Changes in the Transcriptome Caused by Mutations in the Ribosomal Protein uS10 Associated with a Predisposition to Colorectal Cancer
Abstract
:1. Introduction
2. Results
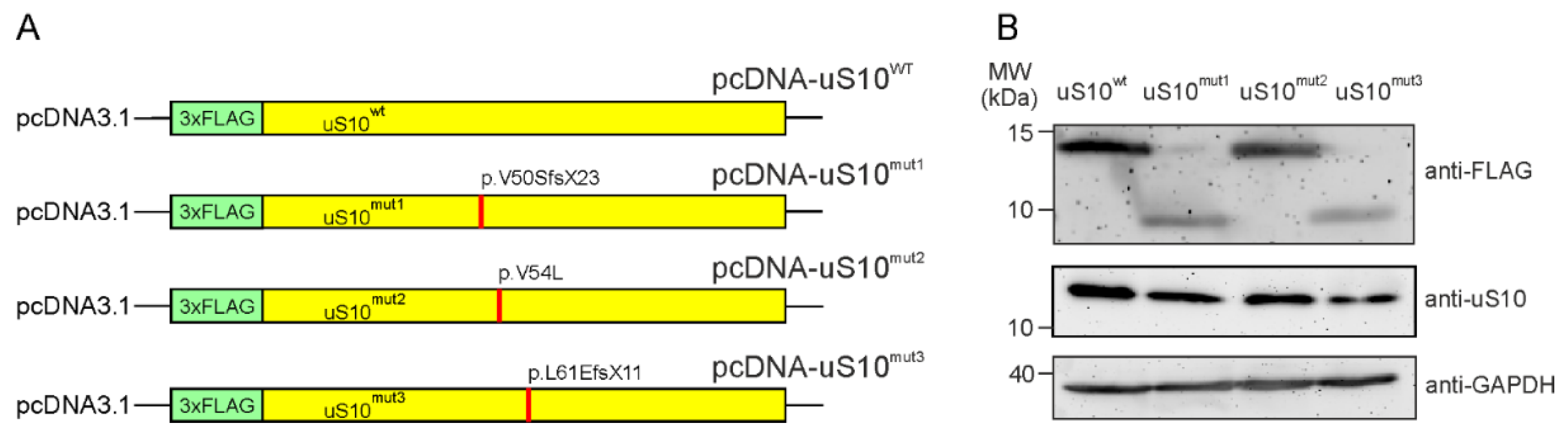
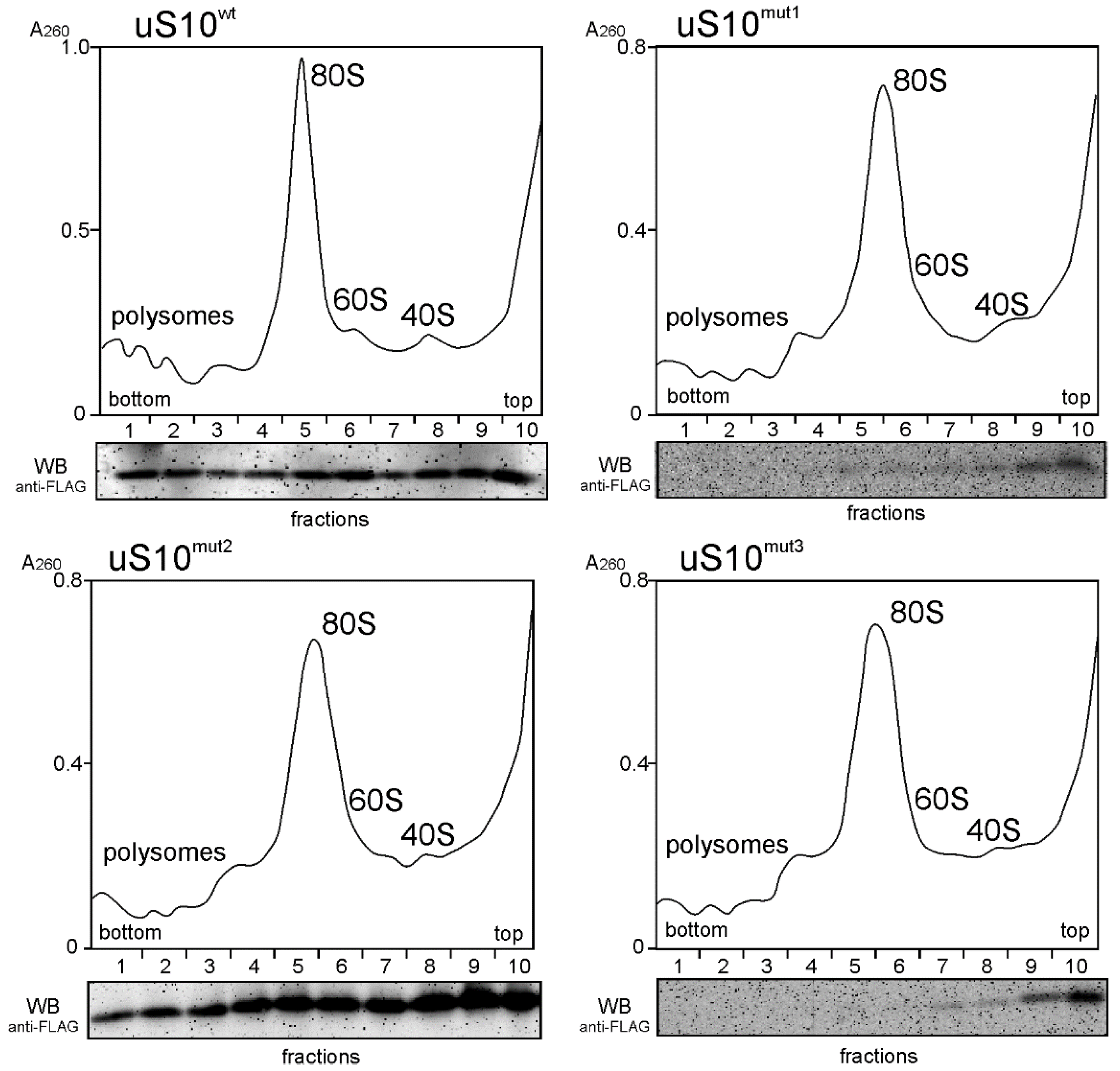
2.1. The Ectopic Production of Mutant Forms of the Ribosomal Protein uS10 in HEK293T Cells
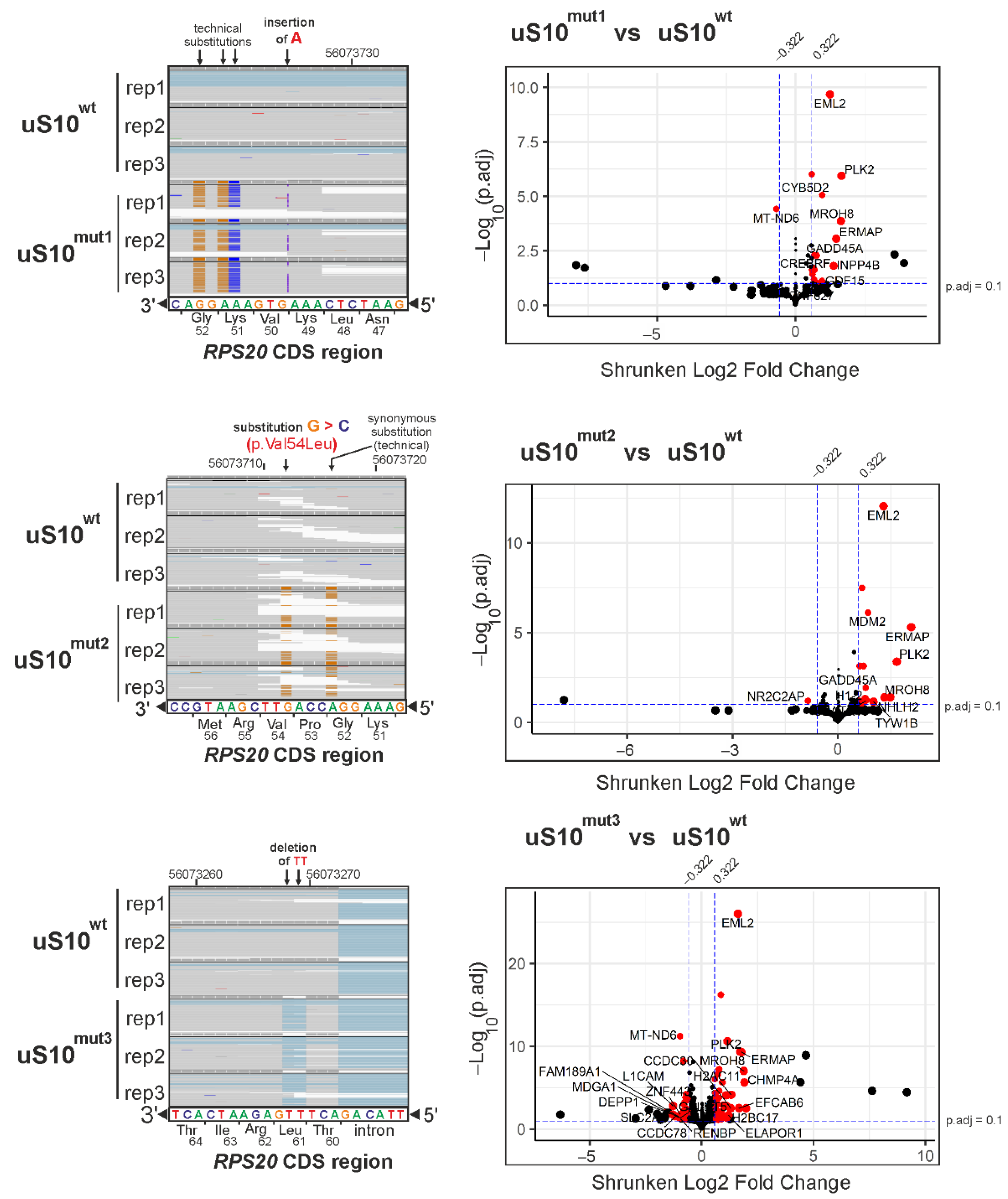
2.2. A Comparative Analysis of RNA-Seq Data from Cells Producing 3×FLAG-Tagged uS10 and Its Mutant Forms
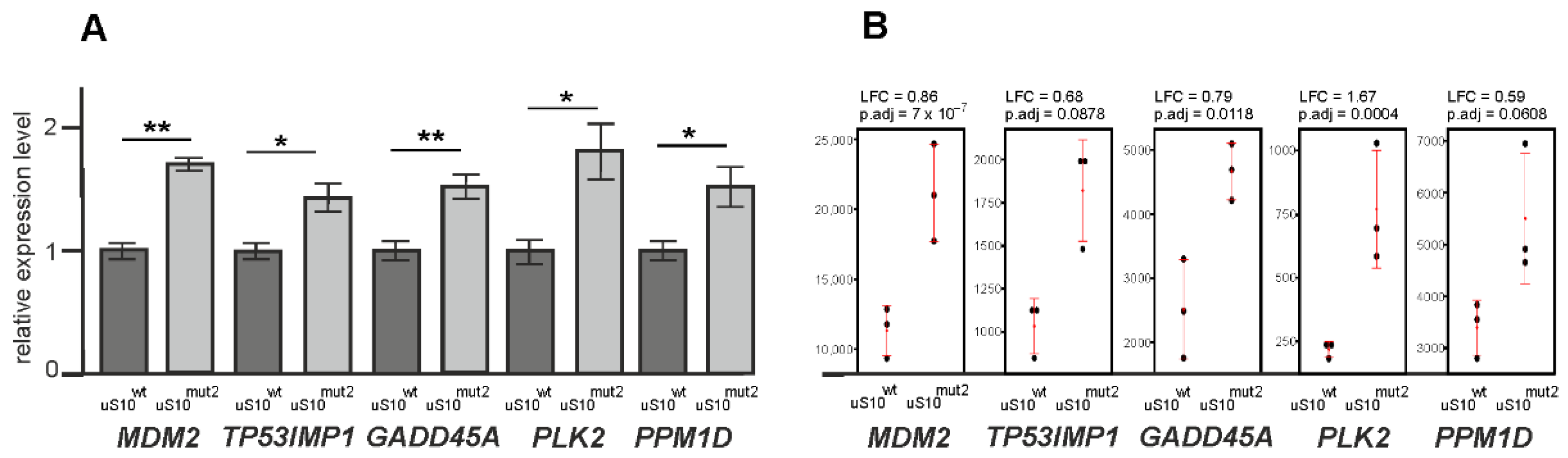
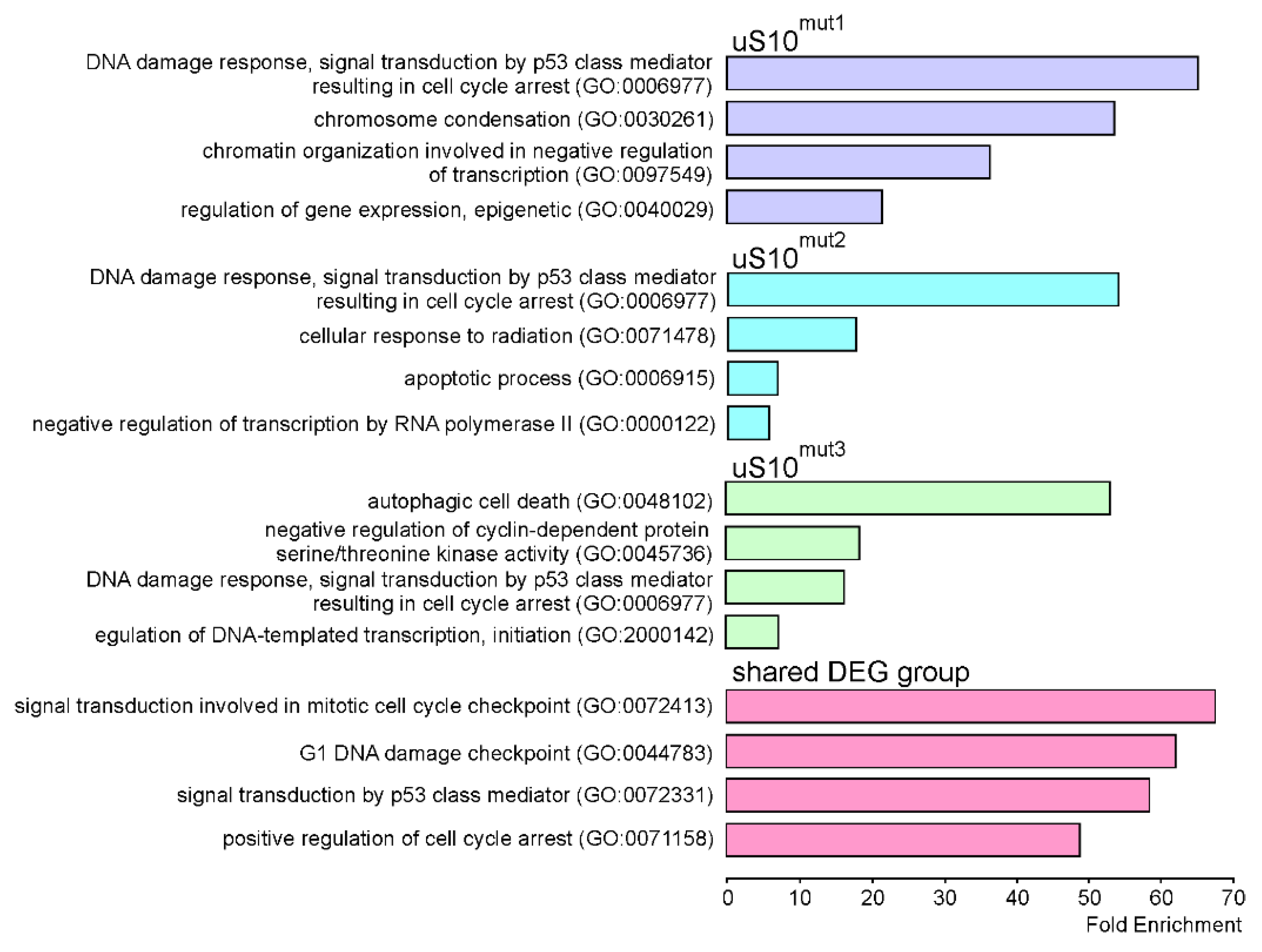
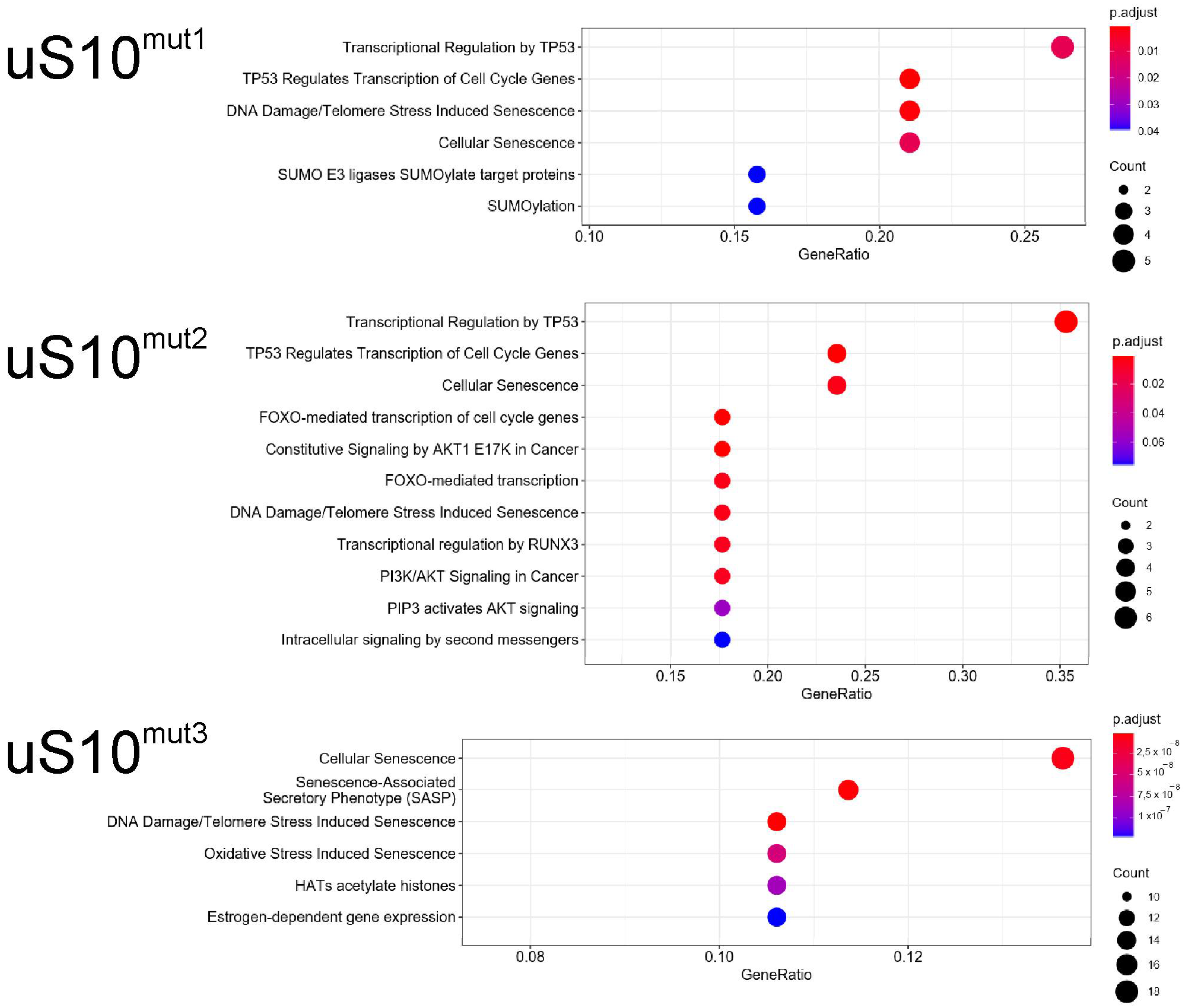
2.3. Processes Associated with Genes Whose Expression Changes When Cells Produce Mutant Forms of uS10wt
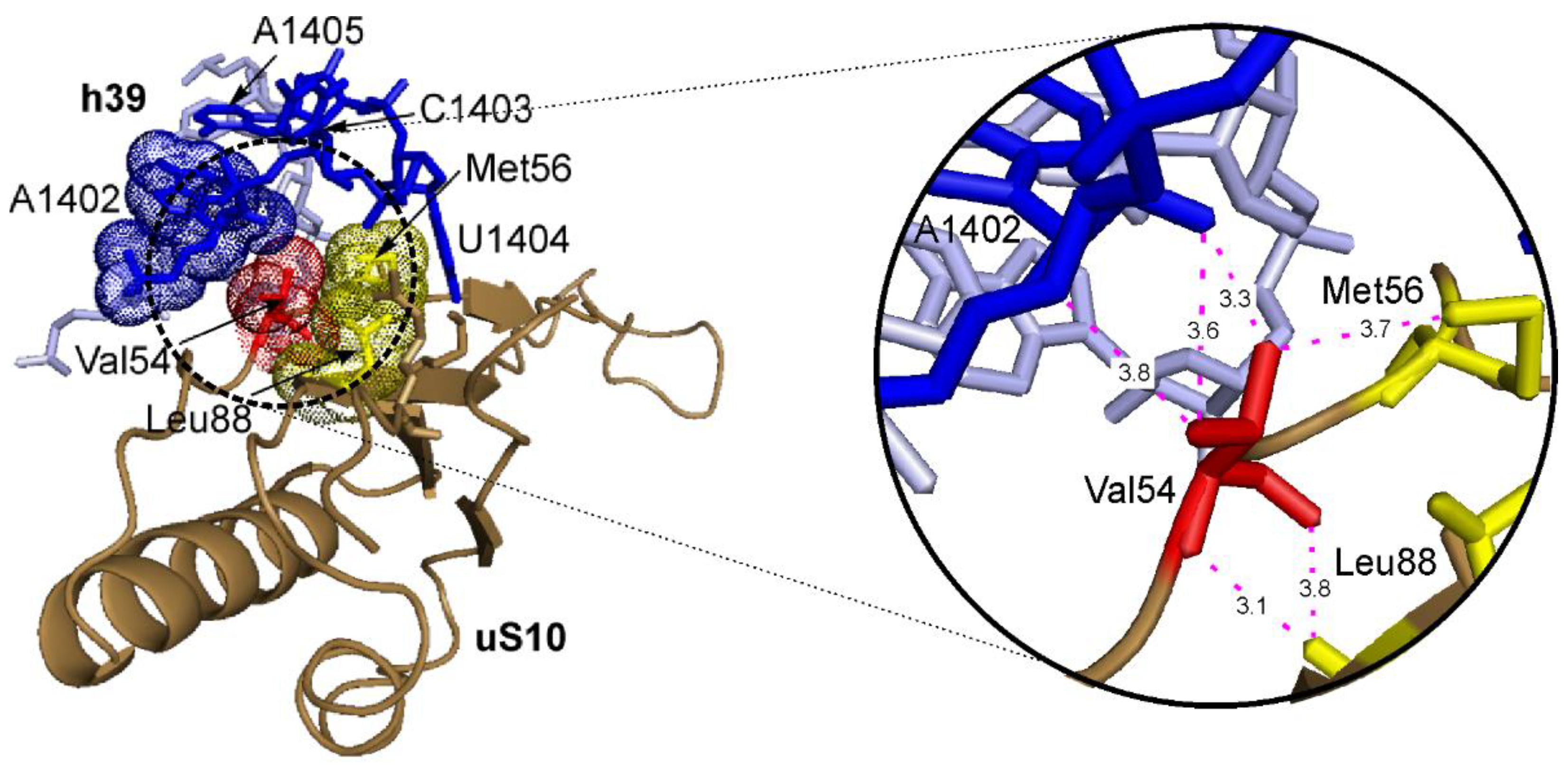
3. Discussion
4. Materials and Methods
4.1. Plasmids Preparation, Cell Culturing and Target Protein Production Analysis
4.2. DNA Libraries Preparation and High-Throughput Sequencing
4.3. Raw NGS Data Processing
4.4. Bioinformatics Analysis of Processed NGS Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomson, E.; Ferreira-Cerca, S.; Hurt, E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013, 126, 4815–4821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassler, J.; Hurt, E. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 2019, 88, 281–306. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Unbehaun, A.; Loerke, J.; Behrmann, E.; Collier, M.; Bürger, J.; Mielke, T.; Spahn, C.M.T. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol. 2014, 21, 721–727. [Google Scholar] [CrossRef] [Green Version]
- Muhs, M.; Hilal, T.; Mielke, T.; Skabkin, M.A.; Sanbonmatsu, K.Y.; Pestova, T.V.; Spahn, C.M. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol. Cell 2015, 57, 422–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonetti, A.; Brito Querido, J.; Myasnikov, A.G.; Mancera-Martinez, E.; Renaud, A.; Kuhn, L.; Hashem, Y. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol. Cell 2016, 63, 206–217. [Google Scholar] [CrossRef] [Green Version]
- Graifer, D.; Karpova, G. Interaction of tRNA with eukaryotic ribosome. Int. J. Mol. Sci. 2015, 16, 7173–7194. [Google Scholar] [CrossRef] [Green Version]
