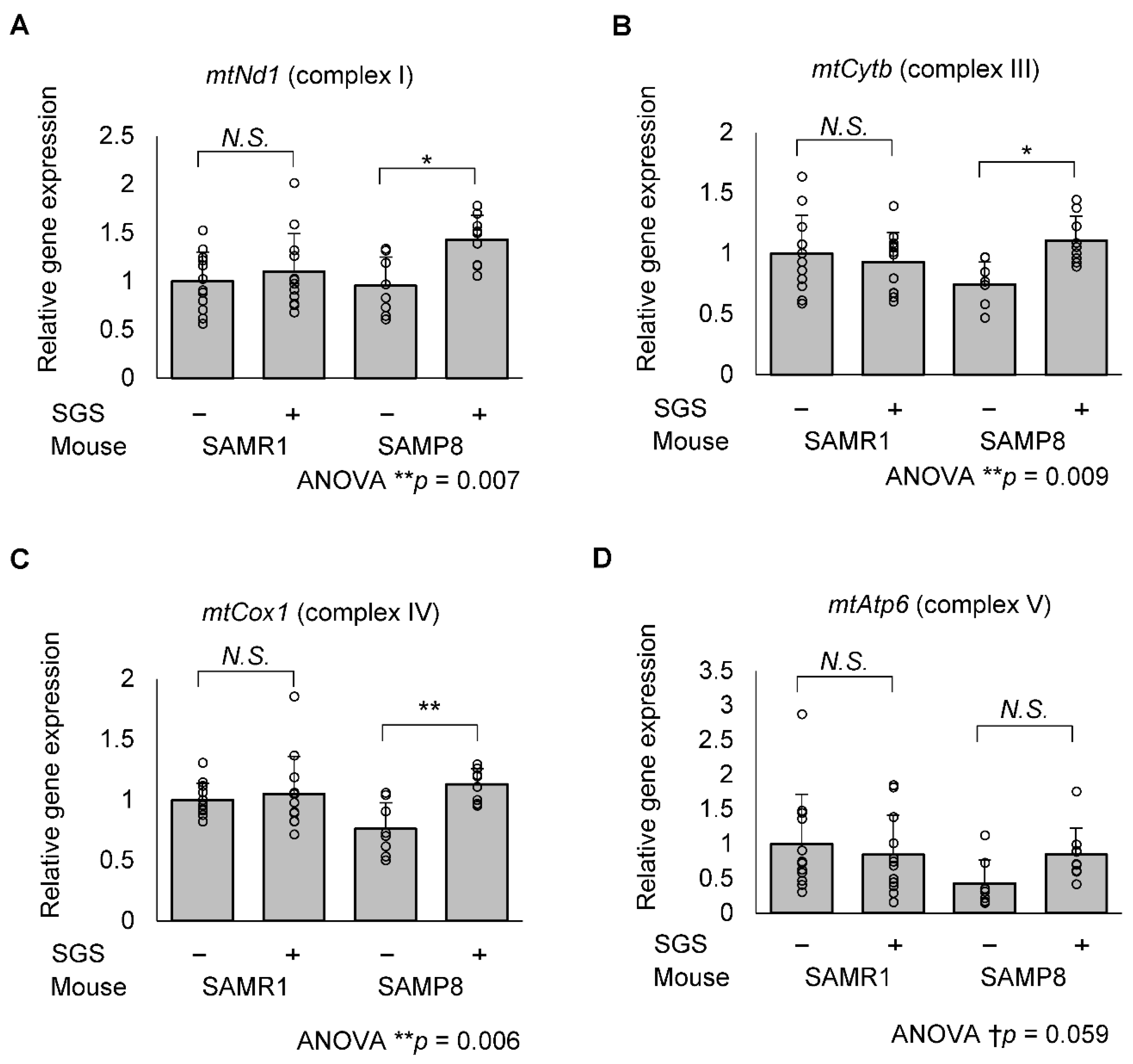
2.5. SGS Increases Mitochondria-Encoded Gene Expression in the Hippocampi of SAMP8 Mice
3. Discussion
4. Materials and Methods
4.1. Glucoraphanin Preparation
4.2. Animals and Glucoraphanin Preparation
4.3. Modified YM Test
4.4. Step-Through PA Test
4.5. Quantitative RT-PCR
4.6. Immunoblot Analysis
4.7. Mitochondrial DNA Copy Number Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tonnies, E.; Trushina, E. Oxidative Stress, Synaptic Dysfunction, and Alzheimer’s Disease. J. Alzheimers Dis. 2017, 57, 1105–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, K.; Mimura, J.; Yamamoto, M. Discovery of the negative regulator of Nrf2, Keap1: A historical overview. Antioxid. Redox Signal 2010, 13, 1665–1678. [Google Scholar] [CrossRef] [PubMed]
- Osama, A.; Zhang, J.; Yao, J.; Yao, X.; Fang, J. Nrf2: A dark horse in Alzheimer’s disease treatment. Ageing Res. Rev. 2020, 64, 101206. [Google Scholar] [CrossRef] [PubMed]
- Uruno, A.; Matsumaru, D.; Ryoke, R.; Saito, R.; Kadoguchi, S.; Saigusa, D.; Saito, T.; Saido, T.C.; Kawashima, R.; Yamamoto, M. Nrf2 Suppresses Oxidative Stress and Inflammation in App Knock-In Alzheimer’s Disease Model Mice. Mol. Cell Biol. 2020, 40, e00467-19. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ren, B.; Guo, R.; Zhang, W.; Ma, S.; Yao, Y.; Yuan, T.; Liu, Z.; Liu, X. Supplementation of lycopene attenuates oxidative stress induced neuroinflammation and cognitive impairment via Nrf2/NF-kappaB transcriptional pathway. Food Chem. Toxicol. 2017, 109, 505–516. [Google Scholar] [CrossRef]
- Maurer, I. A selective defect of cytochrome c oxidase is present in brain of Alzheimer disease patients. Neurobiol. Aging 2000, 21, 455–462. [Google Scholar] [CrossRef]
- Cardoso, S.M.; Santana, I.; Swerdlow, R.H.; Oliveira, C.R. Mitochondria dysfunction of Alzheimer’s disease cybrids enhances Abeta toxicity. J. Neurochem. 2004, 89, 1417–1426. [Google Scholar] [CrossRef] [Green Version]
- Kasai, S.; Shimizu, S.; Tatara, Y.; Mimura, J.; Itoh, K. Regulation of Nrf2 by Mitochondrial Reactive Oxygen Species in Physiology and Pathology. Biomolecules 2020, 10, 320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubli, D.A.; Gustafsson, A.B. Mitochondria and mitophagy: The yin and yang of cell death control. Circ. Res. 2012, 111, 1208–1221. [Google Scholar] [CrossRef] [Green Version]
