Effects of Individual Amino Acids on PPARα Transactivation, mTORC1 Activation, ApoA-I Transcription and pro-ApoA-I Secretion
Abstract
:1. Introduction
2. Results
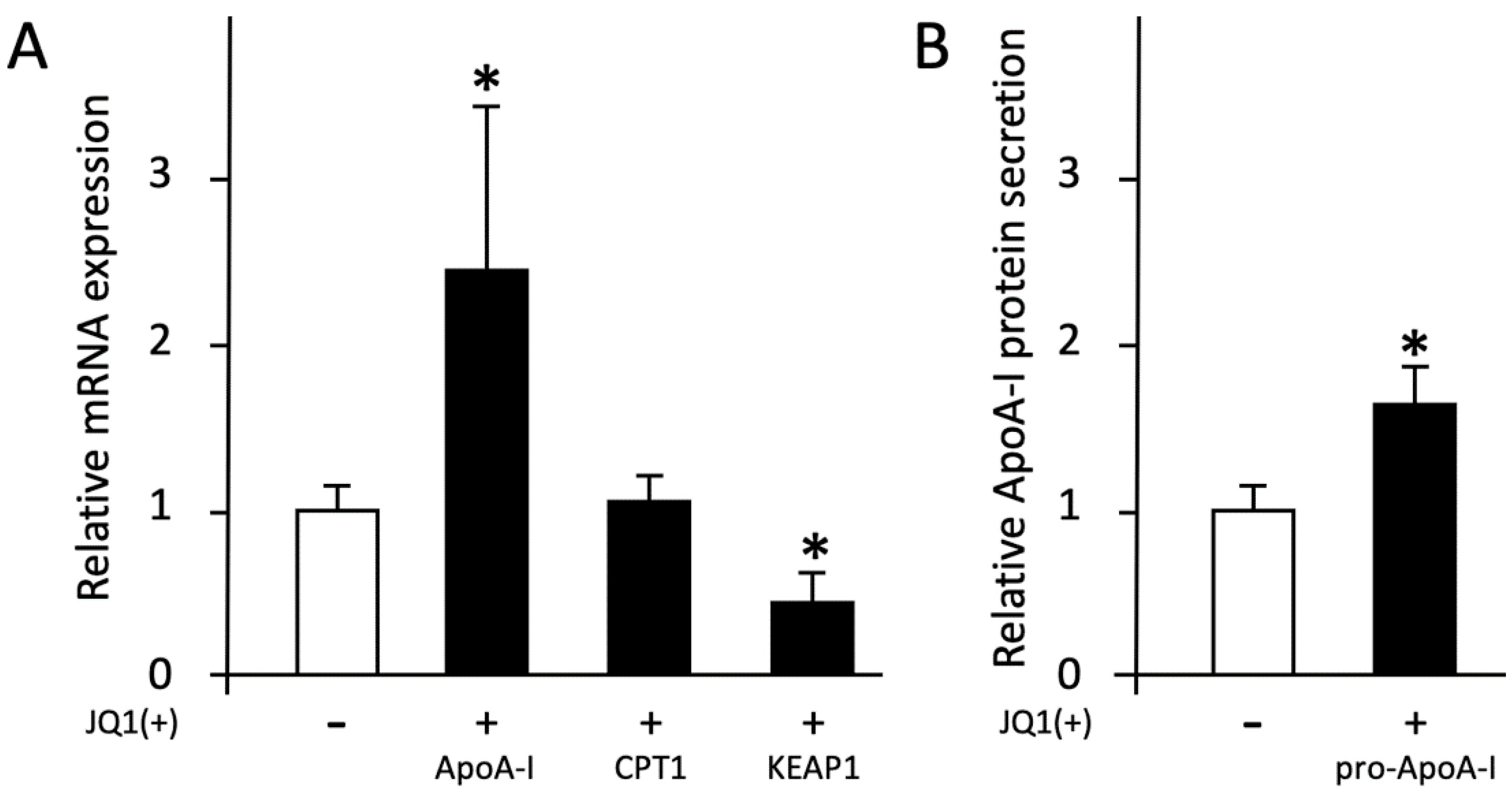
2.1. Effects of BET Inhibitor JQ1(+)
2.2. Effects of Different Amino Acids on ApoA-I mRNA Expression and pro-ApoA-I Secretion
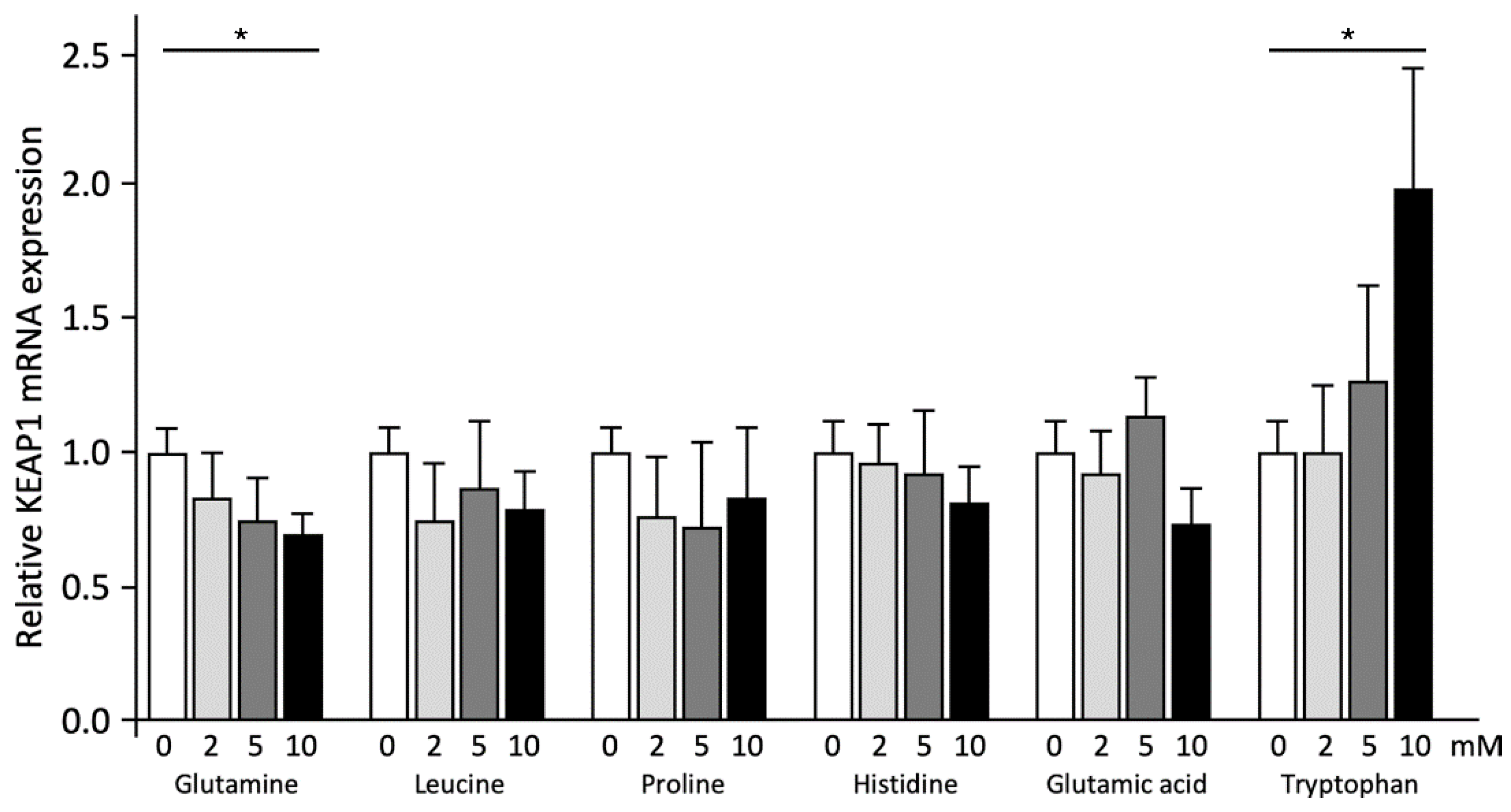
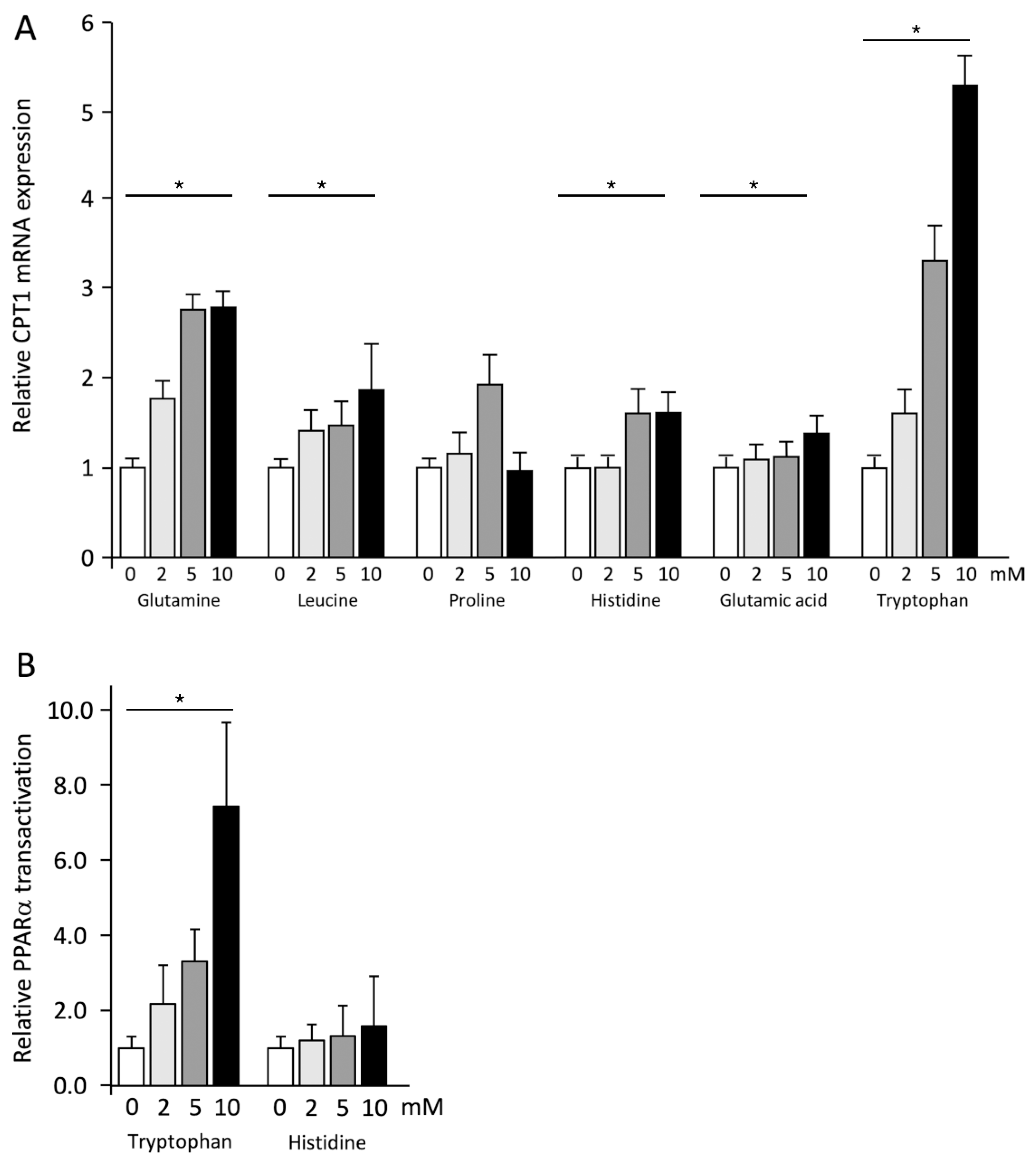
2.3. Effects of Different Amino Acids on BET Inhibition and PPARα Transactivation
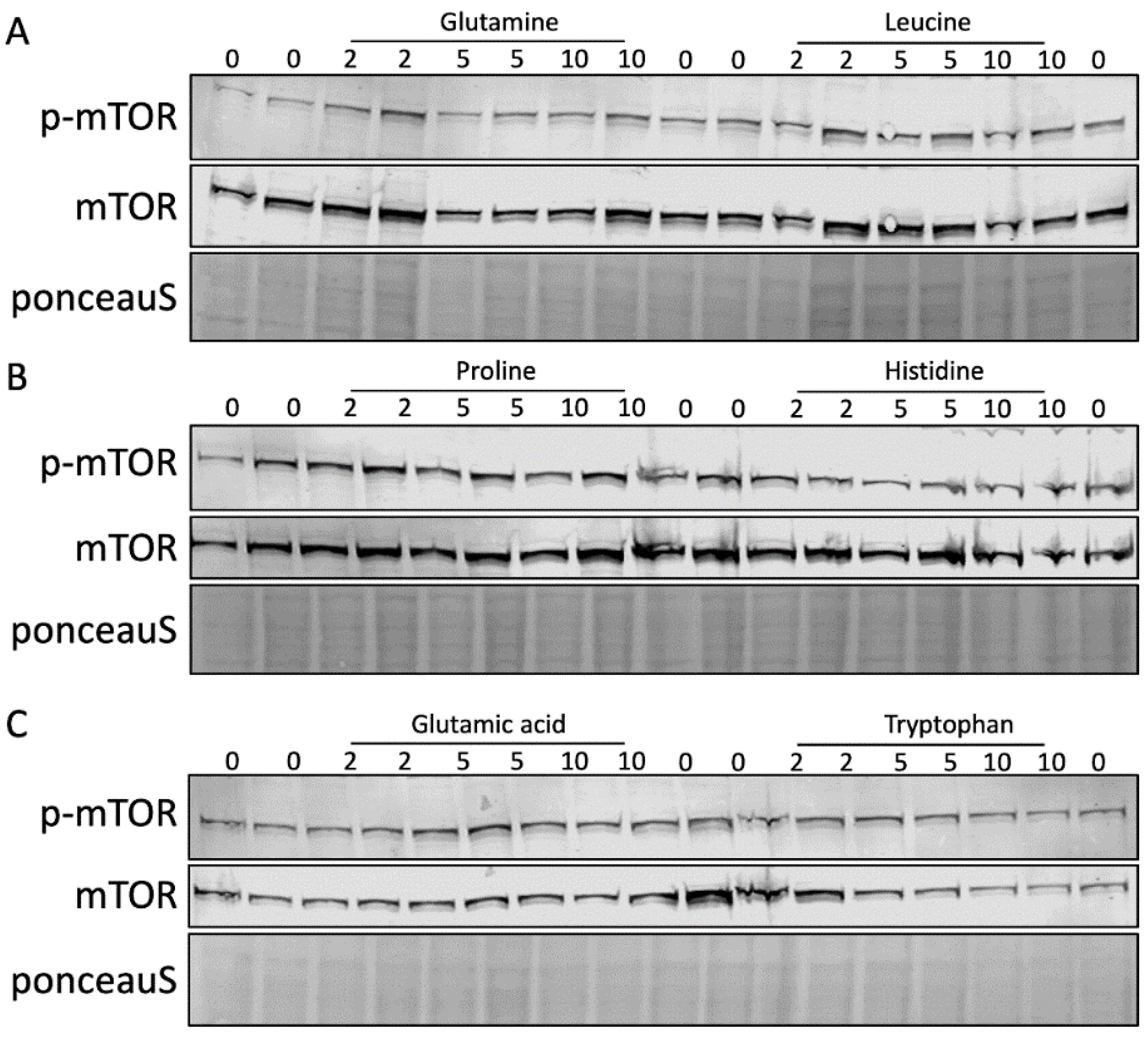
2.4. Effects of Different Amino Acids on mTORC1
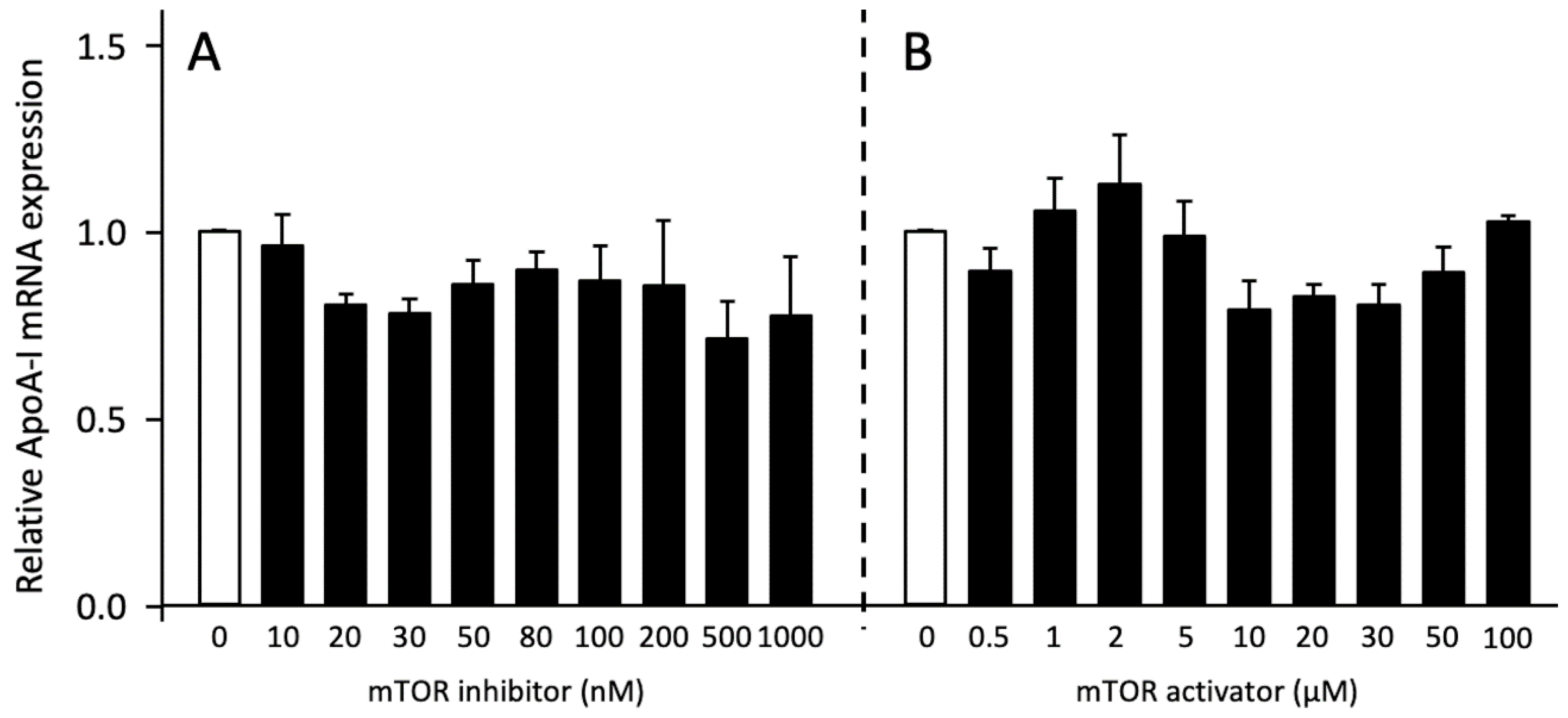
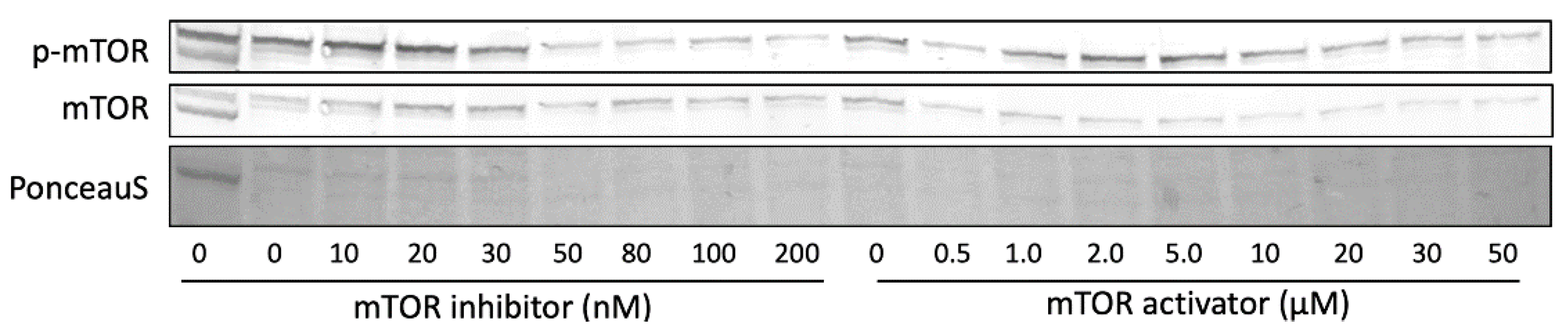
2.5. Role of mTORC1 on ApoA-I Transcription and pro-ApoA-I Secretion
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture and Amino Acids Treatment
4.3. Quantification of Gene mRNA Transcription
4.4. Quantification of pro-ApoA-I Protein Levels in the Culture Medium
4.5. Transfection and Measurement of PPARα Transactivation
4.6. Western Blot for mTORC1 Activity
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malaguarnera, M.; Vacante, M.; Russo, C.; Malaguarnera, G.; Antic, T.; Malaguarnera, L.; Bella, R.; Pennisi, G.; Galvano, F.; Frigiola, A. Lipoprotein(a) in cardiovascular diseases. BioMed Res. Int. 2013, 2013, 650989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chait, A.; Eckel, R.H. Lipids, lipoproteins, and cardiovascular disease: Clinical pharmacology now and in the future. J. Clin. Endocrinol. Metab. 