Molecular Regulation of Arbuscular Mycorrhizal Symbiosis
Abstract
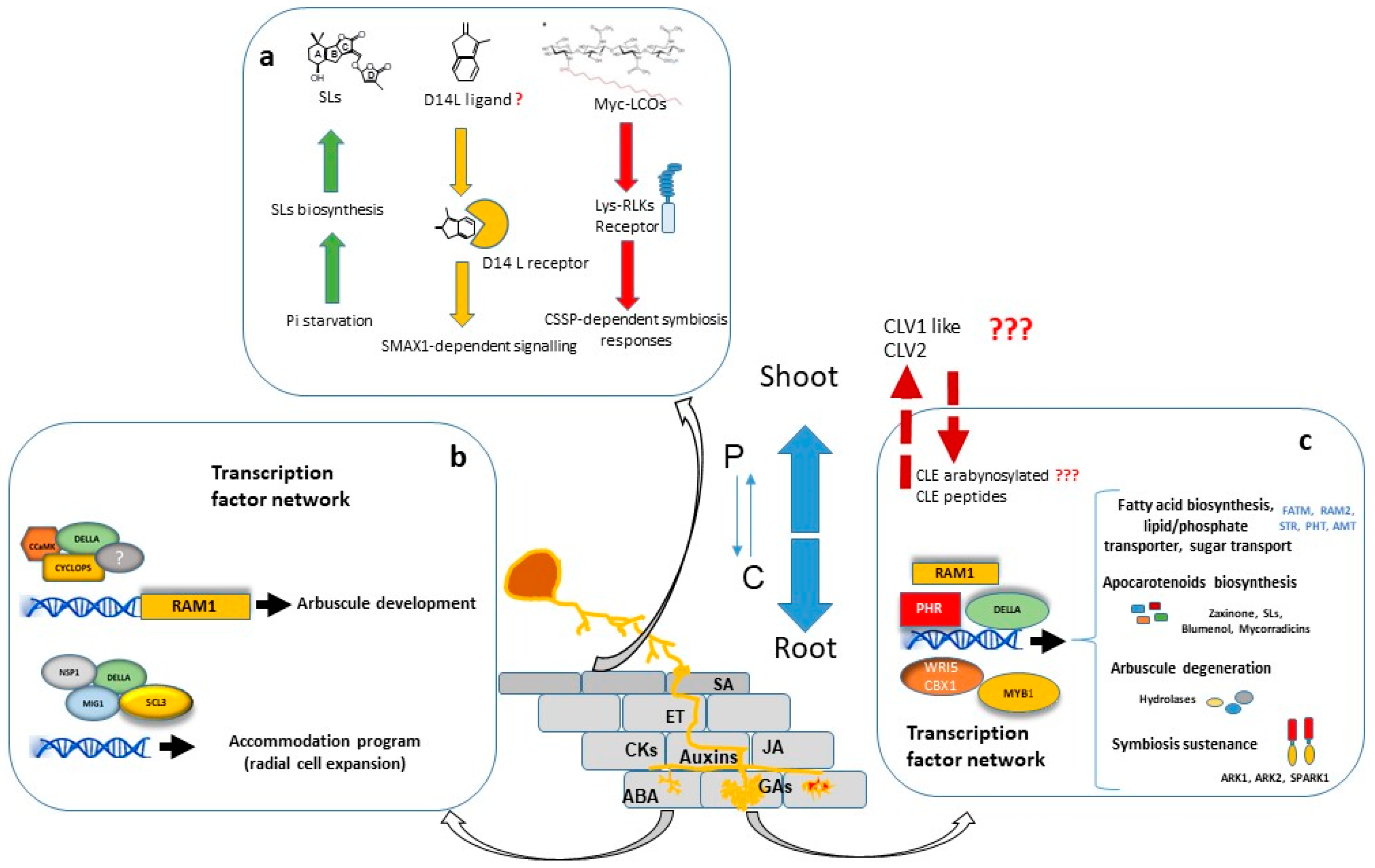
:1. Introduction
2. Nutritional Regulation
3. AM Regulation by Hormones and Other Signalling Molecules
4. Transcriptional Regulation of AM Symbiosis
5. Systemic Autoregulation of AM Symbiosis
6. Post Arbuscule Development Symbiosis Sustenance
7. Challenges and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Xie, K.; Ren, Y.; Chen, A.; Yang, C.; Zheng, Q.; Chen, J.; Wang, D.; Li, Y.; Hu, S.; Xu, G. Plant nitrogen nutrition: The roles of arbuscular mycorrhizal fungi. J. Plant Physiol. 2021, 269, 153591. [Google Scholar] [CrossRef]
- Luginbuehl, L.H.; Oldroyd, G.E. Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Curr. Biol. 2017, 27, R952–R963. [Google Scholar] [CrossRef] [PubMed]
- Bago, B.; Pfeffer, P.E.; Abubaker, J.; Jun, J.; Allen, J.W.; Brouillette, J.; Douds, D.D.; Lammers, P.J.; Shachar-Hill, Y. Carbon export from arbuscular mycorrhizal roots involves the translocation of carbohydrate as well as lipid. Plant Physiol. 2003, 131, 1496–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pozo, M.J.; Azcón-Aguilar, C. Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 2007, 10, 393–398. [Google Scholar] [CrossRef]
- Lenoir, I.; Fontaine, J.; Sahraoui, A.L.-H. Arbuscular mycorrhizal fungal responses to abiotic stresses: A review. Phytochemistry 2016, 123, 4–15. [Google Scholar] [CrossRef]
- Gutjahr, C.; Parniske, M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell. Dev. Biol. 2013, 29, 593–617. [Google Scholar] [CrossRef]
- Smith, S.; Read, D. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: London, UK, 2008. [Google Scholar]
- Balzergue, C.; Puech-Pagès, V.; Bécard, G.; Rochange, S.F. The regulation of arbuscular mycorrhizal symbiosis by phosphate in pea involves early and systemic signalling events. J. Exp. Bot. 2011, 62, 1049–1060. [Google Scholar] [CrossRef] [Green Version]
- Breuillin, F.; Schramm, J.; Hajirezaei, M.; Ahkami, A.; Favre, P.; Druege, U.; Hause, B.; Bucher, M.; Kretzschmar, T.; Bossolini, E.; et al. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 2010, 64, 1002–1017. [Google Scholar] [CrossRef]
- Russo, G.; Spinella, S.; Sciacca, E.; Bonfante, P.; Genre, A. Automated analysis of calcium spiking profiles with CaSA software: Two case studies from root-microbe symbioses. BMC Plant Biol. 2013, 13, 224. [Google Scholar] [CrossRef] [Green Version]
- Balzergue, C.; Chabaud, M.; Barker, D.G.; Bécard, G.; Rochange, S.F. High phosphate reduces host ability to develop arbuscular mycorrhizal symbiosis without affecting root calcium spiking responses to the fungus. Front. Plant Sci. 2013, 4, 426. [Google Scholar] [CrossRef] [Green Version]
- Javot, H.; Penmetsa, R.V.; Terzagi, N.; Cook, D.R.; Harrison, M.J. A Medicago truncatula phosphate transporter indispensabhle for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2007, 104, 1720–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.-Y.; Grønlund, M.; Jakobsen, I.; Grotemeyer, M.S.; Rentsch, D.; Miyao, A.; Hirochika, H.; Kumar, C.S.; Sundaresan, V.; Salamin, N. Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. Plant Cell 2012, 24, 4236–4251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Zhao, B.; Zheng, S.; Zhang, X.; Wang, X.; Dong, W.; Xie, Q.; Wang, G.; Xiao, Y.; Chen, F. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 2021, 184, 5527–5540.e18. [Google Scholar] [CrossRef]
- Das, D.; Paries, M.; Hobecker, K.; Gigl, M.; Dawid, C.; Lam, H.-M.; Zhang, J.; Chen, M.; Gutjahr, C. Phosphate Starvation Response transcription factors enable arbuscular mycorrhiza symbiosis. Nat. Commun. 2022, 13, 477. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Yoneyama, K.; Takeuchi, Y.; Sekimoto, H. Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 2007, 225, 1031–1038. [Google Scholar] [CrossRef]
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone Signaling and Evolution. Annu. Rev. Plant Biol. 2017, 68, 291–322. [Google Scholar] [CrossRef]
- Kobae, Y.; Kameoka, H.; Sugimura, Y.; Saito, K.; Ohtomo, R.; Fujiwara, T.; Kyozuka, J. Strigolactone biosynthesis genes of rice is required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiol. 2018, 59, 544–553. [Google Scholar] [CrossRef]
- Nouri, E.; Surve, R.; Bapaume, L.; Stumpe, M.; Chen, M.; Zhang, Y.; Ruyter-Spira, C.; Bouwmeester, H.; Glauser, G.; Bruisson, S. Phosphate suppression of arbuscular mycorrhizal symbiosis involves gibberellic acid signaling. Plant Cell Physiol. 2021, 62, 959–970. [Google Scholar] [CrossRef]
- Müller, L.M.; Harrison, M.J. Phytohormones, miRNAs, and peptide signals integrate plant phosphorus status with arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 2019, 50, 132–139. [Google Scholar] [CrossRef]
- Nouri, E.; Breuillin-Sessoms, F.; Feller, U.; Reinhardt, D. Phosphorus and nitrogen regulate arbuscular mycorrhizal symbiosis in Petunia hybrida. PLoS ONE 2014, 9, e90841. [Google Scholar] [CrossRef]
