Abstract
Descurainia sophia L. (flixweeds) is a noxious broad-leaf weed infesting winter wheat fields in China that has evolved high resistance to tribenuron-methyl. In this work, a brand new gene CYP77B34 was cloned from tribenuron-methyl-resistant (TR) D. sophia and transferred into Arabidopsis thaliana, and the sensitivities of Arabidopsis with or without the CYP77B34 transgene to herbicides with a different mode of actions (MoAs) were tested. Compared to Arabidopsis expressing pCAMBIA1302-GFP (empty plasmid), Arabidopsis transferring pCAMBIA1302-CYP77B34 (recombinant plasmid) became resistant to acetolactate synthase (ALS)-inhibiting herbicide tribenuron-methyl, protoporphyrinogen oxidase (PPO)-inhibiting herbicides carfentrazone-ethyl and oxyfluorfen. Cytochrome P450 inhibitor malathion could reverse the resistance to tribenuron-methyl, carfentrazone-ethyl and oxyfluorfen in transgenic Arabidopsis plants. In addition, the metabolic rates of tribenuron-methyl in Arabidopsis expressing CYP77B34 were significantly higher than those in Arabidopsis expressing pCAMBIA1302-GFP. Other than that, the transgenic plants showed some tolerance to very-long-chain fatty acid synthesis (VLCFAs)-inhibiting herbicide pretilachlor and photosystem (PS) II-inhibiting herbicide bromoxynil. Subcellular localization revealed that the CYP77B34 protein was located in the endoplasmic reticulum (ER). These results clearly indicated that CYP77B34 mediated D. sophia resistance to tribenuron-methyl and may have been involved in D. sophia cross-resistance to carfentrazone-ethyl, oxyfluorfen, pretilachlor and bromoxynil.
1. Introduction
Herbicides have been the most effective way for controlling weeds for several decades. Weeds have evolved serious resistance to a large number of herbicides due to extensive and intensive use of herbicides, which leads to serious loss of crop yields [1]. The mechanisms of weed resistance are typically classified into two broad categories. Target-site-based resistance (TSR) is endowed by gene mutation or deletion, or (and) gene overexpression or amplification of the herbicide target protein. Meanwhile, non-target-site-based resistance (NTSR) is achieved by enhanced herbicide metabolism (or) and sequestration, reduced penetration and (or) translocation [2,3]. In comparison with the well-known TSR, NTSR is focused on rarely and often covered by TSR or purposely ignored by researchers due to its complexities and difficulties. Metabolic resistance, one of the most common and important NTSRs, is caused by enhanced herbicide metabolism in weeds. Herbicide metabolism in plants usually experiences phases of conversion, conjugation and compartmentation, which are mediated by different metabolic enzymes. These metabolic enzymes include cytochrome P450 monooxygenase (P450s), glucosyltransferases (GTs), glutathione S-transferases (GSTs), aldo-keto reductase, ATP binding cassette transporters (ABC transporters) and esterases. Hence, enhanced metabolism of herbicides is one of the most common and important NTSR mechanisms [4,5,6,7].
Cytochrome P450 is one of the largest families in plant metabolism and represents around 1% of plant protein-coding genes. The P450s are regarded as the nature’s most versatile biological catalysts and catalyze numerous chemical reactions with countless substrates. These reactions refer to the typical mono-oxygenation process and atypical reactions of dimerization, isomerization, dehydration, etc. [8]. The P450s play crucial roles in the synthesis of various endogenous substances with important physiological functions in plants, such as hormones of brassinosteroid (BR), abscisic acid (ABA), gibberellin (GA) [9,10,11], signaling molecules of jasmonic acid (JA) and salicylic acid (SA) [12], plant defense molecules (suberin, lignin, flavonoids, phytoalexins) [13,14,15,16] and structural components (phenylpropanoids, fatty acids, sterols) [17,18]. In addition, P450s can metabolize all kinds of exogenous compounds including polychlorinated biphenyls (PCB), environmental chemicals, drugs and pesticides [8,19,20]. For this reason, P450s not only play important roles in plant growth, development and adaptation, but also are an excellent window to understanding plant evolution. Weed resistance is a good example of plant adaptation to herbicide selection pressures. Therefore, studies on the function of specific P450s are helpful not only to revealing the mechanisms of metabolic resistance in weeds, but also to understanding the evolutionary process of P450s.
Descurainiasophia L. (flixweeds) is a notorious broad-leaf weed infesting winter wheat fields in China that has evolved resistance to ALS-inhibiting herbicide tribenuron-methyl. Our previous work had confirmed that amino acid substitutions at positions Pro197 (substituted by Leu, Ser, Thr or Tyr), Asp376 (Glu) or Trp574 (Leu) in the ALS enzyme are the TSR mechanisms mainly responsible for D. sophia resistance to tribenuron-methyl [21,22,23]. The results of a P450 inhibitor and liquid chromatography−mass spectrometry (LC-MS) analysis confirmed that one or more P450 isoenzymes mediated D. sophia resistance to tribenuron-methyl [24]. Moreover, the results of the RNA-seq experiment indicated that metabolic enzyme families of GSTs, GTs and the ABC transporter may be involved in D. sophia resistance to tribenuron-methyl [25]. Based on the results of the above-mentioned research, five novel P450 genes with full-length sequences were identified and cloned from tribenuron-methyl-resistant (TR) D. sophia, which were named CYP77B34 (NCBI accession number MF152623), CYP81F5 (MF152624), CYP709B8 (MF152625), CYP96A146 (MF152630) and CYP96A147 (MF152626) by the P450 nomenclature committee [24,25]. Although indirect evidence suggested that CYP96A146 and CYP96A147 may mediate D. sophia resistance to tribenuron-methyl, there is no direct evidence on these P450 being involved in D. sophia resistance to tribenuron-methyl [24]. In this work, the TR D. sophia CYP77B34 was transferred into Arabidopsis thaliana, and the sensitivity of A. thaliana transformed with or without CYP77B34 to herbicides with different MoAs was tested. Synergism of the P450 inhibitor malathion and metabolism analysis of tribenuron-methyl by LC-MS were used to further validate the roles of CYP77B34 in herbicide resistance. Subcellular localization of CYP77B34 in Nicotiana benthamiana was observed. In addition, the expression levels of CYP77B34 in D. sophia were determined before and after tribenuron-methyl treatment.