- Graifer, D.; Karpova, G. Ribosomal protein uS3 in cell biology and human disease: Latest insights and prospects. Bioessays 2020, 42, e2000124. [Google Scholar] [CrossRef]
- Graifer, D.; Karpova, G. Eukaryotic protein uS19: A component of the decoding site of ribosomes and a player in human diseases. Biochem. J. 2021, 478, 997–1008. [Google Scholar] [CrossRef]
- Venturi, G.; Montanaro, L. How altered ribosome production can cause or contribute to human disease: The spectrum of ribosomopathies. Cells 2020, 9, 2300. [Google Scholar] [CrossRef]
- Goudarzi, K.M.; Lindstrom, M.S. Role of ribosomal protein mutations in tumor development. Int. J. Oncol. 2016, 48, 1313–1324. [Google Scholar] [CrossRef] [Green Version]
- Warner, J.R.; McIntosh, K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhavsar, R.B.; Makley, L.N.; Tsonis, P.A. The other lives of ribosomal proteins. Hum. Genom. 2010, 4, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Graifer, D.; Malygin, A.; Zharkov, D.O.; Karpova, G. Eukaryotic ribosomal protein S3: A constituent of translational machinery and an extraribosomal player in various cellular processes. Biochimie 2014, 99, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Nieminen, T.T.; O’Donohue, M.F.; Wu, Y.; Lohi, H.; Scherer, S.W.; Paterson, A.D.; Ellonen, P.; Abdel-Rahman, W.M.; Valo, S.; Mecklin, J.-P.; et al. Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology 2014, 147, 595–598. [Google Scholar] [CrossRef] [Green Version]
- Broderick, P.; Dobbins, S.E.; Chubb, D.; Kinnersley, B.; Dunlop, M.G.; Tomlinson, I.; Houlston, R.S. Validation of recently proposed colorectal cancer susceptibility gene variants in an analysis of families and patients—A systematic review. Gastroenterology 2017, 152, 75–77.e4. [Google Scholar] [CrossRef] [Green Version]
- Djursby, M.; Madsen, M.B.; Frederiksen, J.H.; Berchtold, L.A.; Therkildsen, C.; Willemoe, G.L.; Hasselby, J.P.; Wikman, F.; Okkels, H.; Skytte, A.-B.; et al. New pathogenic germline variants in very early onset and familial colorectal cancer patients. Front. Genet. 2020, 11, 566266. [Google Scholar] [CrossRef]
- Anger, A.M.; Armache, J.P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef]