- Maldonado-Mendoza, I.E.; Harrison, M.J. RiArsB and RiMT-11: Two novel genes induced by arsenate in arbuscular mycorrhiza. Fungal Biol. 2017, 122, 121–130. [Google Scholar] [CrossRef]
- Javot, H.; Penmetsa, R.V.; Breuillin, F.; Bhattarai, K.K.; Noar, R.D.; Gomez, S.K.; Zhang, Q.; Cook, D.R.; Harrison, M.J. Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant J. 2011, 68, 954–965. [Google Scholar] [CrossRef]
- Breuillin-Sessoms, F.; Floss, D.S.; Gomez, S.K.; Pumplin, N.; Ding, Y.; Levesque-Tremblay, V.; Noar, R.D.; Daniels, D.A.; Bravo, A.; Eaglesham, J.B. Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. Plant Cell 2015, 27, 1352–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calabrese, S.; Kohler, A.; Niehl, A.; Veneault-Fourrey, C.; Boller, T.; Courty, P.-E. Transcriptome analysis of the Populus trichocarpa–Rhizophagus irregularis mycorrhizal symbiosis: Regulation of plant and fungal transportomes under nitrogen starvation. Plant Cell Physiol. 2017, 58, 1003–1017. [Google Scholar] [CrossRef] [Green Version]
- Pumplin, N.; Mondo, S.J.; Topp, S.; Starker, C.G.; Gantt, J.S.; Harrison, M.J. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 2010, 61, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Floss, D.S.; Levy, J.G.; Lévesque-Tremblay, V.; Pumplin, N.; Harrison, M.J. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2013, 110, E5025–E5034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanke, V.; Renker, C.; Wagner, M.; Füllner, K.; Held, M.; Kuhn, A.J.; Buscot, F. Nitrogen supply affects arbuscular mycorrhizal colonization of Artemisia vulgaris in a phosphate-polluted field site. New Phytol. 2005, 166, 981–992. [Google Scholar] [CrossRef]
- Kiers, E.T.; Duhamel, M.; Beesetty, Y.; Mensah, J.A.; Franken, O.; Verbruggen, E.; Fellbaum, C.R.; Kowalchuk, G.A.; Hart, M.M.; Bago, A. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 2011, 333, 880–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Chen, A.; Chen, C.; Li, C.; Xia, R.; Wang, X. Transcriptomic analysis reveals the possible roles of sugar metabolism and export for positive mycorrhizal growth responses in soybean. Physiol. Plant. 2019, 166, 712–728. [Google Scholar] [CrossRef] [Green Version]
- Schaarschmidt, S.; González, M.-C.; Roitsch, T.; Strack, D.; Sonnewald, U.; Hause, B. Regulation of arbuscular mycorrhization by carbon. The symbiotic interaction cannot be improved by increased carbon availability accomplished by root-specifically enhanced invertase activity. Plant Physiol. 2007, 143, 1827–1840. [Google Scholar] [CrossRef]
- Gabriel-Neumann, E.; Neumann, G.; Leggewie, G.; George, E. Constitutive overexpression of the sucrose transporter SoSUT1 in potato plants increases arbuscular mycorrhiza fungal root colonization under high, but not under low, soil phosphorus availability. J. Plant Physiol. 2011, 168, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Bitterlich, M.; Krügel, U.; Boldt-Burisch, K.; Franken, P.; Kühn, C. The sucrose transporter SlSUT2 from tomato interacts with brassinosteroid functioning and affects arbuscular mycorrhiza formation. Plant J. 2014, 78, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Manck-Götzenberger, J.; Requena, N. Arbuscular mycorrhiza symbiosis induces a major transcriptional reprogramming of the potato SWEET sugar transporter family. Front. Plant Sci. 2016, 7, 487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.; Zeng, T.; Ji, C.; de Graaf, S.; Zheng, Z.; Xiao, T.T.; Deng, X.; Xiao, S.; Bisseling, T.; Limpens, E. A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. New Phytol. 2019, 224, 396–408. [Google Scholar] [CrossRef]
- Helber, N.; Wippel, K.; Sauer, N.; Schaarschmidt, S.; Hause, B.; Requena, N. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 2011, 23, 3812–3823. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Liu, X.; Feng, G.; Zhu, H.; Yao, Q. Linking lipid transfer with reduced arbuscule formation in tomato roots colonized by arbuscular mycorrhizal fungus under low pH stress. Environ. Microbiol. 2020, 22, 1036–1051. [Google Scholar] [CrossRef]
- Bravo, A.; York, T.; Pumplin, N.; Mueller, L.A.; Harrison, M.J. Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat. Plants 2016, 2, 15208. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, W.; Xie, Q.; Liu, N.; Liu, L.; Wang, D.; Zhang, X.; Yang, C.; Chen, X.; Tang, D. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 2017, 356, 1172–1175. [Google Scholar] [CrossRef] [Green Version]
- Brands, M.; Wewer, V.; Keymer, A.; Gutjahr, C.; Dörmann, P. The Lotus japonicus acyl-acyl carrier protein thioesterase FatM is required for mycorrhiza formation and lipid accumulation of Rhizophagus irregularis. Plant J. 2018, 95, 219–232. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.; Schornack, S.; Marsh, J.F.; Gobbato, E.; Schwessinger, B.; Eastmond, P.; Schultze, M.; Kamoun, S.; Oldroyd, G.E. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr. Biol. 2012, 22, 2242–2246. [Google Scholar] [CrossRef] [Green Version]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.; Eastmond, P.J. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.; Dai, H.; Zhang, X.; Zhao, B.; Shi, J.; Zhang, C.; Wang, G.; Yu, N. Colonization of mutualistic mycorrhizal and parasitic blast fungi requires OsRAM2-regulated fatty acid biosynthesis in rice. Mol. Plant-Microbe Interact. 2022, 35, 178–186. [Google Scholar]