2. Results
2.1. Cloning and Characterization of the CYP77B34 Gene
The CYP77B34 gene with full-length sequence was successfully cloned from TR D. sophia (N11) plants (Accession number MF152623) and contained a 1530 bp open reading frame (ORF) encoding a 510 amino acid protein. The deduced molecular weight of CYP77B34 was 57.8 kDa, and the theoretical pI was 8.58. The amino acid sequence of CYP77B34 had typical P450 conserved domains, such as the most diagnostic signature motif (F-x-x-G-x-R-x-C-x-G/A) (where x could represent any amino acid) for a P450 protein surrounding the heme cysteine ligand, less conserved sequences of Helix C (W-x-x-x-R), Helix I (A-G-x-D-T-S), Helix K (E-x-x-R) and meander (P-x-x-F-x-P-x-x-F). These typical structural domains indicated that CYP77B34 was a member of P450 proteins (Figure S1). In addition, CYP77B34 displayed high identity with CYP77A3 in Brassia napus (73.95%), CYP77B1 in A. thaliana (72.56%), CYP77A3 in Eutrema salsugineum (77.67%) and CYP77B1-1 in Isatis tinctoria (90.54%) (Figure S2).
The results of phylogenetic analysis indicated that CYP77B34 showed specific orthologous relationships with other CYP77A and CYP77B subfamilies genes in different cruciferous plants, including I. tinctoria, B. napus, Brassica rapa, Raphanus sativus, E. salsugineum, Camelina sativa, Capsella rubella, Arabidopsis lyrate and A. thaliana (Figure S3).
2.2. Transcript Levels of the CYP77B34 Gene in D. sophia
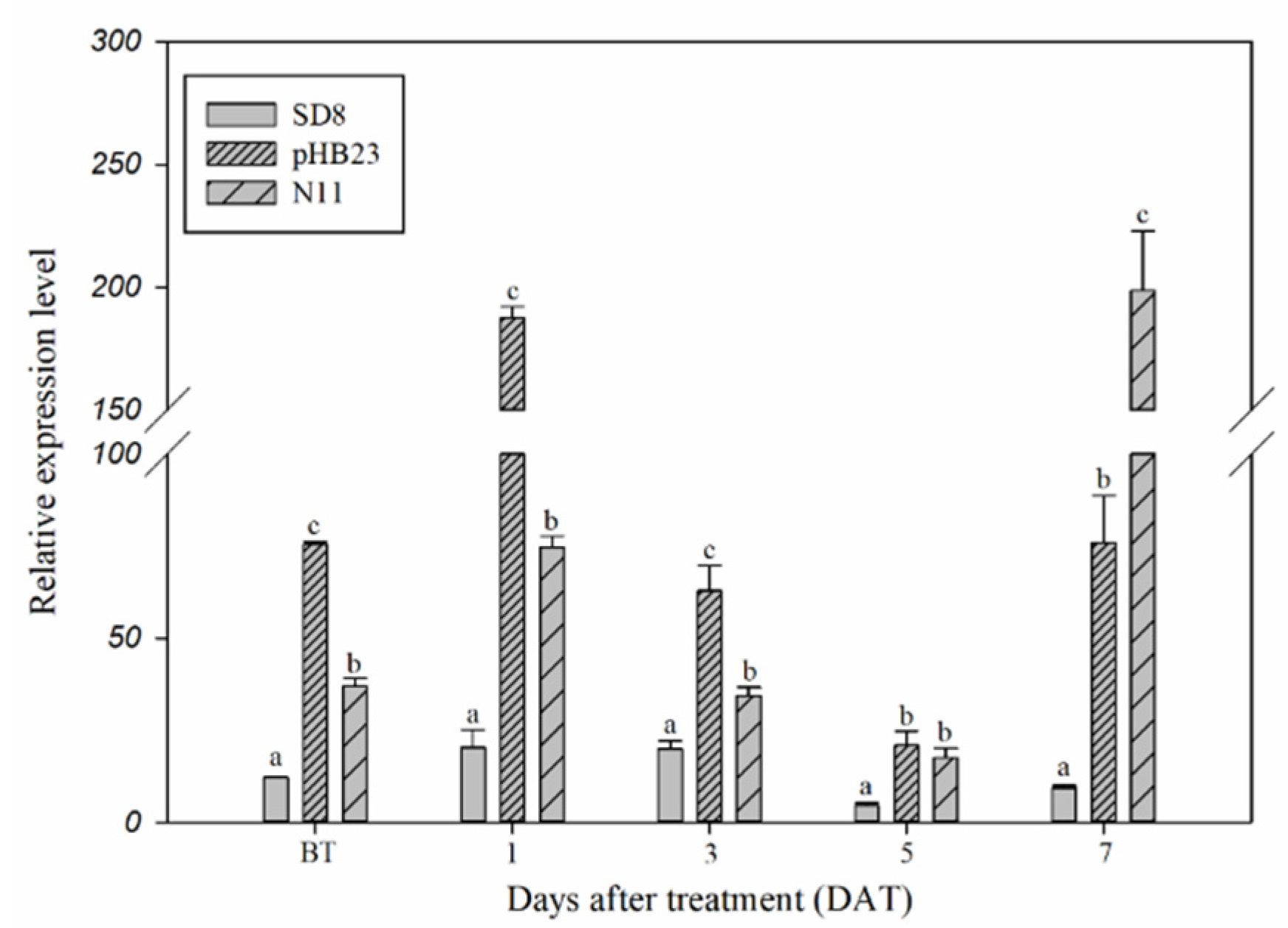
Constitutive and inducible expression of the CYP77B34 gene in D. sophia were determined in order to explore its roles in D. sophia resistance to tribenuron-methyl (Figure 1). The expression level of CYP77B34 in pHB23 and N11 D. sophia populations was 6.21- and 2.16-fold higher, respectively, than that in tribenuron-methyl-susceptible (TS) D. sophia (SD8) before tribenuron-methyl treatment (BT). In addition, CYP77B34 could be induced by tribenuron-methyl, and the expression level in TR D. sophia was always higher than that in TS D. sophia at 1, 3, 5 and 7 days after treatment (DAT). The expression levels increased about 9.00-fold in pHB23 at 1 DAT and 21.25-fold in N11 populations at 7 DAT comparing with that of TS D. sophia. Other than that, we also noted that the expression levels of CYP77B34 in TS and TR D. sophia decreased at 3 and 5 DAT comparing with that at 1 DAT. This may be due to the enhanced concentration of tribenuron-methyl absorption by plants at 3 and 5 DAT, which inhibited the expression of CYP77B34 TS and TR D. sophia. With the metabolism of tribenuron-methyl in two TR D. sophia, the inhibiting effect of tribenuron-methyl on CYP77B34 expression disappeared and even exhibited an inducing effect on CYP77B34 expression at 7 DAT.