- Cuchalova, L.; Kouba, T.; Herrmannova, A.; Danyi, I.; Chiu, W.L.; Valasek, L. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol. 2010, 30, 4671–4686. [Google Scholar] [CrossRef] [Green Version]
- Mitterer, V.; Shayan, R.; Ferreira-Cerca, S.; Murat, G.; Enne, T.; Rinaldi, D.; Weigl, S.; Omanic, H.; Gleizes, P.-E.; Kressler, D.; et al. Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism. Nat. Commun. 2019, 10, 2754. [Google Scholar] [CrossRef] [Green Version]
- Higgins, R.; Gendron, J.M.; Rising, L.; Mak, R.; Webb, K.; Kaiser, S.E.; Zuzow, N.; Riviere, P.; Yang, B.; Fenech, E.; et al. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol. Cell 2015, 59, 35–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuo, Y.; Ikeuchi, K.; Saeki, Y.; Iwasaki, S.; Schmidt, C.; Udagawa, T.; Sato, F.; Tsuchiya, H.; Becker, T.; Tanaka, K.; et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017, 8, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundaramoorthy, E.; Leonard, M.; Mak, R.; Liao, J.; Fulzele, A.; Bennett, E.J. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell 2017, 65, 751–760.e754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeuchi, K.; Tesina, P.; Matsuo, Y.; Sugiyama, T.; Cheng, J.; Saeki, Y.; Tanaka, K.; Becker, T.; Beckmann, R.; Inada, T. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 2019, 38, e100276. [Google Scholar] [CrossRef] [PubMed]
- Daftuar, L.; Zhu, Y.; Jacq, X.; Prives, C. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS ONE 2013, 8, e68667. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, R.; Boddapati, N.; Mahalingam, S. Interplay between human nucleolar GNL1 and RPS20 is critical to modulate cell proliferation. Sci. Rep. 2018, 8, 11421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGowan, K.A.; Li, J.Z.; Park, C.Y.; Beaudry, V.; Tabor, H.K.; Sabnis, A.J.; Zhang, W.; Fuchs, H.; de Angelis, M.H.; Myers, R.M.; et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 2008, 40, 963–970. [Google Scholar] [CrossRef] [Green Version]