- Liu, Y.-N.; Liu, C.-C.; Zhu, A.-Q.; Niu, K.-X.; Guo, R.; Tian, L.; Wu, Y.-N.; Sun, B.; Wang, B. OsRAM2 function in lipid biosynthesis is required for arbuscular mycorrhizal symbiosis in rice. Mol. Plant-Microbe Interact. 2022, 35, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Blaylock, L.A.; Harrison, M.J. Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 2010, 22, 1483–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Xie, Q.; Wang, W.; Yang, J.; Zhang, X.; Yu, N.; Zhou, Y.; Wang, E. Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol. Plant 2018, 11, 1344–1359. [Google Scholar] [CrossRef] [Green Version]
- Pozo, M.J.; López-Ráez, J.A.; Azcón-Aguilar, C.; García-Garrido, J.M. Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 2015, 205, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Bedini, A.; Mercy, L.; Schneider, C.; Franken, P.; Lucic-Mercy, E. Unraveling the initial plant hormone signaling, metabolic mechanisms and plant defense triggering the endomycorrhizal symbiosis behavior. Front. Plant Sci. 2018, 9, 1800. [Google Scholar] [CrossRef] [Green Version]
- Liao, D.; Wang, S.; Cui, M.; Liu, J.; Chen, A.; Xu, G. Phytohormones regulate the development of arbuscular mycorrhizal symbiosis. Int. J. Mol. Sci. 2018, 19, 3146. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.-P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.-C. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189. [Google Scholar] [CrossRef]
- Lopez-Obando, M.; Ligerot, Y.; Bonhomme, S.; Boyer, F.-D.; Rameau, C. Strigolactone biosynthesis and signaling in plant development. Development 2015, 142, 3615–3619. [Google Scholar] [CrossRef] [Green Version]
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J.B.; Reinhardt, D.; Bours, R.; Bouwmeester, H.J.; Martinoia, E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, 341. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, K.; Hayashi, H. Strigolactones: Chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann. Bot. 2006, 97, 925–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besserer, A.; Puech-Pages, V.; Kiefer, P.; Gomez-Roldan, V.; Jauneau, A.; Roy, S.; Portais, J.C.; Roux, C.; Becard, G.; Sejalon-Delmas, N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006, 4, e226. [Google Scholar] [CrossRef] [PubMed]
- Besserer, A.; Bécard, G.; Jauneau, A.; Roux, C.; Séjalon-Delmas, N. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 2008, 148, 402–413. [Google Scholar] [CrossRef] [Green Version]
- Ruyter-Spira, C.; Al-Babili, S.; Van Der Krol, S.; Bouwmeester, H. The biology of strigolactones. Trends Plant Sci. 2013, 18, 72–83. [Google Scholar] [CrossRef]
- MacLean, A.M.; Bravo, A.; Harrison, M.J. Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 2017, 29, 2319–2335. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C. Phytohormone signaling in arbuscular mycorhiza development. Curr. Opin. Plant Biol. 2014, 20, 26–34. [Google Scholar] [CrossRef]
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Koltai, H.; LekKala, S.P.; Bhattacharya, C.; Mayzlish-Gati, E.; Resnick, N.; Wininger, S.; Dor, E.; Yoneyama, K.; Yoneyama, K.; Hershenhorn, J. A tomato strigolactone-impaired mutant displays aberrant shoot morphology and plant interactions. J. Exp. Bot. 2010, 61, 1739–1749. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S.; Kameoka, H.; Tempo, M.; Akiyama, K.; Umehara, M.; Yamaguchi, S.; Hayashi, H.; Kyozuka, J.; Shirasu, K. The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. 2012, 196, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- López-Ráez, J.A.; Fernández, I.; García, J.M.; Berrio, E.; Bonfante, P.; Walter, M.H.; Pozo, M.J. Differential spatio-temporal expression of carotenoid cleavage dioxygenases regulates apocarotenoid fluxes during AM symbiosis. Plant Sci. 2015, 230, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Medina, M.J.; Steinkellner, S.; Vierheilig, H.; Ocampo Bote, J.A.; García Garrido, J.M. Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 2007, 175, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Martín-Rodríguez, J.Á.; León-Morcillo, R.; Vierheilig, H.; Ocampo, J.A.; Ludwig-Müller, J.; García-Garrido, J.M. Ethylene-dependent/ethylene-independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytol. 2011, 190, 193–205. [Google Scholar] [CrossRef]
