Characterization of the Biomass Degrading Enzyme GuxA from Acidothermus cellulolyticus
Abstract
:1. Introduction
2. Results and Discussion
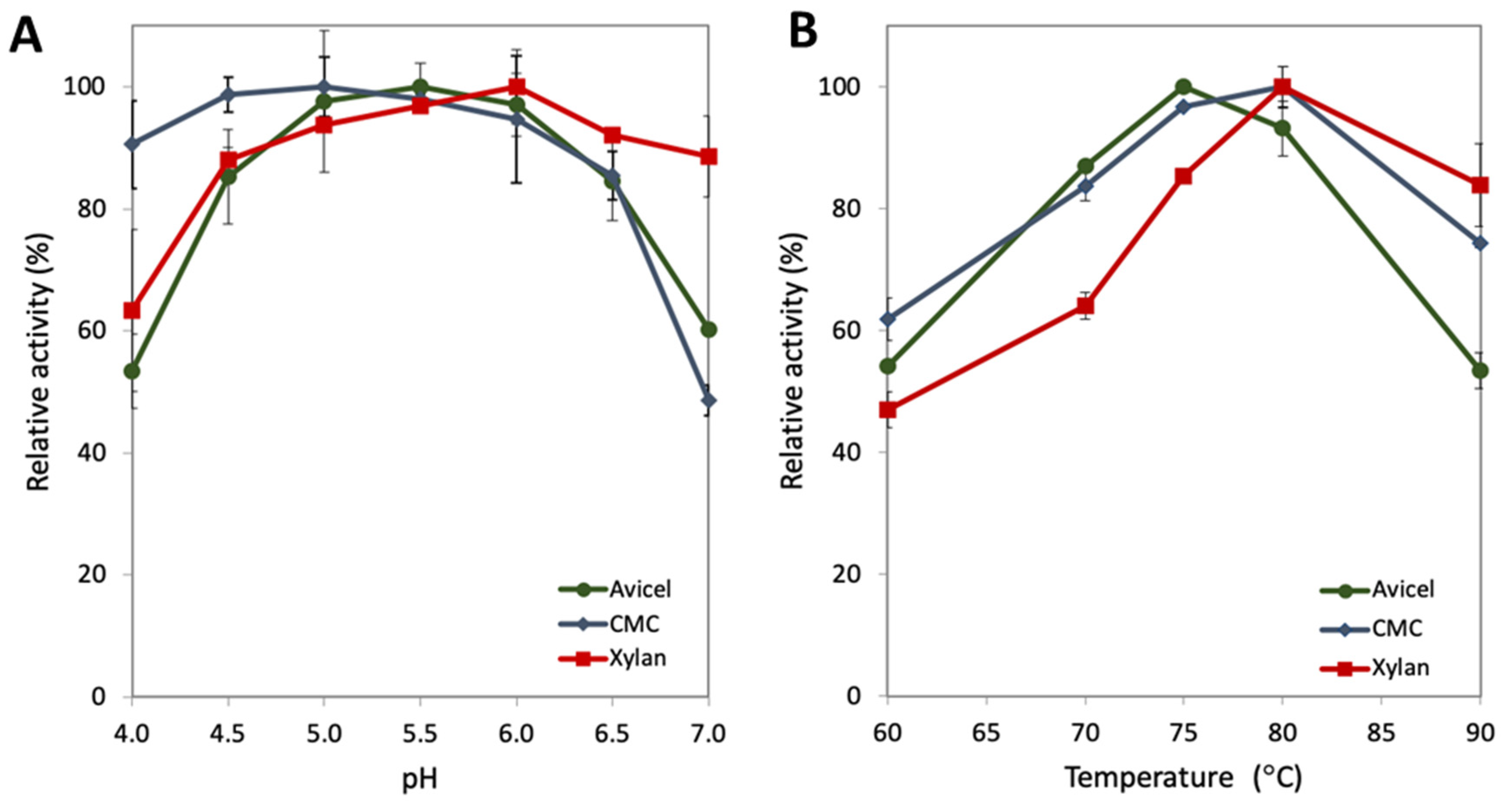
2.1. Characterization of the pH and Temperature Range for GuxA Activity
2.2. Activity of GuxA and Synergy with the Endoglucanase E1 on the Model Substrate Avicel
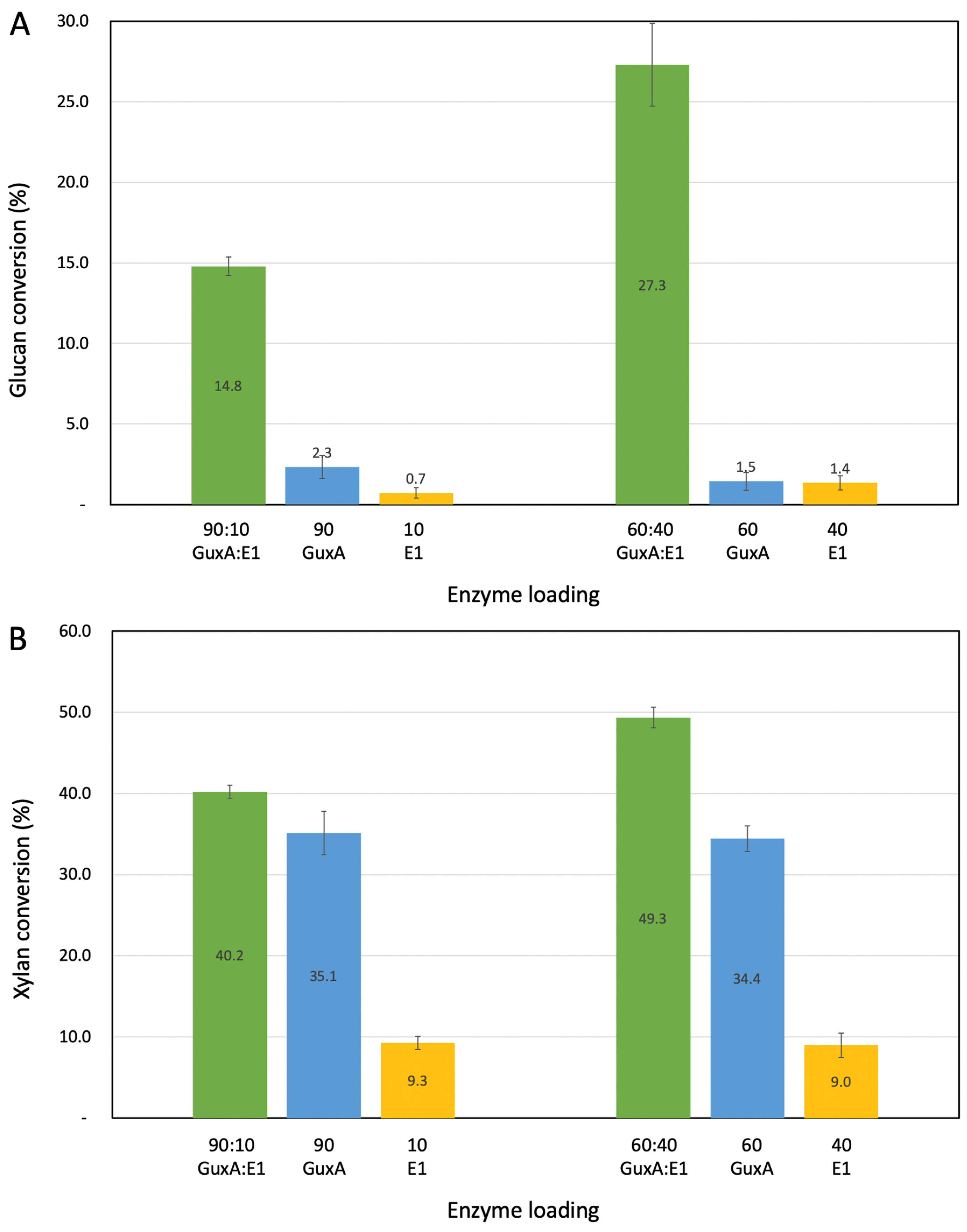
2.3. Activity of GuxA and Synergy with the Endoglucanase E1 on Pretreated Biomass
2.4. Structural Features of the GuxA GH12 Domain
3. Conclusions
4. Materials and Methods
4.1. Construction of C. bescii Strains and Media Composition
4.2. Protein Expression in C. bescii and E. coli
4.3. Protein Purification
4.4. Enzyme Activity Assays
4.5. Crystallization of the GuxA GH12 Domain
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Himmel, M.E.; Ding, S.-Y.; Johnson, D.K.; Adney, W.S.; Nimlos, M.R.; Brady, J.W.; Foust, T.D. Biomass Recalcitrance: Engineering Plants and Enzymes for Biofuels Production. Science 2007, 315, 804–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCann, M.C.; Carpita, N.C. Designing the Deconstruction of Plant Cell Walls. Curr. Opin. Plant Biol. 2008, 11, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Klein-Marcuschamer, D.; Oleskowicz-Popiel, P.; Simmons, B.A.; Blanch, H.W. The Challenge of Enzyme Cost in the Production of Lignocellulosic Biofuels. Biotechnol. Bioeng. 2012, 109, 1083–1087. [Google Scholar] [CrossRef] [PubMed]
- Kun, R.S.; Gomes, A.C.S.; Hildén, K.S.; Salazar Cerezo, S.; Mäkelä, M.R.; de Vries, R.P. Developments and Opportunities in Fungal Strain Engineering for the Production of Novel Enzymes and Enzyme Cocktails for Plant Biomass Degradation. Biotechnol. Adv. 2019, 37, 107361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynd, L.R.; Weimer, P.J.; Zyl, W.H.; van Pretorius, I.S. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [Green Version]
- Bomble, Y.J.; Lin, C.-Y.; Amore, A.; Wei, H.; Holwerda, E.K.; Ciesielski, P.N.; Donohoe, B.S.; Decker, S.R.; Lynd, L.R.; Himmel, M.E. Lignocellulose Deconstruction in the Biosphere. Curr. Opin. Chem. Biol. 2017, 41, 61–70. [Google Scholar] [CrossRef]