- Bhar, S.; Zhou, F.; Reineke, L.C.; Morris, D.K.; Khincha, P.P.; Giri, N.; Mirabello, L.; Bergstrom, K.; Lemon, L.D.; Williams, C.L.; et al. Expansion of germline RPS20 mutation phenotype to include Diamond-Blackfan anemia. Hum. Mutat. 2020, 41, 1918–1930. [Google Scholar] [CrossRef]
- Kampen, K.R.; Sulima, S.O.; Vereecke, S.; De Keersmaecker, K. Hallmarks of ribosomopathies. Nucleic Acids Res. 2020, 48, 1013–1028. [Google Scholar] [CrossRef]
- Salvador, J.M.; Brown-Clay, J.D.; Fornace, A.J., Jr. Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv. Exp. Med. Biol. 2013, 793, 1–19. [Google Scholar] [CrossRef]
- Schafer, A. Gadd45 proteins: Key players of repair-mediated DNA demethylation. Adv. Exp. Med. Biol. 2013, 793, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Tamura, R.E.; de Vasconcellos, J.F.; Sarkar, D.; Libermann, T.A.; Fisher, P.B.; Zerbini, L.F. GADD45 proteins: Central players in tumorigenesis. Curr. Mol. Med. 2012, 12, 634–651. [Google Scholar] [CrossRef]
- Emelyanov, A.; Bulavin, D.V. Wip1 phosphatase in breast cancer. Oncogene 2017, 34, 4429–4438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.; Li, J.; Dorrah, K.; Jimenez-Tapia, D.; Arriaga, B.; Hao, Q.; Cao, W.; Gao, Z.; Vadgama, J.; Wu, Y. The role of PPM1D in cancer and advances in studies of its inhibitors. Biomed. Pharmacother. 2020, 125, 109956. [Google Scholar] [CrossRef]
- Li, K.; Liu, Y.; Xu, S.; Wang, J. PPM1D functions as oncogene and is associated with poor prognosis in psophageal squamous cell carcinoma. Pathol. Oncol. Res. 2020, 26, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Yi, J.; Nie, T.; Yang, Z.; Ding, N.; Du, S.X.; Liu, S.; Peng, T. Silencing of PPMD1 inhibits proliferation of human colon cancer cells via induction of apoptosis and cell cycle arrest. J. Buon. 2019, 24, 1464–1469. [Google Scholar]
- Raab, C.A.; Raab, M.; Becker, S.; Strebhardt, K. Non-mitotic functions of polo-like kinases in cancer cells. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188467. [Google Scholar] [CrossRef]
- Han, J.; Zhang, L.; Zhang, J.; Jiang, Q.; Tong, D.; Wang, X.; Gao, X.; Zhao, L.; Huang, C. CREBRF promotes the proliferation of human gastric cancer cells via the AKT signaling pathway. Cell. Mol. Biol. 2018, 64, 40–45. [Google Scholar] [CrossRef]