- Mercy, L.; Lucic-Mercy, E.; Nogales, A.; Poghosyan, A.; Schneider, C.; Arnholdt-Schmitt, B. A functional approach towards understanding the role of the mitochondrial respiratory chain in an endomycorrhizal symbiosis. Front. Plant Sci. 2017, 8, 417. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Feng, Z.; Zhu, H.; Yao, Q. Exogenous abscisic acid and root volatiles increase sporulation of Rhizophagus irregularis DAOM 197198 in asymbiotic and pre-symbiotic status. Mycorrhiza 2019, 29, 581–589. [Google Scholar] [CrossRef]
- Charpentier, M.; Sun, J.; Wen, J.; Mysore, K.S.; Oldroyd, G.E. Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiol. 2014, 166, 2077–2090. [Google Scholar] [CrossRef] [Green Version]
- García Garrido, J.M.; León Morcillo, R.J.; Martín Rodríguez, J.A.; Ocampo Bote, J.A. Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Mol. Plant-Microbe Interact. 2010, 23, 651–664. [Google Scholar] [CrossRef] [Green Version]
- Martín-Rodríguez, J.Á.; Ocampo, J.A.; Molinero-Rosales, N.; Tarkowská, D.; Ruíz-Rivero, O.; García-Garrido, J.M. Role of gibberellins during arbuscular mycorrhizal formation in tomato: New insights revealed by endogenous quantification and genetic analysis of their metabolism in mycorrhizal roots. Physiol. Plant. 2015, 154, 66–81. [Google Scholar] [CrossRef]
- Takeda, N.; Handa, Y.; Tsuzuki, S.; Kojima, M.; Sakakibara, H.; Kawaguchi, M. Gibberellins interfere with symbiosis signaling and gene expression and alter colonization by arbuscular mycorrhizal fungi in Lotus japonicus. Plant Physiol. 2015, 167, 545–557. [Google Scholar] [CrossRef] [Green Version]
- Foo, E.; Ross, J.J.; Jones, W.T.; Reid, J.B. Plant hormones in arbuscular mycorrhizal symbioses: An emerging role for gibberellins. Ann. Bot. 2013, 111, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Luo, D.; Zhang, X.; Liu, J.; Wang, W.; Jin, Y.; Dong, W.; Liu, J.; Liu, H.; Yang, W. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 2014, 24, 130. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, T.; Miura, C.; Takeda, N.; Kanno, Y.; Takemura, Y.; Seo, M.; Yamato, M.; Kaminaka, H. Gibberellin promotes fungal entry and colonization during Paris-type arbuscular mycorrhizal symbiosis in Eustoma grandiflorum. Plant Cell Physiol. 2020, 61, 565–575. [Google Scholar] [CrossRef]
- Bitterlich, M.; Krügel, U.; Boldt-Burisch, K.; Franken, P.; Kühn, C. Interaction of brassinosteroid functions and sucrose transporter SlSUT2 regulate the formation of arbuscular mycorrhiza. Plant Signal. Behav. 2014, 9, e970426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etemadi, M.; Gutjahr, C.; Couzigou, J.-M.; Zouine, M.; Lauressergues, D.; Timmers, A.; Audran, C.; Bouzayen, M.; Bécard, G.; Combier, J.-P. Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 2014, 166, 281–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanlon, M.T.; Coenen, C. Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytol. 2011, 189, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Foo, E. Auxin influences strigolactones in pea mycorrhizal symbiosis. J. Plant Physiol. 2013, 170, 523–528. [Google Scholar] [CrossRef]
- Chen, X.; Chen, J.; Liao, D.; Ye, H.; Li, C.; Luo, Z.; Yan, A.; Zhao, Q.; Xie, K.; Li, Y. Auxin-mediated regulation of arbuscular mycorrhizal symbiosis: A role of SlGH3. 4 in tomato. Plant Cell Environ. 2022, 45, 955–968. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Martin-Rodriguez, J.A.; Huertas, R.; Ho-Plagaro, T.; Ocampo, J.A.; Tureckova, V.; Tarkowska, D.; Ludwig-Muller, J.; Garcia-Garrido, J.M. Gibberellin-Abscisic Acid Balances during Arbuscular Mycorrhiza Formation in Tomato. Front. Plant Sci. 2016, 7, 1273. [Google Scholar] [CrossRef] [Green Version]
- Cosme, M.; Fernández, I.; Declerck, S.; van der Heijden, M.G.; Pieterse, C.M. A coumarin exudation pathway mitigates arbuscular mycorrhizal incompatibility in Arabidopsis thaliana. Plant Mol. Biol. 2021, 106, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Nadal, M.; Sawers, R.; Naseem, S.; Bassin, B.; Kulicke, C.; Sharman, A.; An, G.; An, K.; Ahern, K.R.; Romag, A. An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat. Plants 2017, 3, 17073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutjahr, C.; Gobbato, E.; Choi, J.; Riemann, M.; Johnston, M.G.; Summers, W.; Carbonnel, S.; Mansfield, C.; Yang, S.Y.; Nadal, M.; et al. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 2015, 350, 1521–1524. [Google Scholar] [CrossRef]
- Ho-Plágaro, T.; Morcillo, R.J.; Tamayo-Navarrete, M.I.; Huertas, R.; Molinero-Rosales, N.; López-Ráez, J.A.; Macho, A.P.; García-Garrido, J.M. DLK2 regulates arbuscule hyphal branching during arbuscular mycorrhizal symbiosis. New Phytol. 2021, 229, 548–562. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Scaffidi, A.; Dun, E.A.; Waters, M.T.; Flematti, G.R.; Dixon, K.W.; Beveridge, C.A.; Ghisalberti, E.L.; Smith, S.M. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 8897–8902. [Google Scholar] [CrossRef] [Green Version]