- Brunecky, R.; Alahuhta, M.; Xu, Q.; Donohoe, B.S.; Crowley, M.F.; Kataeva, I.A.; Yang, S.-J.; Resch, M.G.; Adams, M.W.W.; Lunin, V.V.; et al. Revealing Nature’s Cellulase Diversity: The Digestion Mechanism of Caldicellulosiruptor Bescii CelA. Science 2013, 342, 1513–1516. [Google Scholar] [CrossRef] [Green Version]
- Bayer, E.A.; Belaich, J.-P.; Shoham, Y.; Lamed, R. The Cellulosomes: Multienzyme Machines for Degradation of Plant Cell Wall Polysaccharides. Annu. Rev. Microbiol. 2004, 58, 521–554. [Google Scholar] [CrossRef]
- Brunecky, R.; Chung, D.; Sarai, N.S.; Hengge, N.; Russell, J.F.; Young, J.; Mittal, A.; Pason, P.; Vander Wall, T.; Michener, W.; et al. High Activity CAZyme Cassette for Improving Biomass Degradation in Thermophiles. Biotechnol. Biofuels 2018, 11, 22. [Google Scholar] [CrossRef] [Green Version]
- Conway, J.M.; Crosby, J.R.; Hren, A.P.; Southerland, R.T.; Lee, L.L.; Lunin, V.V.; Alahuhta, P.; Himmel, M.E.; Bomble, Y.J.; Adams, M.W.W.; et al. Novel Multidomain, Multifunctional Glycoside Hydrolases from Highly Lignocellulolytic Caldicellulosiruptor Species. AIChE J. 2018, 64, 4218–4228. [Google Scholar] [CrossRef]
- Brunecky, R.; Subramanian, V.M.; Yarbrough, J.S.; Donohoe, B.B.; Vinzant, T.A.; Vanderwall, T.C.; Knott, B.B.; Chaudhari, Y.J.; Bomble, Y.E.; Himmel, M.; et al. Synthetic Fungal Multifunctional Cellulases for Enhanced Biomass Conversion. Green Chem. 2020, 22, 478–489. [Google Scholar] [CrossRef] [Green Version]
- Chung, D.; Young, J.; Cha, M.; Brunecky, R.; Bomble, Y.J.; Himmel, M.E.; Westpheling, J. Expression of the Acidothermus Cellulolyticus E1 Endoglucanase in Caldicellulosiruptor Bescii Enhances Its Ability to Deconstruct Crystalline Cellulose. Biotechnol. Biofuels 2015, 8, 113. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Chung, D.; Himmel, M.E.; Bomble, Y.J.; Westpheling, J. In Vivo Synergistic Activity of a CAZyme Cassette from Acidothermus Cellulolyticus Significantly Improves the Cellulolytic Activity of the C. Bescii Exoproteome. Biotechnol. Bioeng. 2017, 114, 2474–2480. [Google Scholar] [CrossRef] [PubMed]
- Simpson, P.J.; Xie, H.; Bolam, D.N.; Gilbert, H.J.; Williamson, M.P. The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate-Binding Modules*. J. Biol. Chem. 2000, 275, 41137–41142. [Google Scholar] [CrossRef] [Green Version]