- Ventura-Clapier, R.; Garnier, A.; Veksler, V. Transcriptional control of mitochondrial biogenesis: The central role of PGC-1alpha. Cardiovasc. Res. 2008, 79, 208–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gureev, A.P.; Shaforostova, E.A.; Popov, V.N. Regulation of Mitochondrial Biogenesis as a Way for Active Longevity: Interaction between the Nrf2 and PGC-1alpha Signaling Pathways. Front. Genet. 2019, 10, 435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Athale, J.; Ulrich, A.; MacGarvey, N.C.; Bartz, R.R.; Welty-Wolf, K.E.; Suliman, H.B.; Piantadosi, C.A. Nrf2 promotes alveolar mitochondrial biogenesis and resolution of lung injury in Staphylococcus aureus pneumonia in mice. Free Radic. Biol. Med. 2012, 53, 1584–1594. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Yang, T.; Mao, L.; Zhang, F. Sulforaphane Protects against Brain Diseases: Roles of Cytoprotective Enzymes. Austin J. Cereb. Dis. Stroke 2017, 4, 1054. [Google Scholar] [CrossRef] [Green Version]
- Hou, T.T.; Yang, H.Y.; Wang, W.; Wu, Q.Q.; Tian, Y.R.; Jia, J.P. Sulforaphane Inhibits the Generation of Amyloid-beta Oligomer and Promotes Spatial Learning and Memory in Alzheimer’s Disease (PS1V97L) Transgenic Mice. J. Alzheimers Dis. 2018, 62, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wang, W.; Xu, L.; Li, H.; Wei, Y.; Wu, Q.; Jia, J. Activation of Nrf2/ARE pathway alleviates the cognitive deficits in PS1V97L-Tg mouse model of Alzheimer’s disease through modulation of oxidative stress. J. Neurosci. Res. 2018, 97, 492–505. [Google Scholar] [CrossRef]
- Wang, G.; Fang, H.; Zhen, Y.; Xu, G.; Tian, J.; Zhang, Y.; Zhang, D.; Zhang, G.; Xu, J.; Zhang, Z.; et al. Sulforaphane Prevents Neuronal Apoptosis and Memory Impairment in Diabetic Rats. Cell Physiol. Biochem. 2016, 39, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.; Rajasekar, N.; Hanif, K.; Nath, C.; Shukla, R. Sulforaphane Ameliorates Okadaic Acid-Induced Memory Impairment in Rats by Activating the Nrf2/HO-1 Antioxidant Pathway. Mol. Neurobiol. 2016, 53, 5310–5323. [Google Scholar] [CrossRef] [PubMed]
- Negrette-Guzman, M.; Huerta-Yepez, S.; Vega, M.I.; Leon-Contreras, J.C.; Hernandez-Pando, R.; Medina-Campos, O.N.; Rodriguez, E.; Tapia, E.; Pedraza-Chaverri, J. Sulforaphane induces differential modulation of mitochondrial biogenesis and dynamics in normal cells and tumor cells. Food Chem. Toxicol. 2017, 100, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Kim, J. Pre-Clinical Neuroprotective Evidences and Plausible Mechanisms of Sulforaphane in Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 2929. [Google Scholar] [CrossRef] [PubMed]
- Nouchi, R.; Hu, Q.; Saito, T.; Kawata, N.; Nouchi, H.; Kawashima, R. Brain Training and Sulforaphane Intake Interventions Separately Improve Cognitive Performance in Healthy Older Adults, Whereas a Combination of These Interventions Does Not Have More Beneficial Effects: Evidence from a Randomized Controlled Trial. Nutrients 2021, 13, 352. [Google Scholar] [CrossRef]