2016, 101, 804–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Kothari, V.; Bornfeldt, K.E. High-density lipoprotein function in cardiovascular disease and diabetes mellitus. Arterioscler. Thromb. Vasc. Biol. 2018, 38, e10–e16. [Google Scholar] [CrossRef] [Green Version]
- Penson, P.E.; Pirro, M.; Banach, M. LDL-C: Lower is better for longer—even at low risk. BMC Med. 2020, 18, 320. [Google Scholar] [CrossRef] [PubMed]
- Redondo, S.; Martínez-González, J.; Urraca, C.; Tejerina, T. Emerging therapeutic strategies to enhance HDL function. Lipids Health Dis. 2011, 10, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohashi, R.; Mu, H.; Wang, X.; Yao, Q.; Chen, C. Reverse cholesterol transport and cholesterol efflux in atherosclerosis. QJM 2005, 98, 845–856. [Google Scholar] [CrossRef]
- Brown, W.M.; Chiacchia, F.S. Therapies to Increase ApoA-I and HDL-Cholesterol Levels. Drug Target. Insights 2008, 3, DTI-S447. [Google Scholar] [CrossRef]
- Gilham, D.; Wasiak, S.; Tsujikawa, L.M.; Halliday, C.; Norek, K.; Patel, R.G.; Kulikowski, E.; Johansson, J.; Sweeney, M.; Wong, N.C. RVX-208, a BET-inhibitor for treating atherosclerotic cardiovascular disease, raises ApoA-I/HDL and represses pathways that contribute to cardiovascular disease. Atherosclerosis 2016, 247, 48–57. [Google Scholar] [CrossRef] [Green Version]
- Ooi, E.M.M.; Watts, G.F.; Ng, T.W.K.; Barrett, P.H.R. Effect of dietary Fatty acids on human lipoprotein metabolism: A comprehensive update. Nutrients 2015, 7, 4416–4425. [Google Scholar] [CrossRef] [Green Version]