- Waters, M.T.; Nelson, D.C.; Scaffidi, A.; Flematti, G.R.; Sun, Y.K.; Dixon, K.W.; Smith, S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 2012, 139, 1285–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanga, J.P.; Smith, S.M.; Briggs, W.R.; Nelson, D.C. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 2013, 163, 318–330. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Lee, T.; Cho, J.; Servante, E.K.; Pucker, B.; Summers, W.; Bowden, S.; Rahimi, M.; An, K.; An, G. The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nat. Commun. 2020, 11, 2114. [Google Scholar] [CrossRef]
- Waters, M.T.; Scaffidi, A.; Flematti, G.; Smith, S.M. Substrate-induced degradation of the α/β-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant 2015, 8, 814–817. [Google Scholar] [CrossRef] [Green Version]
- Waters, M.T.; Scaffidi, A.; Sun, Y.K.; Flematti, G.R.; Smith, S.M. The karrikin response system of Arabidopsis. Plant J. 2014, 79, 623–631. [Google Scholar] [CrossRef]
- Conn, C.E.; Nelson, D.C. Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 2016, 6, 1219. [Google Scholar] [CrossRef]
- Fiorilli, V.; Wang, J.Y.; Bonfante, P.; Lanfranco, L.; Al-Babili, S. Apocarotenoids: Old and new mediators of the arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2019, 10, 1186. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Haider, I.; Jamil, M.; Fiorilli, V.; Saito, Y.; Mi, J.; Baz, L.; Kountche, B.A.; Jia, K.-P.; Guo, X. The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat. Commun. 2019, 10, 810. [Google Scholar] [CrossRef] [PubMed]
- Schliemann, W.; Schmidt, J.; Nimtz, M.; Wray, V.; Fester, T.; Strack, D. Accumulation of apocarotenoids in mycorrhizal roots of Ornithogalum umbellatum. Phytochemistry 2006, 67, 1196–1205. [Google Scholar] [CrossRef]
- Fester, T.; Hause, B.; Schmidt, D.; Halfmann, K.; Schmidt, J.; Wray, V.; Hause, G.; Strack, D. Occurrence and localization of apocarotenoids in arbuscular mycorrhizal plant roots. Plant Cell Physiol. 2002, 43, 256–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, E.M.; Robinson, L.A.; Abdul-Sada, A.; Vanbergen, A.J.; Hodge, A.; Hartley, S.E. Arbuscular mycorrhizal fungi and plant chemical defence: Effects of colonisation on aboveground and belowground metabolomes. J. Chem. Ecol. 2018, 44, 198–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Schäfer, M.; Li, D.; Halitschke, R.; Dong, C.; McGale, E.; Paetz, C.; Song, Y.; Li, S.; Dong, J. Blumenols as shoot markers of root symbiosis with arbuscular mycorrhizal fungi. Elife 2018, 7, e37093. [Google Scholar] [CrossRef]
- Fester, T.; Maier, W.; Strack, D. Accumulation of secondary compounds in barley and wheat roots in response to inoculation with an arbuscular mycorrhizal fungus and co-inoculation with rhizosphere bacteria. Mycorrhiza 1999, 8, 241–246. [Google Scholar] [CrossRef]
- Floss, D.S.; Schliemann, W.; Schmidt, J.; Strack, D.; Walter, M.H. RNA interference-mediated repression of MtCCD1 in mycorrhizal roots of Medicago truncatula causes accumulation of C27 apocarotenoids, shedding light on the functional role of CCD1. Plant Physiol. 2008, 148, 1267–1282. [Google Scholar] [CrossRef] [Green Version]
- Feng, F.; Sun, J.; Radhakrishnan, G.V.; Lee, T.; Bozsóki, Z.; Fort, S.; Gavrin, A.; Gysel, K.; Thygesen, M.B.; Andersen, K.R. A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat. Commun. 2019, 10, 5047. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Dong, W.; Sun, J.; Feng, F.; Deng, Y.; He, Z.; Oldroyd, G.E.; Wang, E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015, 81, 258–267. [Google Scholar] [CrossRef]
- Buendia, L.; Wang, T.; Girardin, A.; Lefebvre, B. The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol. 2016, 210, 184–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Yuan, L.; Staehelin, C.; Li, Y.; Ruan, J.; Liang, Z.; Xie, Z.; Wang, W.; Xie, J.; Huang, S. The LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE 1 protein of banana is required for perception of pathogenic and symbiotic signals. New Phytol. 2019, 223, 1530–1546. [Google Scholar] [CrossRef] [PubMed]
- Gibelin-Viala, C.; Amblard, E.; Puech-Pages, V.; Bonhomme, M.; Garcia, M.; Bascaules-Bedin, A.; Fliegmann, J.; Wen, J.; Mysore, K.S.; le Signor, C. The Medicago truncatula LysM receptor-like kinase LYK9 plays a dual role in immunity and the arbuscular mycorrhizal symbiosis. New Phytol. 2019, 223, 1516–1529. [Google Scholar] [CrossRef]
- Leppyanen, I.V.; Pavlova, O.A.; Vashurina, M.A.; Bovin, A.D.; Dolgikh, A.V.; Shtark, O.Y.; Sendersky, I.V.; Dolgikh, V.V.; Tikhonovich, I.A.; Dolgikh, E.A. Lysm receptor-like kinase lyk9 of Pisum Sativum L. may regulate plant responses to chitooligosaccharides differing in structure. Int. J. Mol. Sci. 2021, 22, 711. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, C.; Dai, H.; Liu, H.; Zhang, X.; Yang, J.; Chen, X.; Zhu, Y.; Wang, D.; Qi, X. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol. Plant 2019, 12, 1561–1576. [Google Scholar] [CrossRef] [Green Version]