- Xue, H.; Zhang, J.; Guo, X.; Wang, J.; Li, J.; Gao, X.; Guo, X.; Li, T.; Xu, S.; Zhang, P.; et al. CREBRF is a potent tumor suppressor of glioblastoma by blocking hypoxia-induced autophagy via the CREB3/ATG5 pathway. Int. J. Oncol. 2016, 49, 519–528. [Google Scholar] [CrossRef]
- Fahraeus, R.; Olivares-Illana, V. MDM2’s social network. Oncogene 2014, 33, 4365–4376. [Google Scholar] [CrossRef]
- Wade, M.; Li, Y.C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliner, J.D.; Saiki, A.Y.; Caenepeel, S. The Role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb. Perspect. Med. 2016, 6, a026336. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, N.; Nakamura, Y.; Miyamoto, Y.; Miyamoto, T.; Kabu, K.; Yoshida, M.; Futamura, M.; Ichinose, S.; Arakawa, H. Mieap, a p53-inducible protein, controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLoS ONE 2011, 6, e16060. [Google Scholar] [CrossRef] [PubMed]
- Dan, X.; Babbar, M.; Moore, A.; Wechter, N.; Tian, J.; Mohanty, J.G.; Croteau, D.L.; Bohr, V.A. DNA damage invokes mitophagy through a pathway involving Spata18. Nucleic Acids Res. 2021, 48, 6611–6623. [Google Scholar] [CrossRef]
- Tomasini, R.; Samir, A.A.; Pebusque, M.J.; Calvo, E.L.; Totaro, S.; Dagorn, J.C.; Dusetti, N.J.; Iovanna, J.L. P53-dependent expression of the stress-induced protein (SIP). Eur. J. Cell Biol. 2002, 81, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Motoo, Y.; Yoshida, H.; Iovanna, J.L.; Nakamura, Y.; Kuma, K.; Miyauchi, A. High level of tumour protein p53-induced nuclear protein 1 (TP53INP1) expression in anaplastic carcinoma of the thyroid. Pathology 2006, 38, 545–547. [Google Scholar] [CrossRef]
- Taïeb, D.; Giusiano, S.; Sebag, F.; Marcy, M.; De Micco, C.; Palazzo, F.F.; Dusetti, N.J.; Iovanna, J.L.; Henry, J.F.; Garcia, S.; et al. Tumor protein p53-induced nuclear protein (TP53INP1) expression in medullary thyroid carcinoma: A molecular guide to the optimal extent of surgery? World J. Surg. 2010, 34, 830–835. [Google Scholar] [CrossRef]
- Giusiano, S.; Garcia, S.; Andrieu, C.; Dusetti, N.J.; Bastide, C.; Gleave, M.; Taranger-Charpin, C.; Iovanna, J.L.; Rocchi, P. TP53INP1 overexpression in prostate cancer correlates with poor prognostic factors and is predictive of biological cancer relapse. Prostate 2012, 72, 117–128. [Google Scholar] [CrossRef]