- Vélez-Carrasco, W.; Lichtenstein, A.H.; Welty, F.K.; Li, Z.; Lamon-Fava, S.; Dolnikowski, G.G.; Schaefer, E.J. Dietary restriction of saturated fat and cholesterol decreases HDL ApoA-I secretion. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 918–924. [Google Scholar] [CrossRef] [Green Version]
- Noakes, M.; Foster, P.R.; Keogh, J.B.; James, A.P.; Mamo, J.C.; Clifton, P.M. Comparison of isocaloric very low carbohydrate/high saturated fat and high carbohydrate/low saturated fat diets on body composition and cardiovascular risk. Nutr. Metab. 2006, 3, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jegatheesan, P.; Beutheu, S.; Ventura, G.; Sarfati, G.; Nubret, E.; Kapel, N.; Waligora-Dupriet, A.-J.; Bergheim, I.; Cynober, L.; De-Bandt, J.-P. Effect of specific amino acids on hepatic lipid metabolism in fructose-induced non-alcoholic fatty liver disease. Clin. Nutr. 2016, 35, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.R.; Pesti, G.M. The influence of dietary tryptophan on broiler chick growth and lipid metabolism as mediated by dietary protein levels. Poult. Sci. 1990, 69, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dai, X.-y.; Zhou, Z.; Zhao, G.-x.; Wang, X.; Xu, M.-j. Leucine supplementation via drinking water reduces atherosclerotic lesions in apoE null mice. Acta Pharmacol. Sin. 2016, 37, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Yanni, A.E.; Perrea, D.N.; Yatzidis, H.A. Effect of antiatherogenic L-aspartate and L-glutamate on serum lipoproteins cholesterol