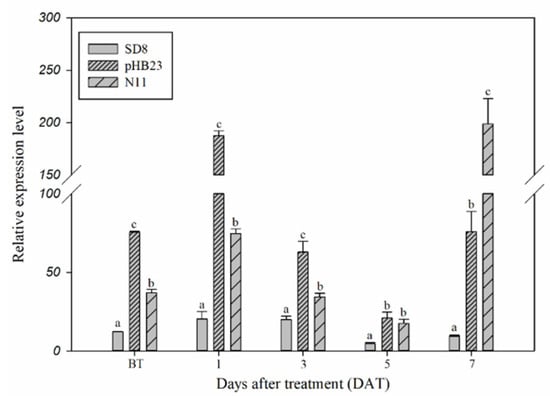
Figure 1.
Expression levels of CYP77B34 in TS (SD8) and TR (pHB23 and N11) D. sophia plants before (BT) or 1, 3, 5 and 7 days after treatment (DAT) with or without tribenuron-methyl. 18sRNA was the reference gene. All data are mean ± SE of three replicates. Columns with different letters indicate that the relative expression level had a significant difference at the same time point (using one-way analysis of variance, combined with Dunnett’s post-test comparison, p < 0.05).
2.3. Subcellular Localization of the CYP77B34 Protein
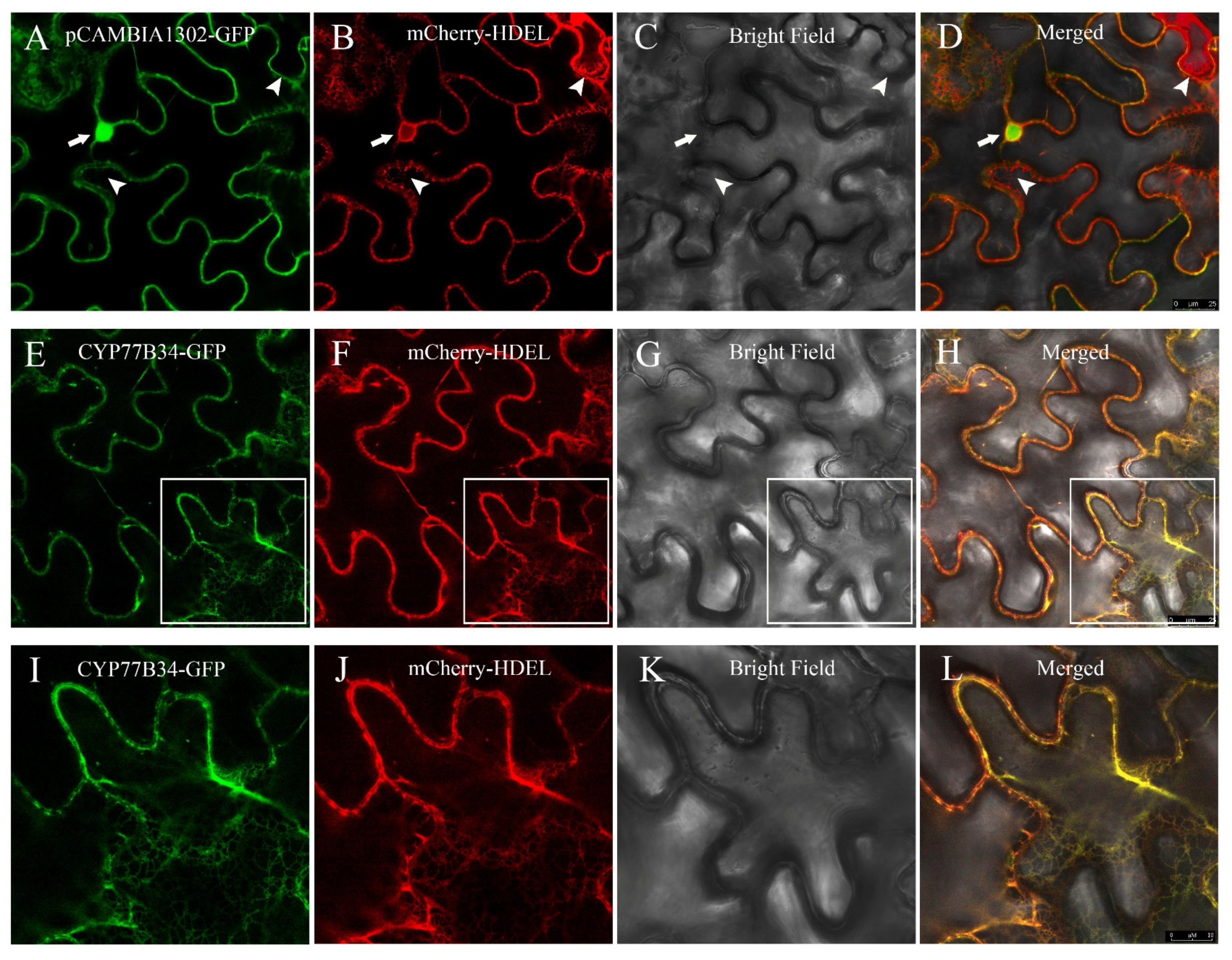
The results of transient expression indicated that the green fluorescence associated with the empty vector pCAMBIA1302-GFP distributed throughout the nucleus and cytoplasm (Figure 2A), and the red fluorescence emitted by the endoplasmic reticulum (ER) marker mCherry-HDEL was observed in the ER (Figure 2B). Merging of the green and red fluorescence was not unanimous, which suggested that the empty vector pCAMBIA1302-GFP was localized in the cytoplasm and the nucleus rather than in the ER (Figure 2D). When pCAMBIA1302-CYP77B34-GFP and mCherry-HDEL constructs were co-infiltrated in N. benthamiana leaves, the co-localization signals were observed in the ER network (Figure 2E–H). Pearson’s correlation coefficients between mCherry-HDEL and CYP77B34-GFP or mCherry-HDEL and pCAMBIA1302-GFP were 0.84 ± 0.021 and 0.27 ± 0.067, respectively (Figure S4). These results demonstrated that CYP77B34 was localized in the ER, which is similar to most P450s.