- Lin, Y.C.; Boone, M.; Meuris, L.; Lemmens, I.; Roy, N.V.; Soete, A.; Reumers, J.; Moisse, M.; Plaisance, S.; Drmanac, R.T.; et al. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun. 2014, 5, 4767. [Google Scholar] [CrossRef] [Green Version]
- Sheppard, H.M.; Corneillie, S.I.; Espiritu, C.; Gatti, A.; Liu, X. New insights into the mechanism of inhibition of p53 by Simian virus 40 large T antigen. Mol. Cell. Biol. 1999, 19, 2746–2753. [Google Scholar] [CrossRef] [Green Version]
- Van Drie, J.H. Protein folding, protein homeostasis, and cancer. Chin. J. Cancer 2011, 30, 124–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pluquet, O.; Pourtier, A.; Abbadie, C. The unfolded protein response and cellular senescence. A review in the theme: Cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. Am. J. Physiol. Cell Physiol. 2011, 308, C415–C425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorgoulis, V.G.; Pefani, D.E.; Pateras, I.S.; Trougakos, I.P. Integrating the DNA damage and protein stress responses during cancer development and treatment. J. Pathol. 2018, 246, 12–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufey, E.; Pedro, J.M.B.-S.; Eggers, C.; González-Quiroz, M.; Urra, H.; Sagredo, A.I.; Sepulveda, D.; Pihán, P.; Carreras-Sureda, A.; Hazari, Y.; et al. Genotoxic stress triggers the activation of IRE1alpha-dependent RNA decay to modulate the DNA damage response. Nat. Commun. 2020, 11, 2401. [Google Scholar] [CrossRef] [PubMed]
- Bolland, H.; Ma, T.S.; Ramlee, S.; Ramadan, K.; Hammond, E.M. Links between the unfolded protein response and the DNA damage response in hypoxia: A systematic review. Biochem. Soc. Trans. 2021, 49, 1251–1263. [Google Scholar] [CrossRef]
- Scott, M.D.; Frydman, J. Aberrant protein folding as the molecular basis of cancer. Methods Mol. Biol. 2003, 232, 67–76. [Google Scholar] [CrossRef]
- Kato, H.; Nishitoh, H. Stress responses from the endoplasmic reticulum in cancer. Front. Oncol. 2015, 5, 93. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Brewer, M.D.; Guo, L.; Wang, R.; Jiang, P.; Yang, X. Enhanced degradation of misfolded proteins promotes tumorigenesis. Cell Rep. 2017, 18, 3143–3154. [Google Scholar] [CrossRef]