and apolipoproteins A-1 and B in rabbits fed with high cholesterol diet. Nutr. Metab. Cardiovasc. Dis. 2005, 15, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Van der Krieken, S.E.; Popeijus, H.E.; Mensink, R.P.; Plat, J. Link between ER-stress, PPAR-alpha activation, and BET inhibition in relation to apolipoprotein A-I transcription in HepG2 cells. J. Cell Biochem. 2017, 118, 2161–2167. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Duez, H.; Blanquart, C.; Berezowski, V.; Poulain, P.; Fruchart, J.C.; Najib-Fruchart, J.; Glineur, C.; Staels, B. Statin-induced inhibition of the Rho-signaling pathway activates PPARalpha and induces HDL apoA-I. J. Clin. Investig. 2001, 107, 1423–1432. [Google Scholar] [CrossRef] [Green Version]
- Georgila, K.; Gounis, M.; Havaki, S.; Gorgoulis, V.G.; Eliopoulos, A.G. mTORC1-dependent protein synthesis and autophagy uncouple in the regulation of Apolipoprotein AI expression. Metabolism 2020, 105, 154186. [Google Scholar] [CrossRef]
- Tayyeb, J.Z.; Popeijus, H.E.; Mensink, R.P.; Konings, M.; Mulders, K.H.R.; Plat, J. The effects of short-chain fatty acids on the transcription and secretion of apolipoprotein A-I in human hepatocytes in vitro. J. Cell. Biochem. 2019, 120, 17219–17227. [Google Scholar] [CrossRef] [Green Version]
- Popeijus, H.E.; Van Otterdijk, S.D.; Van der Krieken, S.E.; Konings, M.; Serbonij, K.; Plat, J.; Mensink, R.P. Fatty acid chain length and saturation influences PPARα transcriptional activation and repression in HepG2 cells. Mol. Nutr. Food Res. 2014, 58, 2342–2349. [Google Scholar] [CrossRef]