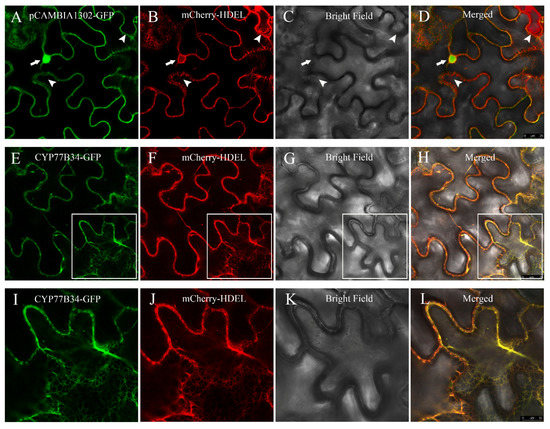
Figure 2.
Subcellular localization of CYP77B34. In (A–D), pCAMBIA1302-GFP shown in green (A) co-expressed with the endoplasmic reticulum (ER) marker mCherry-HDEL shown in red; (B–D) are the bright-field and merged images, respectively. The arrows and arrowheads show the nucleus and the reticulate-structure ER. In (E–H), CYP77B34-GFP shown in green (E) co-expressed with mCherry-HDEL shown in red; (F–H) are the bright-field and merged images, respectively. Images (I–L) are the magnifications of the selected areas in (E–H). Scale bars: 25 μm in (A–H) and 10 μm in (I–L).
2.4. Herbicide Sensitivity of Transgenic Arabidopsis Expressing CYP77B34
In order to characterize the metabolic ability of CYP77B34, the herbicide sensitivities of transgenic Arabidopsis expressing CYP77B34-GFP or GFP were compared. Four individual transgenic lines were obtained with different transcript levels of CYP77B34 (Figure S5). Obviously, the transgenic Arabidopsis carrying CYP77B34-GFP displayed different tolerance of herbicides with different MoAs. Compared with Arabidopsis expressing GFP, three lines of Arabidopsis carrying CYP77B34-GFP exhibited higher tolerance to tribenuron-methyl (ALS), carfentrazone-ethyl (PPO) and oxyfluorfen (PPO) (Table 1, Figures S6 and S7).

Table 1.
Herbicide sensitivity of different transgenic Arabidopsis lines expressing CYP77B34 to ALS-inhibiting herbicide (tribenuron-methyl) and PPO-inhibiting herbicides (carfentrazone-ethyl and oxyfluorfen) with or without malathion treatment.
Among the three lines, the line of 77B34#7 displayed the highest tolerance to tribenuron-methyl, carfentrazone-ethyl and oxyfluorfen with RIs of 3.08, 1.82 and 2.13, respectively. In addition, Arabidopsis with CYP77B34-GFP had tolerance of herbicides of bromoxynil (PSII) and pretilachlor (ALCFAs) (Figure S7F,G). Most seedlings of Arabidopsis with GFP grown in 20 μM bromoxynil were etiolated, while there were few etiolated seedlings for Arabidopsis with CYP77B34-GFP (Figure S7F). The seedlings’ growth of Arabidopsis carrying GFP was completely inhibited by pretilachlor at the concentration of 80 μM, while that of the Arabidopsis carrying CYP77B34-GFP was much better (Figure S7G).
In addition, the sensitivities of Arabidopsis with or without the CYP77B34 gene to other herbicides with different concentrations were tested in this work. These herbicides (MoAs) included imazamox and flumetsulam (ALS), napropamide (unknown MoA), glyphosate (enolpyruvyl shikimate phosphate synthase, EPSPS), glufosinate ammonium (glutamine synthetase, GS), diclofop-methyl (acetyl-coenzyme A carboxylase, ACCase), atrazine (PSII), propyzamide and pendimethalin (microtubule assembly), fluroxypyr (auxin mimics) and fomesafen (PPO). However, Arabidopsis with or without CYP77B34 gene showed no obvious differences to the herbicides mentioned above (Figure S8).
More importantly, the P450 inhibitor malathion could reverse the resistance to those three herbicides. Treatment with malathion (55 μM) had no response to the control line CK-GFP and three 77B34 lines. Synergism of herbicides and malathion could reduce the GR50 of three 77B34 lines, while it had no effect on CK-GFP (Table 1, Figure S6).