- Lochner, A.; Giannone, R.J.; Rodriguez, M.; Shah, M.B.; Mielenz, J.R.; Keller, M.; Antranikian, G.; Graham, D.E.; Hettich, R.L. Use of Label-Free Quantitative Proteomics To Distinguish the Secreted Cellulolytic Systems of Caldicellulosiruptor Bescii and Caldicellulosiruptor Obsidiansis. Appl. Environ. Microbiol. 2011, 77, 4042–4054. [Google Scholar] [CrossRef] [Green Version]
- Tucker, M.P.; Mohagheghi, A.; Grohmann, K.; Himmel, M.E. Ultra-Thermostable Cellulases From Acidothermus Cellulolyticus: Comparison of Temperature Optima with Previously Reported Cellulases. Bio/Technology 1989, 7, 817–820. [Google Scholar] [CrossRef]
- Maheshwari, R.; Bharadwaj, G.; Bhat, M.K. Thermophilic Fungi: Their Physiology and Enzymes. Microbiol. Mol. Biol. Rev. 2000, 64, 461–488. [Google Scholar] [CrossRef] [Green Version]
- Pradeep, G.C.; Choi, Y.H.; Choi, Y.S.; Seong, C.N.; Cho, S.S.; Lee, H.J.; Yoo, J.C. A Novel Thermostable Cellulase Free Xylanase Stable in Broad Range of PH from Streptomyces Sp. CS428. Process Biochem. 2013, 48, 1188–1196. [Google Scholar] [CrossRef]
- Kamble, R.D.; Jadhav, A.R. Production, Purification and Characterisation of Alkali Stable Xylanase from Cellulosimicrobium Sp. MTCC 10645. Asian Pac. J. Trop. Biomed. 2012, 2, S1790–S1797. [Google Scholar] [CrossRef]
- Karp, E.M.; Donohoe, B.S.; O’Brien, M.H.; Ciesielski, P.N.; Mittal, A.; Biddy, M.J.; Beckham, G.T. Alkaline Pretreatment of Corn Stover: Bench-Scale Fractionation and Stream Characterization. ACS Sustain. Chem. Eng. 2014, 2, 1481–1491. [Google Scholar] [CrossRef]
- Kim, H.-W.; Kataoka, M.; Ishikawa, K. Atomic Resolution of the Crystal Structure of the Hyperthermophilic Family 12 Endocellulase and Stabilizing Role of the DxDxDG Calcium-Binding Motif in Pyrococcus Furiosus. FEBS Lett. 2012, 586, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.W.; Driskill, L.E.; Callen, W.; Snead, M.A.; Mathur, E.J.; Kelly, R.M. An Endoglucanase, EglA, from the Hyperthermophilic ArchaeonPyrococcus Furiosus Hydrolyzes β-1,4 Bonds in Mixed-Linkage (1 → 3),(1 → 4)-β-d-Glucans and Cellulose. J. Bacteriol. 