- Stec, D.E.; John, K.; Trabbic, C.J.; Luniwal, A.; Hankins, M.W.; Baum, J.; Hinds, T.D., Jr. Bilirubin Binding to PPARα Inhibits Lipid Accumulation. PLoS ONE 2016, 11, e0153427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, D.M.; Hong, S.H.; Kipp, Z.A.; Hinds, T.D., Jr. Identification of Binding Regions of Bilirubin in the Ligand-Binding Pocket of the Peroxisome Proliferator-Activated Receptor-A (PPARalpha). Molecules 2021, 26, 2975. [Google Scholar] [CrossRef] [PubMed]
- Stec, D.E.; Gordon, D.M.; Hipp, J.A.; Hong, S.; Mitchell, Z.L.; Franco, N.R.; Robison, J.W.; Anderson, C.D.; Stec, D.F.; Hinds, T.D.J. Loss of hepatic PPARalpha promotes inflammation and serum hyperlipidemia in diet-induced obesity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 317, R733–R745. [Google Scholar] [CrossRef] [PubMed]
- Mogilenko, D.A.; Dizhe, E.B.; Shavva, V.S.; Lapikov, I.A.; Orlov, S.V.; Perevozchikov, A.P. Role of the nuclear receptors HNF4 alpha, PPAR alpha, and LXRs in the TNF alpha-mediated inhibition of human apolipoprotein A-I gene expression in HepG2 cells. Biochemistry 2009, 48, 11950–11960. [Google Scholar] [CrossRef]
- Schnuck, J.K.; Sunderland, K.L.; Gannon, N.P.; Kuennen, M.R.; Vaughan, R.A. Leucine Stimulates Peroxisome Proliferator Activator Receptors in Skeletal Muscle: 1603 Board# 256 June 2, 800 AM-930 AM. Med. Sci. Sports Exerc. 2016, 48, 441–442. [Google Scholar] [CrossRef]