2.5. Absorption and Metabolism of Tribenuron-Methyl by Transgenic Arabidopsis Expressing CYP77B34
The Retention time of tribenuron-methyl was 1.366 min (Figure S9). The determination coefficients (R2) of two linear curves were 0.9951 (elution matrix) and 0.9914 (extract matrix) (Figure S10). Average recoveries were 90.93–105.19% (elution matrix) and 81.36–91.80% (extract matrix) at three different addition levels (Table S1). Relative standard deviation (RSD) of repeatability was below 5% (n = 4) (Table S1). The result showed that this method had high accuracy and good repeatability and was suitable for the absorption and metabolism analysis of tribenuron-methyl in Arabidopsis.
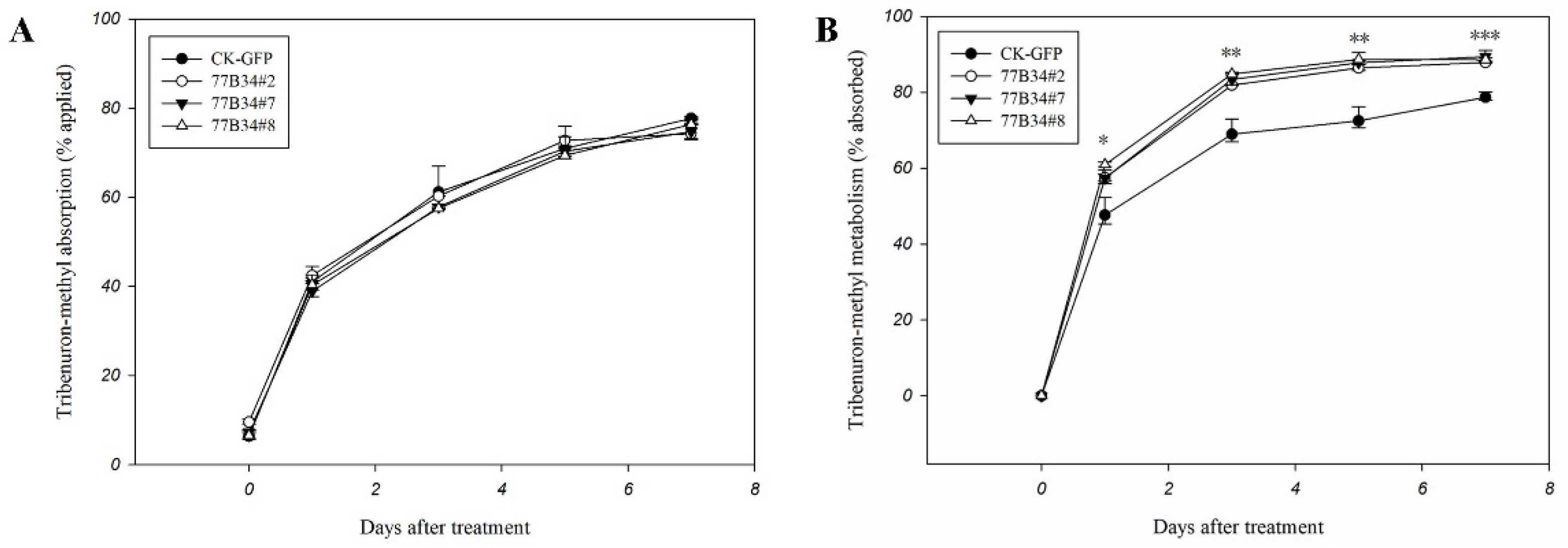
The reduced dosage between what was applied to and eluted from leaf surface was considered the absorption dosage of tribenuron-methyl by transgenic Arabidopsis plants. The absorption of tribenuron-methyl by CK-GFP and 77B34 lines (77B34#2, 77B34#7 and 77B34#8) displayed no significant differences at 0, 1, 3, 5 and 7 DAT (Figure 3A).
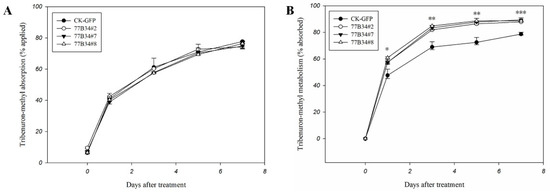
Figure 3.
Tribenuron-methyl absorption (A) and metabolism (B) in control (CK-GFP) and transgenic Arabidopsis lines (77B34#2, 77B34#7 and 77B34#8). Each data point is the mean ± SE of four replicates. *, ** and *** indicate the mean of tribenuron-methyl metabolized in CK-GFP, and three 77B34 lines displayed significant differences at p < 0.05, 0.01 and 0.001, respectively.
The difference between the absorption and residue in transgenic Arabidopsis plants was counted as tribenuron-methyl metabolism. The metabolic rate of tribenuron-methyl in three transgenic lines (77B34#2, 77B34#7 and 77B34#8) was significantly higher than that in line CK-GFP at 1, 3, 5 and 7 DAT (Figure 3B). Three transgenic Arabidopsis lines exhibited no significant differences in tribenuron-methyl metabolism at 1, 3 and 5 DAT. However, the metabolic ability for tribenuron-methyl of 77B34#7 and 77B34#8 was significantly higher than that in 77B34#2 at 7 DAT (Table S2).