- Liao, D.; Sun, X.; Wang, N.; Song, F.; Liang, Y. Tomato LysM receptor-like kinase SlLYK12 is involved in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2018, 9, 1004. [Google Scholar] [CrossRef] [Green Version]
- Kloppholz, S.; Kuhn, H.; Requena, N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 2011, 21, 1204–1209. [Google Scholar] [CrossRef] [Green Version]
- Tsuzuki, S.; Handa, Y.; Takeda, N.; Kawaguchi, M. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol. Plant-Microbe Interact. 2016, 29, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Voß, S.; Betz, R.; Heidt, S.; Corradi, N.; Requena, N. RiCRN1, a crinkler effector from the arbuscular mycorrhizal fungus Rhizophagus irregularis, functions in arbuscule development. Front. Microbiol. 2018, 9, 2068. [Google Scholar] [CrossRef]
- Prasad Singh, P.; Srivastava, D.; Jaiswar, A.; Adholeya, A. Effector proteins of Rhizophagus proliferus: Conserved protein domains may play a role in host-specific interaction with different plant species. Braz. J. Microbiol. 2019, 50, 593–601. [Google Scholar] [CrossRef]
- Toro, K.S.; Brachmann, A. The effector candidate repertoire of the arbuscular mycorrhizal fungus Rhizophagus clarus. BMC Genom. 2016, 17, 101. [Google Scholar]
- Chen, E.C.; Morin, E.; Beaudet, D.; Noel, J.; Yildirir, G.; Ndikumana, S.; Charron, P.; St-Onge, C.; Giorgi, J.; Krüger, M. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 2018, 220, 1161–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, T.; Holmer, R.; Hontelez, J.; Te Lintel-Hekkert, B.; Marufu, L.; de Zeeuw, T.; Wu, F.; Schijlen, E.; Bisseling, T.; Limpens, E. Host- and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J. 2018, 94, 411–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rich, M.K.; Courty, P.E.; Roux, C.; Reinhardt, D. Role of the GRAS transcription factor ATA/RAM1 in the transcriptional reprogramming of arbuscular mycorrhiza in Petunia hybrida. BMC Genom. 2017, 18, 589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, L.; Cui, H.; Buer, B.; Vijayakumar, V.; Delaux, P.-M.; Junkermann, S.; Bucher, M. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 2015, 167, 854–871. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, R.M.; Schaepe, S.; Nübel, D.; Petersen, A.C.; Bertolini, M.; Vasilev, J.; Küster, H.; Hohnjec, N. Insights into the complex role of GRAS transcription factors in the arbuscular mycorrhiza symbiosis. Sci. Rep. 2019, 9, 3360. [Google Scholar] [CrossRef] [Green Version]
- Ho-Plágaro, T.; Molinero-Rosales, N.; Flores, D.F.; Díaz, M.V.; García-Garrido, J.M. Identification and expression analysis of GRAS transcription factor genes involved in the control of arbuscular mycorrhizal development in tomato. Front. Plant Sci. 2019, 10, 268. [Google Scholar] [CrossRef]
- García-Garrido, J.M.; Ho Plágaro, T. Multifarious and interactive roles of GRAS transcription factors during arbuscular mycorrhiza development. Front. Plant Sci. 2022, 13, 908. [Google Scholar]
- Pimprikar, P.; Gutjahr, C. Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol. 2018, 59, 673–690. [Google Scholar] [CrossRef]
- Miller, J.B.; Pratap, A.; Miyahara, A.; Zhou, L.; Bornemann, S.; Morris, R.J.; Oldroyd, G.E. Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 2013, 25, 5053–5066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yano, K.; Yoshida, S.; Müller, J.; Singh, S.; Banba, M.; Vickers, K.; Markmann, K.; White, C.; Schuller, B.; Sato, S. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 2008, 105, 20540–20545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Katzer, K.; Lambert, J.; Cerri, M.; Parniske, M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 2014, 15, 139–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimprikar, P.; Carbonnel, S.; Paries, M.; Katzer, K.; Klingl, V.; Bohmer, M.J.; Karl, L.; Floss, D.S.; Harrison, M.J.; Parniske, M. A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr. Biol. 2016, 26, 987–998. [Google Scholar] [CrossRef] [Green Version]
- Camps, C.; Jardinaud, M.F.; Rengel, D.; Carrère, S.; Hervé, C.; Debellé, F.; Gamas, P.; Bensmihen, S.; Gough, C. Combined genetic and transcriptomic analysis reveals three major signalling pathways activated by Myc-LCOs in Medicago truncatula. New Phytol. 2015, 208, 224–240. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Miller, J.B.; Granqvist, E.; Wiley-Kalil, A.; Gobbato, E.; Maillet, F.; Cottaz, S.; Samain, E.; Venkateshwaran, M.; Fort, S. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell 2015, 27, 823–838. [Google Scholar] [CrossRef] [Green Version]
- Delaux, P.M.; Bécard, G.; Combier, J.P. NSP1 is a component of the Myc signaling pathway. New Phytol. 2013, 199, 59–65. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, H.; Luo, D.; Yu, N.; Dong, W.; Wang, C.; Zhang, X.; Dai, H.; Yang, J.; Wang, E. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 2016, 7, 12433. [Google Scholar] [CrossRef]
- Das, D.; Gutjahr, C. Old dog, new trick: The PHR-SPX system regulates arbuscular mycorrhizal symbiosis. Mol. Plant 2022, 15, 225–227. [Google Scholar] [CrossRef]