1999, 181, 284–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gloster, T.M.; Ibatullin, F.M.; Macauley, K.; Eklöf, J.M.; Roberts, S.; Turkenburg, J.P.; Bjørnvad, M.E.; Jørgensen, P.L.; Danielsen, S.; Johansen, K.S.; et al. Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families GH5 and GH12. J. Biol. Chem. 2007, 282, 19177–19189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karim, A.; Nawaz, M.A.; Aman, A.; Ul Qader, S.A. Role of Anionic Polysaccharide (Alginate) on Activity, Stability and Recycling Efficiency of Bacterial Endo (1 → 4) β-d-Glucanase of GH12 Family. Catal. Lett. 2017, 147, 1792–1801. [Google Scholar] [CrossRef]
- Crennell, S.J.; Hreggvidsson, G.O.; Nordberg Karlsson, E. The Structure of Rhodothermus Marinus Cel12A, A Highly Thermostable Family 12 Endoglucanase, at 1.8Å Resolution. J. Mol. Biol. 2002, 320, 883–897. [Google Scholar] [CrossRef]
- Halldórsdóttir, S.; Thórólfsdóttir, E.T.; Spilliaert, R.; Johansson, M.; Thorbjarnardóttir, S.H.; Palsdottir, A.; Hreggvidsson, G.Ó.; Kristjánsson, J.K.; Holst, O.; Eggertsson, G. Cloning, Sequencing and Overexpression of a Rhodothermus Marinus Gene Encoding a Thermostable Cellulase of Glycosyl Hydrolase Family 12. Appl. Microbiol. Biotechnol. 1998, 49, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Sulzenbacher, G.; Shareck, F.; Morosoli, R.; Dupont, C.; Davies, G.J. The Streptomyces Lividans Family 12 Endoglucanase: Construction of the Catalytic Cre, Expression, and X-Ray Structure at 1.75 A Resolution. Biochemistry 1997, 36, 16032–16039. [Google Scholar] [CrossRef]
- Kluepfel, D.; Shareck, F.; Mondou, F.; Morosoli, R. Characterization of Cellulase and Xylanase Activities of Streptomyces Lividans. Appl. Microbiol. Biotechnol. 1986, 24, 230–234. [Google Scholar] [CrossRef]
- Sandgren, M.; Gualfetti, P.J.; Shaw, A.; Gross, L.S.; Saldajeno, M.; Day, A.G.; Jones, T.A.; Mitchinson, C. Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability. Protein Sci. 2003, 12, 848–860. [Google Scholar] [CrossRef]
- Cheng, Y.-S.; Ko, T.-P.; Wu, T.-H.; Ma, Y.; Huang, C.-H.; Lai, H.-L.; Wang, A.H.-J.; Liu, J.-R.; Guo, R.-T. Crystal Structure and Substrate-Binding Mode of Cellulase 12A from Thermotoga Maritima. Proteins 2011, 79, 1193–1204. [Google Scholar] [CrossRef]