- Van Meijl, L.E.; Popeijus, H.E.; Mensink, R.P. Amino acids stimulate Akt phosphorylation, and reduce IL-8 production and NF-κB activity in HepG2 liver cells. Mol. Nutr. Food Res. 2010, 54, 1568–1573. [Google Scholar] [CrossRef]
- Zhang, N.; Chu, E.S.; Zhang, J.; Li, X.; Liang, Q.; Chen, J.; Chen, M.; Teoh, N.; Farrell, G.; Sung, J.J. Peroxisome proliferator activated receptor alpha inhibits hepatocarcinogenesis through mediating NF-κB signaling pathway. Oncotarget 2014, 5, 8330. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, P.; Tollervey, D. mRNA turnover. Curr. Opin. Cell Biol. 2001, 13, 320–325. [Google Scholar] [CrossRef]
- Wang, S.; Schianchi, F.; Neumann, D.; Wong, L.Y.; Sun, A.; Van Nieuwenhoven, F.A.; Zeegers, M.P.; Strzelecka, A.; Col, U.; Glatz, J.F.C.; et al. Specific amino acid supplementation rescues the heart from lipid overload-induced insulin resistance and contractile dysfunction by targeting the endosomal mTOR-v-ATPase axis. Mol. Metab. 2021, 53, 101293. [Google Scholar] [CrossRef]
- Aramburu, J.; Ortells, M.; Tejedor, S.; Buxadé, M.; López-Rodríguez, C. Transcriptional regulation of the stress response by mTOR. Sci. Signal. 2014, 7, re2. [Google Scholar] [CrossRef]
- Yecies, J.; Manning, B. Transcriptional control of cellular metabolism by mTOR signaling. Cancer Res. 2011, 71, 2815–2820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laplante, M.; Sabatini, D.M. Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 2013, 126, 1713–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimball, S.; Jefferson, L. Role of amino acids in the translational control of protein synthesis in mammals. Semin. Cell Dev. Biol. 2005, 16, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; McGrath, B.; Reinert, J.; Olsen, D.; Lei, L.; Gill, S.; Wek, S.; Vattem, K.; Wek, R.; Kimball, S.; et al. The GCN2 eIF2alpha Kinase Is Required for Adaptation to Amino Acid Deprivation in Mice. Mol. Cell. Biol. 2002, 22, 6681–6688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anthony, T.; Reiter, A.; Anthony, J.; Kimball, S.; Jefferson, L. Deficiency of dietary EAA preferentially inhibits mRNA translation of ribosomal proteins in liver of meal-fed rats. Am. J. Physiol. Endocrinol. Metab. 2001, 281, E430–E439. [Google Scholar] [CrossRef]
- Anthony, T.; McDaniel, B.; Byerley, R.; McGrath, B.; Cavener, D.; McNurlan, M.; Wek, R. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J. Biol. Chem. 2004, 279, 36553–36561. [Google Scholar] [CrossRef] [Green Version]
- Gilde, A.J.; Van der Lee, K.A.; Willemsen, P.H.; Chinetti, G.; Van der Leij, F.R.; Van der Vusse, G.J.; Staels, B.; Van Bilsen, M. Peroxisome proliferator-activated receptor (PPAR) alpha and PPARbeta/delta, but not PPARgamma, modulate the expression of genes involved in cardiac lipid metabolism. Circ. Res. 2003, 92, 518–524. [Google Scholar] [CrossRef] [Green Version]