- Gobbato, E.; Marsh, J.F.; Vernié, T.; Wang, E.; Maillet, F.; Kim, J.; Miller, J.B.; Sun, J.; Bano, S.A.; Ratet, P. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 2012, 22, 2236–2241. [Google Scholar] [CrossRef] [Green Version]
- Park, H.-J.; Floss, D.S.; Levesque-Tremblay, V.; Bravo, A.; Harrison, M.J. Hyphal branching during arbuscule development requires RAM1. Plant Physiol. 2015, 169, 2774–2788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rich, M.K.; Schorderet, M.; Bapaume, L.; Falquet, L.; Morel, P.; Vandenbussche, M.; Reinhardt, D. The petunia GRAS transcription factor ATA/RAM1 regulates symbiotic gene expression and fungal morphogenesis in arbuscular mycorrhiza. Plant Physiol. 2015, 168, 788–797. [Google Scholar] [CrossRef] [Green Version]
- Keymer, A.; Pimprikar, P.; Wewer, V.; Huber, C.; Brands, M.; Bucerius, S.L.; Delaux, P.-M.; Klingl, V.; von Roepenack-Lahaye, E.; Wang, T.L. Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 2017, 6, e29107. [Google Scholar] [CrossRef]
- Xue, L.; Klinnawee, L.; Zhou, Y.; Saridis, G.; Vijayakumar, V.; Brands, M.; Dörmann, P.; Gigolashvili, T.; Turck, F.; Bucher, M. AP2 transcription factor CBX1 with a specific function in symbiotic exchange of nutrients in mycorrhizal Lotus japonicus. Proc. Natl. Acad. Sci. USA 2018, 115, E9239–E9246. [Google Scholar] [CrossRef] [Green Version]
- Müller, L.M.; Campos-Soriano, L.; Levesque-Tremblay, V.; Bravo, A.; Daniels, D.A.; Pathak, S.; Park, H.-J.; Harrison, M.J. Constitutive overexpression of RAM1 leads to an increase in arbuscule density in Brachypodium distachyon. Plant Physiol. 2020, 184, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Heck, C.; Kuhn, H.; Heidt, S.; Walter, S.; Rieger, N.; Requena, N. Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr. Biol. 2016, 26, 2770–2778. [Google Scholar] [CrossRef] [Green Version]
- Seemann, C.; Heck, C.; Voß, S.; Schmoll, J.; Enderle, E.; Schwarz, D.; Requena, N. Root cortex development is fine-tuned by the interplay of MIGs, SCL3 and DELLAs during arbuscular mycorrhizal symbiosis. New Phytol. 2022, 233, 948–965. [Google Scholar] [CrossRef] [PubMed]
- Kobae, Y.; Hata, S. Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol. 2010, 51, 341–353. [Google Scholar] [CrossRef] [Green Version]
- Alexander, T.; Toth, R.; Meier, R.; Weber, H.C. Dynamics of arbuscule development and degeneration in onion, bean, and tomato with reference to vesicular–arbuscular mycorrhizae in grasses. Can. J. Bot. 1989, 67, 2505–2513. [Google Scholar] [CrossRef]
- Floss, D.S.; Gomez, S.K.; Park, H.-J.; MacLean, A.M.; Müller, L.M.; Bhattarai, K.K.; Lévesque-Tremblay, V.; Maldonado-Mendoza, I.E.; Harrison, M.J. A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr. Biol. 2017, 27, 1206–1212. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Snijders, R.; Kohlen, W.; Liu, J.; Bisseling, T.; Limpens, E. Medicago SPX1 and SPX3 regulate phosphate homeostasis, mycorrhizal colonization, and arbuscule degradation. Plant Cell 2021, 33, 3470–3486. [Google Scholar] [CrossRef]
- Lv, Q.; Zhong, Y.; Wang, Y.; Wang, Z.; Zhang, L.; Shi, J.; Wu, Z.; Liu, Y.; Mao, C.; Yi, K. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 2014, 26, 1586–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Ruan, W.; Shi, J.; Zhang, L.; Xiang, D.; Yang, C.; Li, C.; Wu, Z.; Liu, Y.; Yu, Y. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 14953–14958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauressergues, D.; Delaux, P.M.; Formey, D.; Lelandais-Brière, C.; Fort, S.; Cottaz, S.; Bécard, G.; Niebel, A.; Roux, C.; Combier, J.P. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012, 72, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Couzigou, J.-M.; Lauressergues, D.; André, O.; Gutjahr, C.; Guillotin, B.; Bécard, G.; Combier, J.-P. Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell Host Microbe 2017, 21, 106–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shtark, O.Y.; Sulima, A.S.; Zhernakov, A.I.; Kliukova, M.S.; Fedorina, J.V.; Pinaev, A.G.; Kryukov, A.A.; Akhtemova, G.A.; Tikhonovich, I.A.; Zhukov, V.A. Arbuscular mycorrhiza development in pea (Pisum sativum L.) mutants impaired in five early nodulation genes including putative orthologs of NSP1 and NSP2. Symbiosis 2016, 68, 129–144. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; André, O.; Puech-Pagès, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Gresshoff, P.M. Plant genetic control of nodulation. Annu. Rev. Microbiol. 1991, 45, 345–382. [Google Scholar] [CrossRef]
- Vierheilig, H.; Maier, W.; Wyss, U.; Samson, J.; Strack, D.; Piché, Y. Cyclohexenone derivative-and phosphate-levels in split-root systems and their role in the systemic suppression of mycorrhization in precolonized barley plants. J. Plant Physiol. 2000, 157, 593–599. [Google Scholar] [CrossRef]