- Cheng, Y.-S.; Ko, T.-P.; Huang, J.-W.; Wu, T.-H.; Lin, C.-Y.; Luo, W.; Li, Q.; Ma, Y.; Huang, C.-H.; Wang, A.H.-J.; et al. Enhanced Activity of Thermotoga Maritima Cellulase 12A by Mutating a Unique Surface Loop. Appl. Microbiol. Biotechnol. 2012, 95, 661–669. [Google Scholar] [CrossRef]
- Okano, H.; Ozaki, M.; Kanaya, E.; Kim, J.-J.; Angkawidjaja, C.; Koga, Y.; Kanaya, S. Structure and Stability of Metagenome-Derived Glycoside Hydrolase Family 12 Cellulase (LC-CelA) a Homolog of Cel12A from Rhodothermus Marinus. FEBS Open Bio 2014, 4, 936–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.-W.; Liu, W.; Lai, H.-L.; Cheng, Y.-S.; Zheng, Y.; Li, Q.; Sun, H.; Kuo, C.-J.; Guo, R.-T.; Chen, C.-C. Crystal Structure and Genetic Modifications of FI-CMCase from Aspergillus Aculeatus F-50. Biochem. Biophys. Res. Commun. 2016, 478, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T.; Shimizu, T.; Hirano, H.; Sato, M.; Hashimoto, H. Structural Basis for Inhibition of Xyloglucan-Specific Endo-β-1,4-Glucanase (XEG) by XEG-Protein Inhibitor. J. Biol. Chem. 2012, 287, 18710–18716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashid, M.H.; Javed, M.R.; Kawaguchi, T.; Sumitani, J.; Arai, M. Improvement of Aspergillus Oryzae for Hyperproduction of Endoglucanase: Expression Cloning of Cmc-1 Gene of Aspergillus Aculeatus. Biotechnol. Lett. 2008, 30, 2165. [Google Scholar] [CrossRef]
- Khademi, S.; Zhang, D.; Swanson, S.M.; Wartenberg, A.; Witte, K.; Meyer, E.F. Determination of the Structure of an Endoglucanase from Aspergillus Niger and Its Mode of Inhibition by Palladium Chloride. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 660–667. [Google Scholar] [CrossRef] [Green Version]
- Master, E.R.; Zheng, Y.; Storms, R.; Tsang, A.; Powlowski, J. A Xyloglucan-Specific Family 12 Glycosyl Hydrolase from Aspergillus Niger: Recombinant Expression, Purification and Characterization. Biochem. J. 2008, 411, 161–170. [Google Scholar] [CrossRef] [Green Version]
- RCSB PDB—4NPR: Crystal Structure of the Family 12 Xyloglucanase from Aspergillus Niveus. Available online: https://www.rcsb.org/structure/4NPR (accessed on 8 March 2021).