3. Discussion
The roles of P450s in herbicide metabolism and weed resistance are well-known and have been reviewed in detail elsewhere [7,26]. Nevertheless, most of the existing evidence on P450’s involvement in herbicide resistance is indirect according to the results of synergism of P450 inhibitors, RNA-seq, transcript level or contents of P450s [27,28,29]. The strongest evidence on P450 conferring herbicide resistance is the expressing P450 allele in a herbicide-sensitive individual or a substituted organism (such as Arabidopsis), and observing a decrease of susceptibility to herbicides [3]. However, identification of herbicide-metabolizing and resistance-endowing genes is slow, and only a few P450 genes have been identified in grass weeds of Echinochloa phyllopogon, Echinochloa crus-galli and Lolium rigidum. The metabolisms of these P450s to herbicides were confirmed by heterologous expression. For example, Arabidopsis or Oryza sativa L. expressing E. phyllopogon P450 genes (CYP81A12, CYP81A14, CYP81A15, CYP81A18, CYP81A21, CYP81A24, CYP81A63) display metabolic ability for herbicides of ALS-inhibiting herbicides (pyrazosulfuron-ethyl, bensulfuron-methyl, penoxsulam, pyriftalid, pyrimisulfan, propyrisulfuron, chlorsulfuron, azimsulfuron or propoxycarbazone-sodium), ACCase-inhibiting herbicides (diclofop-methyl, tralkoxydim, pinoxaden), Deoxy-D-xyulose phosphate synthase-inhibiting herbicide (clomazone), PSII-inhibiting herbicide (bentazone), HPPD-inhibiting herbicide (mesotrione), PPO-inhibiting herbicide (pyraclonil) or (and) PDS-inhibiting herbicide (norflurazon) [30,31,32,33]. O. sativa transferring the E. crus-galli CYP81A68 gene has shown resistance to an ALS-inhibiting herbicide (penoxsulam) and ACCase-inhibiting herbicides (cyhalofop-butyl and metamifop) [34]. Another O. sativa transferring the L. rigidum CYP81A10v7 gene became highly resistant to ACCase- and ALS-inhibiting herbicides (diclofop-methyl, tralkoxydim, chlorsulfuron) and moderately resistant to HPPD-inhibiting herbicide trifluralin [35].
We noticed that studies on metabolic herbicide resistance in broad-leaf weed species are limited, and genes related to herbicide metabolism almost remain unknown [24,36,37,38,39]. In recent years, our team has proved the important role of P450s in D. sophia resistance to tribenuron-methyl through a series of experiments, such as synergism of P450 inhibitors, absorption/metabolism, RNA- seq and qRT-PCR. A batch of metabolic enzyme genes including P450s (CYP77B34, CYP709B8, CYP81F5, CYP96A146 and CYP96A147) were identified in TR D. sophia plants [24,25]. These P450s of CYP77B34, CYP709B8, CYP81F5, CYP96A146 and CYP96A147 may confer D. sophia resistance to tribenuron-methyl. Nevertheless, the function of a specific P450 gene in the metabolism and resistance to tribenuron-methyl is still ambiguous. In this work, the CYP77B34 gene was transferred into Arabidopsis, and the sensitivities to tribenuron-methyl and other herbicides with different MoAs were observed. The results indicated that P450 of CYP77B34 exhibited metabolic ability for herbicides with MoAs of an ALS-inhibiting herbicide (tribenuron-methyl), PPO-inhibiting herbicides (carfentrazone-ethyl and oxyfluorfen), a VLCFAs-inhibiting herbicide (pretilachlor) and a PSII-inhibiting herbicide (bromoxynil) (Figures S6 and S7). For three lines of Arabidopsis expressing CYP77B34, the RIs to tribenuron-methyl were 1.87, 3.08 and 2.17, respectively (Table 1). The P450 inhibitor malathion reduced the RI values of the three lines to 1.38, 1.62 and 1.67, which demonstrated that P450 (CYP77B34) mediated tribenuron-methyl metabolism (Table 1). In addition, tribenuron-methyl metabolism in transgenic Arabidopsis expressing CYP77B34 was faster than that in CK-GFP (Figure 3). The present results indicated that CYP77B34 mediated the metabolism and resistance to tribenuron-methyl in D. sophia. However, the proportion of resistant contribution caused by CYP77B34 is unclear because of the enormous quantity of P450s and their unclear function in D. sophia. In the meantime, both the constitutive and induced expression levels of CYP77B34 in TR D. sophia were significantly higher than that in TS D. sophia (Figure 1). This demonstrated that CYP77B34 could be involved in resistance evolution to tribenuron-methyl in TR D. sophia. What we can not ignore is that the TR D. sophia had potential cross-resistance risk to carfentrazone-ethyl, oxyfluorfen, pretilachlor and bromoxynil due to its metabolism of these herbicides. Compared with those of TSR, the biggest threat is that NTSR causes weeds to evolve unpredictable cross-resistance to herbicides with different MoAs, even including herbicides that are not yet in the market. In addition, resistance management strategies of herbicide mixtures and rotations which are effective in managing TSR may have little or no effects on NTSR metabolic resistance [7,40]. This situation has been confirmed in many resistance cases mediated by P450. For example, O. sativa transferring the L. rigidum CYP81A10v7 gene became highly resistant to ACCase- and ALS-inhibiting herbicides (diclofop-methyl, tralkoxydim, chlorsulfuron) and moderately resistant to HPPD-inhibiting herbicide trifluralin [35].
The evolution of new functions in plant metabolism usually experiences a very long process. However, the comparative analysis of plant P450s is helpful to understanding P450 function and evolution in plant metabolism [41]. Phylogenetic analysis of nucleotide sequences of CYP77B34 was inferred using the neighbor-joining method employing MEGA7 software. The CYP77B34 gene exhibited maximum similarity with members of theCYP77A and CYP77B subfamilies (Figure S3). The results of recombinant enzymes demonstrate that CYP77s can either in-chain hydroxylate or epoxidize fatty acids, in particular with 16- and 18-carbon chain lengths to form multiple precursors of cutin and suberin [42,43]. For example, Arabidopsis CYP77A6 was confirmed to be involved in the synthesis of flower cutin monomers [44], and Arabidopsis CYP77A4 is involved in signaling defense with epoxidation and in-chain hydroxylation [45,46]. CYP77A1 and CYP77B1 in petunia were related to the development of ornamental organs [42,47]. In P450-catalyzed herbicide metabolism, alkyl- or aryl-hydroxylation is one of the most common reactions [48]. Hydrophobic herbicide molecules can be oxidized to more hydrophilic metabolites by the hydroxylation reaction catalyzed by P450, which is the key step of herbicide degradation. CYP77B34 may then play this pivotal role in the metabolism of herbicides. Meanwhile, the CYP77B34 protein is localized in the endoplasmic reticulum, which encompasses various kinds of molecular machines of proteins, including folding, quality control, signal transduction and degradation [49]. CYP77B34 may play one or more roles in that.