- Churchward-Venne, T.A.; Pinckaers, P.J.M.; Smeets, J.S.J.; Peeters, W.M.; Zorenc, A.H.; Schierbeek, H.; Rollo, I.; Verdijk, L.B.; van Loon, L.J.C. Myofibrillar and Mitochondrial Protein Synthesis Rates Do Not Differ in Young Men Following the Ingestion of Carbohydrate with Whey, Soy, or Leucine-Enriched Soy Protein after Concurrent Resistance- and Endurance-Type Exercise. J. Nutr. 2019, 149, 210–220. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tayyeb, J.Z.; Popeijus, H.E.; van de Sanden, J.; Zwaan, W.; Mensink, R.P.; Plat, J. Effects of Individual Amino Acids on PPARα Transactivation, mTORC1 Activation, ApoA-I Transcription and pro-ApoA-I Secretion. Int. J. Mol. Sci. 2022, 23, 6071. https://doi.org/10.3390/ijms23116071
Tayyeb JZ, Popeijus HE, van de Sanden J, Zwaan W, Mensink RP, Plat J. Effects of Individual Amino Acids on PPARα Transactivation, mTORC1 Activation, ApoA-I Transcription and pro-ApoA-I Secretion. International Journal of Molecular Sciences. 2022; 23(11):6071. https://doi.org/10.3390/ijms23116071
Chicago/Turabian StyleTayyeb, Jehad Z., Herman E. Popeijus, Janna van de Sanden, Willem Zwaan, Ronald P. Mensink, and Jogchum Plat. 2022. "Effects of Individual Amino Acids on PPARα Transactivation, mTORC1 Activation, ApoA-I Transcription and pro-ApoA-I Secretion" International Journal of Molecular Sciences 23, no. 11: 6071. https://doi.org/10.3390/ijms23116071
APA StyleTayyeb, J. Z., Popeijus, H. E., van de Sanden, J., Zwaan, W., Mensink, R. P., & Plat, J. (2022). Effects of Individual Amino Acids on PPARα Transactivation, mTORC1 Activation, ApoA-I Transcription and pro-ApoA-I Secretion. International Journal of Molecular Sciences, 23(11), 6071. https://doi.org/10.3390/ijms23116071