- Damásio, A.R.L.; Ribeiro, L.F.C.; Ribeiro, L.F.; Furtado, G.P.; Segato, F.; Almeida, F.B.R.; Crivellari, A.C.; Buckeridge, M.S.; Souza, T.A.C.B.; Murakami, M.T.; et al. Functional Characterization and Oligomerization of a Recombinant Xyloglucan-Specific Endo-β-1,4-Glucanase (GH12) from Aspergillus Niveus. Biochim. Biophys. Acta BBA—Proteins Proteom. 2012, 1824, 461–467. [Google Scholar] [CrossRef]
- Prates, É.T.; Stankovic, I.; Silveira, R.L.; Liberato, M.V.; Henrique-Silva, F.; Pereira, N.; Polikarpov, I.; Skaf, M.S. X-Ray Structure and Molecular Dynamics Simulations of Endoglucanase 3 from Trichoderma Harzianum: Structural Organization and Substrate Recognition by Endoglucanases That Lack Cellulose Binding Module. PLoS ONE 2013, 8, e59069. [Google Scholar] [CrossRef]
- Generoso, W.C.; Malagó, W., Jr.; Pereira, N., Jr.; Henrique-Silva, F. Recombinant Expression and Characterization of an Endoglucanase III (Cel12a) from Trichoderma Harzianum (Hypocreaceae) in the Yeast Pichia Pastoris. Genet. Mol. Res. 2012, 11, 1544–1557. [Google Scholar] [CrossRef]
- Sandgren, M.; Shaw, A.; Ropp, T.H.; Wu, S.; Bott, R.; Cameron, A.D.; Ståhlberg, J.; Mitchinson, C.; Jones, T.A. The X-Ray Crystal Structure of the Trichoderma Reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution. J. Mol. Biol. 2001, 308, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Sandgren, M.; Ståhlberg, J.; Mitchinson, C. Structural and Biochemical Studies of GH Family 12 Cellulases: Improved Thermal Stability, and Ligand Complexes. Prog. Biophys. Mol. Biol. 2005, 89, 246–291. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Young, J.; Bomble, Y.J.; Wall, T.A.V.; Groom, J.; Himmel, M.E.; Westpheling, J. Homologous Expression of the Caldicellulosiruptor Bescii CelA Reveals That the Extracellular Protein Is Glycosylated. PLoS ONE 2015, 10, e0119508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunecky, R.; Donohoe, B.S.; Yarbrough, J.M.; Mittal, A.; Scott, B.R.; Ding, H.; Ii, L.E.T.; Russell, J.F.; Chung, D.; Westpheling, J.; et al. The Multi Domain Caldicellulosiruptor Bescii CelA Cellulase Excels at the Hydrolysis of Crystalline Cellulose. Sci. Rep. 2017, 7, 9622. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular Replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef] [PubMed]
- RCSB PDB—3B7M: Crystal Structure of a Meso-Active Thermo-Stable Cellulase (MT Cel12A) Derived by Making Non-Contiguous Mutations in the Active Surface of the Cel12A Cellulase of Rhodothermus Marinus. Available online: https://www.rcsb.org/structure/3B7M (accessed on 5 March 2021).
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- PyMOL|Pymol.Org. Available online: https://pymol.org/2/ (accessed on 2 March 2021).
- YASARA—Yet Another Scientific Artificial Reality Application. Available online: http://www.yasara.org/ (accessed on 2 March 2021).