4. Materials and Methods
4.1. Plant Materials
The tribenuron-methyl-susceptible (TS) D. sophia population (SD8) was originally collected from remote areas at Linyi city of Shandong province in China (35°05′45.00″ N, 118°09′3.78″ E), and two tribenuron-methyl-resistant (TR) D. sophia populations (pHB23, N11) were harvested from winter wheat fields at Baoding city of Hebei province (38°48′55.4″ N, 115°23′23.0″ E for pHB23; 38°36′32.80″ N, 115°01′52.50″ E for N11) in 2013. All the seeds were purified by individual plant propagation to ensure the consistency of seeds’ genetic backgrounds. D. sophia of pHB23 and N11 evolved 258.3- and 116.3-fold resistance to tribenuron-methyl, respectively [25,50].
Seeds of D. sophia were sterilized with 20% H2O2 for 30 min, then soaked in a 0.3% gibberellin solution overnight after rinsing with distilled water. Next, these seeds were germinated on wet paper for 3–5 days in a climate chamber, then transplanted into plastic pots containing moist loam soil, then grown in a climate chamber under conditions of 25 °C/23 °C (day/night) temperature, 16 h photoperiod with light intensity of 20,000 lux.
Arabidopsis thaliana ecotype Columbia (Col-0) and Nicotiana benthamiana seeds were sterilized with 75% ethyl alcohol for 30 s and further sterilized in a 5% sodium hypochlorite solution for 8 min after rinsing with sterile water. Finally, the seeds were washed with sterile water and germinated on Murashige and Skoog solid medium (MS) for 2–3 weeks in a climate chamber with the same conditions as above after vernalization in 4 °C for 12–24 h [51]. These operations were performed under sterile conditions.
4.2. Expression Levels Determination of CYP77B34 in D. sophia
TS and TR D. sophia populations at the 6-leaf stage were treated with 15 μL tribenuron-methyl acetone solutions with concentration of 20 mg L−1 (0.3 μg per plant) by a micro applicator (Hamilton PB600 dispenser, Hamilton Co., Reno, NV, USA). The seedlings above the ground were collected for qRT-PCR at 0, 1, 3, 5 and 7 days after treatment (DAT). At each time point, four plants were harvested as one replicate, and three replicates were applied. Total RNA was extracted according to the instructions of an RNApre Pure Plant Kit (TIANGEN, Beijing, China). cDNA was synthesized using RNA as a template according to the instructions of a TIANScriptII RT Kit (TIANGEN, Beijing, China).
The expression levels were determined according to the instructions of a KAPA SYBR FAST qPCR Kit by an ABI Prism 7500 Real-Time PCR System (Applied Biosystems, Foster city, CA, USA). A total of 20 μL of reactions mixtures contained 10 μL 2 × KAPA SYBR FAST qPCR Master Mix Universal, 0.4 μL 50 × ROX Row, 0.6 μL primer pairs (10 μM), 1 μL cDNA template and 7.4 μL ddH2O. Primer information of CYP77B34 and reference gene 18sRNA were listed in Table 2. The qPCR program was as follows: 95 °C for 3 min and 40 cycles of (95 °C for 3 s, 60 °C for 20 s and 72 °C for 32 s). Data was collected at the stage of 72 °C for 32 s, and melting analysis was carried out to verify the absence of nonspecific amplification at the end of the cycles. The data of gene expression was analyzed with 7500 Software v2.3 (Applied Biosystems, Foster City, CA, USA). Relative expression ratio (as 2−ΔΔCt) was calculated by the comparative CT method, where ΔCT = [CT target gene-CT internal control gene]. One-way analysis of variance (ANOVA) with Dunnett’s post-test (α = 5%) was conducted to assess significant differences by IBM SPSS Statistics 21 (International Business Machines Corporation, Armonk, NY, USA). Three biological replicates and four technical replicates were performed for each time point of TS and TR D. sophia populations [52].

Table 2.
Information of primers for cloning, vector construction and qRT-PCR of the CYP77B34 gene.
4.3. Cloning and Phylogenetic Analysis
Five plants of TR D. sophia (N11) at the four-leaf stage were used for RNA extraction. Total RNA extraction and cDNAs synthesis were the same as in the above-mentioned methods. The amplification of the CYP77B34 gene was performed using TIANSeq HiFi Amplification Mix (TIANGEN, Beijing, China). The PCR reaction mixtures contained 100 ng of cDNA, 0.25 μM of forward and reverse primers (Table 2) and 1 × HiFi amplification mix. The PCR conditions were as follows: 94 °C for 2 min, 35 cycles (98 °C for 10 s, 60 °C for 30 s and 68 °C for 45 s) and an extension step of 68 °C for 5 min. The PCR products were sequenced (Sangon Biotech, Shanghai, China) and aligned with DNAMAN 6.0 software (Lynnon Biosoft, San Ramon, CA, USA).
The phylogenetic analyses of CYP77B34 were performed with the Mega 7 program using available nucleotide sequences of plant P450s in the CYP77 family by the neighbor-joining method [53]. Bootstrap analysis with 1000 replicates was conducted in order to obtain confidence levels for the branches. In order to facilitate and simplify the result, only major groups were shown.
4.4. pCAMBIA1302-CYP77B34-GFP Vector Construction and Agrobacterium Transformation
The cDNA of CYP77B34 with a full-length sequence was inserted in the pCAMBIA1302-GFP vector with 35S promoters according to the instructions of a Clone ExpresII One-Step Cloning Kit (Vazyme Biotech, Nanjing, China). The pCAMBIA1302-CYP77B34-GFP vector was transformed into Escherichia coli, and the insert fragment sequence was confirmed by sequencing. The pCAMBIA1302-CYP77B34-GFP plasmid was purified and then transferred into the Agrobacterium tumefaiciens strain GV3101 (Biomed, Beijing, China).