- Ren, H.L.; Lv, C.N.; Xing, Y.; Geng, Y.; Zhang, F.; Bu, W.; Wang, M.W. Downregulated Nuclear Factor E2-Related Factor 2 (Nrf2) Aggravates Cognitive Impairments via Neuroinflammation and Synaptic Plasticity in the Senescence-Accelerated Mouse Prone 8 (SAMP8) Mouse: A Model of Accelerated Senescence. Med. Sci. Monit. 2018, 24, 1132–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagata, N.; Xu, L.; Kohno, S.; Ushida, Y.; Aoki, Y.; Umeda, R.; Fuke, N.; Zhuge, F.; Ni, Y.; Nagashimada, M.; et al. Glucoraphanin Ameliorates Obesity and Insulin Resistance through Adipose Tissue Browning and Reduction of Metabolic Endotoxemia in Mice. Diabetes 2017, 66, 1222–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svenningsson, A.L.; Stomrud, E.; Insel, P.S.; Mattsson, N.; Palmqvist, S.; Hansson, O. beta-amyloid pathology and hippocampal atrophy are independently associated with memory function in cognitively healthy elderly. Sci. Rep. 2019, 9, 11180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorbach, T.; Pudas, S.; Lundquist, A.; Oradd, G.; Josefsson, M.; Salami, A.; de Luna, X.; Nyberg, L. Longitudinal association between hippocampus atrophy and episodic-memory decline. Neurobiol. Aging 2017, 51, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Kaup, A.R.; Mirzakhanian, H.; Jeste, D.V.; Eyler, L.T. A review of the brain structure correlates of successful cognitive aging. J. Neuropsychiatry Clin. Neurosci. 2011, 23, 6–15. [Google Scholar] [CrossRef] [Green Version]
- Brunetti, D.; Bottani, E.; Segala, A.; Marchet, S.; Rossi, F.; Orlando, F.; Malavolta, M.; Carruba, M.O.; Lamperti, C.; Provinciali, M.; et al. Targeting Multiple Mitochondrial Processes by a Metabolic Modulator Prevents Sarcopenia and Cognitive Decline in SAMP8 Mice. Front. Pharm. 2020, 11, 1171. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, X.; Song, Q.; Liu, J.; Ji, X.; Wang, P. POLD1 deficiency is involved in cognitive function impairment in AD patients and SAMP8 mice. Biomed. Pharm. 2019, 114, 108833. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Tian, S.; Teng, C.; Huang, L.; Liu, X.; Wang, J.; Zhang, Y.; Li, B.; Shan, Y. Sulforaphane Improves Lipid Metabolism by Enhancing Mitochondrial Function and Biogenesis In Vivo and In Vitro. Mol. Nutr. Food Res. 2019, 63, e1800795. [Google Scholar] [CrossRef]