| GH12 Source Organism | PDB | Cα RMSD w/7MKR (Å) | Temperature Optimum (°C) | Hydrogen Bonds | Hydrophobic Interactions | Salt Bridges | Disulfide Bridges | Ala35 Equivalent |
|---|---|---|---|---|---|---|---|---|
| Acidothermus cellulolyticus | 7MKR | 0.000 | 80 | 176 | 1034 | 3 | 2 | L |
| Pyrococcus furiosus | 3VGI [21] | 1.397 | 100 [22] | 212 | 1652 | 20 | 0 | Y |
| Bacillus licheniformis | 2JEM [23] | 1.37 | 60 [24] | 187 | 1206 | 9 | 0 | H |
| Rhodothermus marinus ITI378 | 2BWA [25] | 0.967 | 100 [26] | 173 | 1157 | 9 | 2 | V |
| Streptomyces lividans | 2NLR [27] | 1.188 | 60 [28] | 171 | 1054 | 5 | 2 | A |
| Streptomyces sp 11AG8 | 1OA4 [29] | 1.237 | ≥60 [29] | 174 | 1022 | 7 | 2 | V |
| Thermotoga maritima | 3AMH [30] | 1.425 | 100 [31] | 202 | 1624 | 20 | 0 | F |
| Uncultured bacterium | 3WX5 [32] | 0.98 | 90 [32] | 173 | 1211 | 9 | 2 | I |
| Aspergillus aculeatus F-50 | 5GM4 [33] | 1.27 | 50 [33] | 169 | 1044 | 4 | 1 | V |
| Aspergillus aculeatus KSM 510 | 3VL8 [34] | 1.344 | 50 [35] | 159 | 1037 | 2 | 1 | V |
| Aspergillus niger CBS 120.49/N400 | 1KS4 [36] | 1.324 | 60 [37] | 141 | 1002 | 4 | 1 | V |
| Aspergillus niveus PR-2 | 4NPR [38] | 1.279 | 60 [39] | 156 | 1061 | 2 | 1 | F |
| Hypocrea schweinitzii ATCC 66965 | 1OA3 [29] | 1.253 | 50 [29] | 168 | 1032 | 3 | 1 | S |
| Trichoderma harzianum IOC-3844 | 4H7M [40] | 1.293 | 48 [41] | 166 | 1044 | 4 | 1 | V |
| Trichoderma reesei QM9414 | 1H8V [42] | 1.264 | 50 [29] | 157 | 991 | 3 | 1 | A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hengge, N.N.; Mallinson, S.J.B.; Pason, P.; Lunin, V.V.; Alahuhta, M.; Chung, D.; Himmel, M.E.; Westpheling, J.; Bomble, Y.J. Characterization of the Biomass Degrading Enzyme GuxA from Acidothermus cellulolyticus. Int. J. Mol. Sci. 2022, 23, 6070. https://doi.org/10.3390/ijms23116070
Hengge NN, Mallinson SJB, Pason P, Lunin VV, Alahuhta M, Chung D, Himmel ME, Westpheling J, Bomble YJ. Characterization of the Biomass Degrading Enzyme GuxA from Acidothermus cellulolyticus. International Journal of Molecular Sciences. 2022; 23(11):6070. https://doi.org/10.3390/ijms23116070
Chicago/Turabian StyleHengge, Neal N., Sam J. B. Mallinson, Patthra Pason, Vladimir V. Lunin, Markus Alahuhta, Daehwan Chung, Michael E. Himmel, Janet Westpheling, and Yannick J. Bomble. 2022. "Characterization of the Biomass Degrading Enzyme GuxA from Acidothermus cellulolyticus" International Journal of Molecular Sciences 23, no. 11: 6070. https://doi.org/10.3390/ijms23116070
APA StyleHengge, N. N., Mallinson, S. J. B., Pason, P., Lunin, V. V., Alahuhta, M., Chung, D., Himmel, M. E., Westpheling, J., & Bomble, Y. J. (2022). Characterization of the Biomass Degrading Enzyme GuxA from Acidothermus cellulolyticus. International Journal of Molecular Sciences, 23(11), 6070. https://doi.org/10.3390/ijms23116070