- Meixner, C.; Vegvari, G.; Ludwig-Müller, J.; Gagnon, H.; Steinkellner, S.; Staehelin, C.; Gresshoff, P.; Vierheilig, H. Two defined alleles of the LRR receptor kinase GmNARK in supernodulating soybean govern differing autoregulation of mycorrhization. Physiol. Plant. 2007, 130, 261–270. [Google Scholar] [CrossRef]
- Catford, J.G.; Staehelin, C.; Lerat, S.; Piché, Y.; Vierheilig, H. Suppression of arbuscular mycorrhizal colonization and nodulation in split-root systems of alfalfa after pre-inoculation and treatment with Nod factors. J. Exp. Bot. 2003, 54, 1481–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khaosaad, T.; Staehelin, C.; Steinkellner, S.; Hage-Ahmed, K.; Ocampo, J.A.; Garcia-Garrido, J.M.; Vierheilig, H. The Rhizobium sp. strain NGR234 systemically suppresses arbuscular mycorrhizal root colonization in a split-root system of barley (Hordeum vulgare). Physiol. Plant. 2010, 140, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Tsikou, D.; Yan, Z.; Holt, D.B.; Abel, N.B.; Reid, D.E.; Madsen, L.H.; Bhasin, H.; Sexauer, M.; Stougaard, J.; Markmann, K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 2018, 362, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Suzaki, T.; Soyano, T.; Kojima, M.; Sakakibara, H.; Kawaguchi, M. Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 2014, 5, 4983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Reid, J.B.; Foo, E. The art of self-control–autoregulation of plant–microbe symbioses. Front. Plant Sci. 2018, 9, 988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirakawa, Y.; Sawa, S. Diverse function of plant peptide hormones in local signaling and development. Curr. Opin. Plant Biol. 2019, 51, 81–87. [Google Scholar] [CrossRef]
- Handa, Y.; Nishide, H.; Takeda, N.; Suzuki, Y.; Kawaguchi, M.; Saito, K. RNA-seq transcriptional profiling of an arbuscular mycorrhiza provides insights into regulated and coordinated gene expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol. 2015, 56, 1490–1511. [Google Scholar] [CrossRef] [Green Version]
- Funayama-Noguchi, S.; Noguchi, K.; Yoshida, C.; Kawaguchi, M. Two CLE genes are induced by phosphate in roots of Lotus japonicus. J. Plant Res. 2011, 124, 155–163. [Google Scholar] [CrossRef]
- Le Marquer, M.; Bécard, G.; Frei dit Frey, N. Arbuscular mycorrhizal fungi possess a CLAVATA3/embryo surrounding region-related gene that positively regulates symbiosis. New Phytol. 2019, 222, 1030–1042. [Google Scholar] [CrossRef]
- Wang, C.; Velandia, K.; Kwon, C.-T.; Wulf, K.E.; Nichols, D.S.; Reid, J.B.; Foo, E. The role of CLAVATA signalling in the negative regulation of mycorrhizal colonization and nitrogen response of tomato. J. Exp. Bot. 2021, 72, 1702–1713. [Google Scholar] [CrossRef]
- Müller, L.M.; Flokova, K.; Schnabel, E.; Sun, X.; Fei, Z.; Frugoli, J.; Bouwmeester, H.J.; Harrison, M.J. A CLE–SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat. Plants 2019, 5, 933–939. [Google Scholar] [CrossRef]
- Karlo, M.; Boschiero, C.; Landerslev, K.G.; Blanco, G.S.; Wen, J.; Mysore, K.S.; Dai, X.; Zhao, P.X.; de Bang, T.C. The CLE53–SUNN genetic pathway negatively regulates arbuscular mycorrhiza root colonization in Medicago truncatula. J. Exp. Bot. 2020, 71, 4972–4984. [Google Scholar] [CrossRef] [PubMed]
- De Bang, T.C.; Lundquist, P.K.; Dai, X.; Boschiero, C.; Zhuang, Z.; Pant, P.; Torres-Jerez, I.; Roy, S.; Nogales, J.; Veerappan, V. Genome-wide identification of Medicago peptides involved in macronutrient responses and nodulation. Plant Physiol. 2017, 175, 1669–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, R.; Chiapello, M.; Montero, H.; Gehrig, P.; Grossmann, J.; O’Holleran, K.; Hartken, D.; Walters, F.; Yang, S.-Y.; Hillmer, S. A rice Serine/Threonine receptor-like kinase regulates arbuscular mycorrhizal symbiosis at the peri-arbuscular membrane. Nat. Commun. 2018, 9, 4677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montero, H.; Lee, T.; Pucker, B.; Ferreras-Garrucho, G.; Oldroyd, G.; Brockington, S.F.; Miyao, A.; Paszkowski, U. A mycorrhiza-associated receptor-like kinase with an ancient origin in the green lineage. Proc. Natl. Acad. Sci. USA 2021, 118, e2105281118. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ho-Plágaro, T.; García-Garrido, J.M. Molecular Regulation of Arbuscular Mycorrhizal Symbiosis. Int. J. Mol. Sci. 2022, 23, 5960. https://doi.org/10.3390/ijms23115960
Ho-Plágaro T, García-Garrido JM. Molecular Regulation of Arbuscular Mycorrhizal Symbiosis. International Journal of Molecular Sciences. 2022; 23(11):5960. https://doi.org/10.3390/ijms23115960
Chicago/Turabian StyleHo-Plágaro, Tania, and José Manuel García-Garrido. 2022. "Molecular Regulation of Arbuscular Mycorrhizal Symbiosis" International Journal of Molecular Sciences 23, no. 11: 5960. https://doi.org/10.3390/ijms23115960
APA StyleHo-Plágaro, T., & García-Garrido, J. M. (2022). Molecular Regulation of Arbuscular Mycorrhizal Symbiosis. International Journal of Molecular Sciences, 23(11), 5960. https://doi.org/10.3390/ijms23115960