- Lindeboom, R.G.; Supek, F.; Lehner, B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 2016, 48, 1112–1118. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Franklin, D.A.; Dong, J.; Zhang, Y. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res. 2014, 74, 7161–7167. [Google Scholar] [CrossRef] [Green Version]
- Liebl, M.C.; Hofmann, T.G. The role of p53 signaling in colorectal cancer. Cancers 2021, 13, 2125. [Google Scholar] [CrossRef] [PubMed]
- Karni-Schmidt, O.; Lokshin, M.; Prives, C. The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 2016, 11, 617–644. [Google Scholar] [CrossRef] [PubMed]
- Ochkasova, A.S.; Meschaninova, M.I.; Venyaminova, A.G.; Ivanov, A.V.; Graifer, D.M.; Karpova, G.G. The human ribosome can interact with the abasic site in mRNA via a specific peptide of the uS3 protein located near the mRNA entry channel. Biochimie 2019, 158, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Natchiar, S.K.; Myasnikov, A.G.; Hazemann, I.; Klaholz, B.P. Visualizing the role of 2’-OH rRNA methylations in the human ribosome structure. Biomolecules 2018, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Peng, T.-S.; He, Y.-H.; Nie, T.; Hu, X.-D.; Lu, H.-Y.; Yi, J.; Shuai, Y.-F.; Luo, M. PPM1D is a prognostic marker and therapeutic target in colorectal cancer. Exp. Ther. Med. 2014, 8, 430–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [Green Version]
- Ou, B.; Zhao, J.; Guan, S.; Wangpu, X.; Zhu, C.; Zong, Y.; Ma, J.; Sun, J.; Zheng, M.; Feng, H.; et al. Plk2 promotes tumor growth and inhibits apoptosis by targeting Fbxw7/Cyclin E in colorectal cancer. Cancer Lett. 2016, 380, 457–466. [Google Scholar] [CrossRef]
- Yanshina, D.D.; Gopanenko, A.V.; Karpova, G.G.; Malygin, A.A. Replacement of hydroxylated His39 in ribosomal protein uL15 with Ala or Thr impairs the translational activity of human ribosomes. Mol. Biol. 2020, 54, 512–521. [Google Scholar] [CrossRef]