- Moreno-Loshuertos, R.; Acin-Perez, R.; Fernandez-Silva, P.; Movilla, N.; Perez-Martos, A.; Rodriguez de Cordoba, S.; Gallardo, M.E.; Enriquez, J.A. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 2006, 38, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, H.; Yokota, M.; Mori, M.; Shimizu, A.; Nakada, K.; Hayashi, J. Nuclear but not mitochondrial DNA involvement in respiratory complex I defects found in senescence-accelerated mouse strain, SAMP8. Exp. Anim. 2011, 60, 397–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakahara, H.; Kanno, T.; Inai, Y.; Utsumi, K.; Hiramatsu, M.; Mori, A.; Packer, L. Mitochondrial Dysfunction in the Senescence Accelerated Mouse (SAM). Free Radic. Biol. Med. 1998, 24, 85–92. [Google Scholar] [CrossRef]
- Okatani, Y.; Wakatsuki, A.; Reiter, R.J. Melatonin protects hepatic mitochondrial respiratory chain activity in senescence-accelerated mice. J. Pineal Res. 2002, 32, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.; Hsu, A.; Williams, D.E.; Dashwood, R.H.; Stevens, J.F.; Yamamoto, M.; Ho, E. Metabolism and tissue distribution of sulforaphane in Nrf2 knockout and wild-type mice. Pharm. Res. 2011, 28, 3171–3179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomobe, K.; Shinozuka, T.; Kuroiwa, M.; Nomura, Y. Age-related changes of Nrf2 and phosphorylated GSK-3beta in a mouse model of accelerated aging (SAMP8). Arch. Gerontol. Geriatr. 2012, 54, e1–e7. [Google Scholar] [CrossRef]
- Whitman, S.A.; Long, M.; Wondrak, G.T.; Zheng, H.; Zhang, D.D. Nrf2 modulates contractile and metabolic properties of skeletal muscle in streptozotocin-induced diabetic atrophy. Exp. Cell Res. 2013, 319, 2673–2683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piantadosi, C.A.; Carraway, M.S.; Babiker, A.; Suliman, H.B. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ. Res. 2008, 103, 1232–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldelli, S.; Aquilano, K.; Ciriolo, M.R. Punctum on two different transcription factors regulated by PGC-1alpha: Nuclear factor erythroid-derived 2-like 2 and nuclear respiratory factor 2. Biochim. Biophys. Acta 2013, 1830, 4137–4146. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.C. Nuclear Factor-Erythroid 2-Related Factor 2 (Nrf2) and Mitochondrial Dynamics/Mitophagy in Neurological Diseases. Antioxidants 2020, 9, 617. [Google Scholar] [CrossRef] [PubMed]
- Vanduchova, A.; Anzenbacher, P.; Anzenbacherova, E. Isothiocyanate from Broccoli, Sulforaphane, and Its Properties. J. Med. Food 2019, 22, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Konar, A.; Singh, P.; Thakur, M.K. Age-associated Cognitive Decline: Insights into Molecular Switches and Recovery Avenues. Aging Dis. 2016, 7, 121–129. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, M.; Ushida, Y.; Shiozawa, H.; Umeda, R.; Tsuruya, K.; Aoki, Y.; Suganuma, H.; Nishizaki, Y. Sulforaphane-rich broccoli sprout extract improves hepatic abnormalities in male subjects. World J. Gastroenterol. 2015, 21, 12457–12467. [Google Scholar] [CrossRef] [PubMed]
- Okunade, O.; Niranjan, K.; Ghawi, S.K.; Kuhnle, G.; Methven, L. Supplementation of the Diet by Exogenous Myrosinase via Mustard Seeds to Increase the Bioavailability of Sulforaphane in Healthy Human Subjects after the Consumption of Cooked Broccoli. Mol. Nutr. Food Res. 2018, 62, e1700980. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Hayashida, M.; Zhao, Q.; Shibahara, N.; Tanaka, K.; Miyata, T.; Matsumoto, K. Ameliorative effects of yokukansan on learning and memory deficits in olfactory bulbectomized mice. J. Ethnopharmacol. 2011, 135, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Sithisarn, P.; Rojsanga, P.; Jarikasem, S.; Tanaka, K.; Matsumoto, K. Ameliorative Effects of Acanthopanax trifoliatus on Cognitive and Emotional Deficits in Olfactory Bulbectomized Mice: An Animal Model of Depression and Cognitive Deficits. Evid. Based Complement. Altern. Med. 2013, 2013, 701956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meguro, K. Effects of thioperamide, a histamine H3 antagonist, on the step-through passive avoidance response and histidine decarboxylase activity in senescence-accelerated mice. Pharmacol. Biochem. Behav. 1995, 50, 321–325. [Google Scholar] [CrossRef]
- Wang, Y.J.; Lu, J.; Wu, D.M.; Zheng, Z.H.; Zheng, Y.L.; Wang, X.H.; Ruan, J.; Sun, X.; Shan, Q.; Zhang, Z.F. Ursolic acid attenuates lipopolysaccharide-induced cognitive deficits in mouse brain through suppressing p38/NF-kappaB mediated inflammatory pathways. Neurobiol. Learn. Mem. 2011, 96, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Quiros, P.M.; Goyal, A.; Jha, P.; Auwerx, J. Analysis of mtDNA/nDNA Ratio in Mice. Curr. Protoc. Mouse Biol. 2017, 7, 47–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Nfe2l2 | CAGCACATCCAGACAGACACCA | CGTATTAAGACACTGTAACTCGGGAATGG |
| Hmox1 | TGACACCTGAGGTCAAGCAC | TCCTCTGTCAGCATCACCTG |
| Nqo1 | AGCGTTCGGTATTACGATCC | AGTACAATCAGGGCTCTTCTCG |
| Gpx3 | CGAGTATGGAGCCCTCACCA | GCCCAGAATGACCAAGCCAA |
| Gclm | GGGAACCTGCTCAACTGGGG | CTGCATGGGCATGGTGCATT |
| Txn1 | CCCTTCTTCCATTCCCTCT | TCCACATCCACTTCAAGGAAC |
| Pgc1α | CACCGCAATTCTCCCTTGTA | TGCGGTATTCATCCCTCTTG |
| Nrf1 | CCTCTGATGCTTGCGTCGTCT | TTACTCTGCTGTGGCTGATGG |
| Tfam | TGGAGGGAGCTACAGAAGCAG | GCCTCCTTCTCCATACCCATCAGC |
| Cat | GGCAAAGGTGTTTGAGCATATT | GAGTCTGTGGGTTTCTCTTCTG |
| Gsta1/3 | GGCAGAATGGAGTGCATCA | TCCAAATCTTCCGGACTCTG |
| Gstm3 | GCACAACCTGTGTGGAGAGAC | ACTCTGGCTTCTGCTTCTCAA |
| Gstp1 | TGTCACCCTCATCTACACCAAC | GGACAGCAGGGTCTCAAAAG |
| Gpx1 | ATGCCTTAGGGGTTGCTAGG | CGACATCGAACCCGATATAGA |
| Gpx2 | CAGCTTCCAGACCATCAACA | CACTGAGCCCTGAGGAAGAC |
| Sod1 | AACCAGTTGTGTTGTCAGGAC | CCACCATGTTTCTTAGAGTGAGG |