- Babaylova, E.S.; Gopanenko, A.V.; Bulygin, K.N.; Tupikin, A.E.; Kabilov, M.R.; Malygin, A.A.; Karpova, G.G. mRNA regions where 80S ribosomes pause during translation elongation in vivo interact with protein uS19, a component of the decoding site. Nucleic Acids Res. 2020, 48, 912–923. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Conesa, A.; Garcia-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Gopanenko, A.V.; Kolobova, A.V.; Meschaninova, M.I.; Venyaminova, A.G.; Tupikin, A.E.; Kabilov, M.R.; Malygin, A.A.; Karpova, G.G. Knockdown of the mRNA encoding the ribosomal protein eL38 in mammalian cells causes a substantial reorganization of genomic transcription. Biochimie 2021, 184, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [Green Version]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Yu, G.; He, Q.Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Babaylova, E.S.; Gopanenko, A.V.; Tupikin, A.E.; Kabilov, M.R.; Malygin, A.A.; Karpova, G.G. Changes in the Transcriptome Caused by Mutations in the Ribosomal Protein uS10 Associated with a Predisposition to Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 6174. https://doi.org/10.3390/ijms23116174
Tian Y, Babaylova ES, Gopanenko AV, Tupikin AE, Kabilov MR, Malygin AA, Karpova GG. Changes in the Transcriptome Caused by Mutations in the Ribosomal Protein uS10 Associated with a Predisposition to Colorectal Cancer. International Journal of Molecular Sciences. 2022; 23(11):6174. https://doi.org/10.3390/ijms23116174
Chicago/Turabian StyleTian, Yueming, Elena S. Babaylova, Alexander V. Gopanenko, Alexey E. Tupikin, Marsel R. Kabilov, Alexey A. Malygin, and Galina G. Karpova. 2022. "Changes in the Transcriptome Caused by Mutations in the Ribosomal Protein uS10 Associated with a Predisposition to Colorectal Cancer" International Journal of Molecular Sciences 23, no. 11: 6174. https://doi.org/10.3390/ijms23116174
APA StyleTian, Y., Babaylova, E. S., Gopanenko, A. V., Tupikin, A. E., Kabilov, M. R., Malygin, A. A., & Karpova, G. G. (2022). Changes in the Transcriptome Caused by Mutations in the Ribosomal Protein uS10 Associated with a Predisposition to Colorectal Cancer. International Journal of Molecular Sciences, 23(11), 6174. https://doi.org/10.3390/ijms23116174