| Sod2 | CAGACCTGCCTTACGACTATGG | CTCGGTGGCGTTGAGATTGTT |
| Gclc | TGGCCACTATCTGCCCAATT | GTCTGACACGTAGCCTCGGTAA |
| Txnrd1 | GTGGCGACTTGGCTAATC | ACCAGGAGAGACACTCAC |
| Srxn1 | CCCACTGGACCAACTTCTGT | GTGGCTAGCTCAGACCAAGG |
| Mfn1 | GCAGACAGCACATGGAGAGA | GATCCGATTCCGAGCTTCCG |
| Mfn2 | TGCACCGCCATATAGAGGAAG | TCTGCAGTGAACTGGCAATG |
| Opa1 | ACCTTGCCAGTTTAGCTCCC | TTGGGACCTGCAGTGAAGAA |
| Drp1 | ATGCCAGCAAGTCCACAGAA | TGTTCTCGGGCAGACAGTTT |
| mtNd1 | TACGAGCCGTAGCCCAAACA | GATCGTAACGGAAGCGTGGA |
| mtCytb | ATTCCTTCATGTCGGACGAG | ACTGAGAAGCCCCCTCAAAT |
| mtCox1 | CTGAGCGGGAATAGTGGGTA | TGGGGCTCCGATTATTAGTG |
| mtAtp6 | TCCCATCCTCAAAACGCCTA | CCAGCTCATAGTGGAATGGC |
| Gapdh | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| mt16s | CCGCAAGGGAAAGATGAAAGAC | TCGTTTGGTTTCGGGGTTTC |
| mtNd1 | CTAGCAGAAACAAACCGGGC | CCGGCTGCGTATTCTACGTT |
| mtCox2 | GTTGATAACCGAGTCGTT | CCTGGGATGGCATCAGTT |
| mtCytb | AGACAAAGCCACCTTGACCCGAT | ACGATTGCTAGGGCCGCGAT |
| mtD-Loop | TGCCCCTCTTCTCGCTCCGG | GGCGATAACGCATTTGATGGCC |
| HK2 | GCCAGCCTCTCCTGATTTTAGTGT | GGGAACACAAAAGACCTCTTCTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimizu, S.; Kasai, S.; Yamazaki, H.; Tatara, Y.; Mimura, J.; Engler, M.J.; Tanji, K.; Nikaido, Y.; Inoue, T.; Suganuma, H.; et al. Sulforaphane Increase Mitochondrial Biogenesis-Related Gene Expression in the Hippocampus and Suppresses Age-Related Cognitive Decline in Mice. Int. J. Mol. Sci. 2022, 23, 8433. https://doi.org/10.3390/ijms23158433
Shimizu S, Kasai S, Yamazaki H, Tatara Y, Mimura J, Engler MJ, Tanji K, Nikaido Y, Inoue T, Suganuma H, et al. Sulforaphane Increase Mitochondrial Biogenesis-Related Gene Expression in the Hippocampus and Suppresses Age-Related Cognitive Decline in Mice. International Journal of Molecular Sciences. 2022; 23(15):8433. https://doi.org/10.3390/ijms23158433
Chicago/Turabian StyleShimizu, Sunao, Shuya Kasai, Hiromi Yamazaki, Yota Tatara, Junsei Mimura, Máté János Engler, Kunikazu Tanji, Yoshikazu Nikaido, Takuro Inoue, Hiroyuki Suganuma, and et al. 2022. "Sulforaphane Increase Mitochondrial Biogenesis-Related Gene Expression in the Hippocampus and Suppresses Age-Related Cognitive Decline in Mice" International Journal of Molecular Sciences 23, no. 15: 8433. https://doi.org/10.3390/ijms23158433
APA StyleShimizu, S., Kasai, S., Yamazaki, H., Tatara, Y., Mimura, J., Engler, M. J., Tanji, K., Nikaido, Y., Inoue, T., Suganuma, H., Wakabayashi, K., & Itoh, K. (2022). Sulforaphane Increase Mitochondrial Biogenesis-Related Gene Expression in the Hippocampus and Suppresses Age-Related Cognitive Decline in Mice. International Journal of Molecular Sciences, 23(15), 8433. https://doi.org/10.3390/ijms23158433