4.5. Subcellular Localization
Tobacco leaves’ transient expression systems were used to determine the subcellular localization of the CYP77B34 protein. The subcellular localization was conducted according to the methods described by Pang et al. [54]. pCAMBIA1302-CYP77B34-GFP, the empty vector pCAMBIA1302-GFP and endoplasmic-reticulum (ER)-specific marker (mCherry-HDEL) were co-expressed in tobacco leaves. A. tumefaciens (GV3101 strain) transformed with pCAMBIA1302-CYP77B34-GFP, pCAMBIA1302-GFP or mCherry-HDEL was incubated at 28 °C with shaking (200 rpm) for 18 h. Bacteria were collected by centrifugation at 4000 rpm for 4 min and were resuspended to a final OD600 of 1.5 in an MES buffer (10 mM of MgCl2 and 10 mM of MES, pH 5.6) containing 150 nM of acetosyringone and were rested for 3 h. The bacteria suspensions mixed with a p19 silencing suppressor were infiltrated into leaves of N. benthamiana. Three days later, leaves of N. benthamiana were collected for microscopic observation using a Leica Stellaris5 confocal microscope (Leica, Heidelberg GmbH, Heidelberg, Germany). The green fluorescent protein (GFP) signal was excited at 488 nm and detected in the 500–550 nm range. The mCherry signal was excited at 561 nm and detected at 590–640 nm. The ImageJ plugin coloc 2 was used to measure Pearson’s correlation coefficient. More than 10 pictures were quantified for each co-localization.
4.6. Arabidopsis Transformation and Transcript Analysis of CYP77B34
A. tumefaiciens-transformed pCAMBIA1302-CYP77B34-GFP or pCAMBIA1302-GFP were transformed into A. thaliana by the standard floral dip method [55]. Transgenic Arabidopsis were selected on MS plates containing 25 μg mL−1 of hygromycin B, and homozygous plants were obtained by single plant reproduction for 3 consecutive generations. At least 4 independent transgenic lines, which was confirmed by PCR, were collected for each construct. The expression levels of CYP77B34 in each line of transgenic Arabidopsis were determined by qRT-PCR.
Transgenic Arabidopsis at the 6-leaf stage was used for transcript analysis. The experiment was carried out on three biological and three technical replications. The GAPDH gene was selected as a reference gene, and the primer information is shown in Table 2. The qRT-PCR program was the same as that in expression levels’ determination of CYP77B34 in D. sophia.
4.7. Herbicide Sensitivity of Transgenic Arabidopsis Expressing CYP77B34
The transgenic Arabidopsis was cultivated in MS solid medium containing herbicides (or/and 55 μM of malathion) with different concentrations and kept in a climate chamber under conditions of 25 °C/23 °C (day/night) temperature, 16 h photoperiod with light intensity of 20,000 lux. The plant growth or fresh weight of transgenic Arabidopsis were observed or weighed. These herbicides included inhibitors of ALS (tribenuron-methyl, imazamox and flumetsulam), VLCFAs (pretilachlor), EPSPS (glyphosate), GS (glufosinate ammonium), ACCase (diclofop-methyl), PSII (bromoxynil and atrazine), microtubule assembly (propyzamide and pendimethalin), auxin mimics (fluroxypyr), PPO (fomesafen, carfentrazone-ethyl and oxyfluorfen) and an unknown MoA (napropamide).
GR50, 50% inhibition of plant growth reduction, was calculated using the four-parameter log-logistic equation by SigmaPlot 12.0 (Systat Software Inc., San Jose, CA, USA).
where b is the slope at GR50, C and D are the lower and upper limits, respectively. The resistance index (RI) was calculated by GR50 of the transgenic Arabidopsis expressing CYP77B34-GFP divided by that of the transgenic Arabidopsis expressing GFP to estimate the resistance levels.
4.8. Tribenuron-Methyl Metabolism Analysis in Transgenic Arabidopsis Using LC-MS
Transgenic Arabidopsis plants expressing GFP and CYP77B34 were treated with tribenuron-methyl (0.16 μg/plant) at the 6-leaf stage. Above-ground parts of transgenic Arabidopsis plants were harvested at 0, 1, 3, 5 and 7 DAT. For each time point, five plants were harvested as one replicate per line, and four replicates were applied.
The residual tribenuron-methyl on the surfaces of transgenic Arabidopsis plants was washed with 8 mL of acetonitrile. The elution of acetonitrile was filtered with a 0.22 μm syringe filter, and tribenuron-methyl was quantified by LC-MS (Agilent 1290 InfinityII-6470, Palo Alto, CA, USA). Above-mentioned plant tissues were used for tribenuron-methyl metabolism measurement. The plants were ground into powder in liquid nitrogen and sonicated for 10 min with 5 mL of acetonitrile (with 1% acetic acid). After adding 0.5 g of NaCl, the homogenate was vibrated by a vortex for 2 min and centrifuged at 5000 rpm for 5 min. A 1 mL supernatant acetonitrile layer was added into a 2 mL centrifuge tube (containing 50 mg of anhydrous MgSO4, 25 mg of PSA and 7 mg of GCB) and vibrated by a vortex for 1 min. Then, the supernatant was filtered with a 0.22 μm syringe filter after centrifugation at 12,000 rpm for 3 min and analyzed by LC-MS.
The analysis of tribenuron-methyl was conducted by LC-MS with Agilent HPLC packed Shim-pack GIST C18 (50 × 2.1 mm i.d., 2 μm). The mobile phase was composed of 80% water and 20% acetonitrile, and the flow rate was 0.4 mL/min. Injection volume was 2 μL, and run time was 2.5 min. MS ran at conditions of DL temperature of 250 °C, heat block temperature of 300 °C, nebulizing gas flow of 5.0 L/min and drying gas flow of 11.0 L/min. The multiple reaction monitoring mode (MRM) for tribenuron-methyl was optimized at 396.2 > 155.1 under the above-mentioned conditions.
Linearity, recovery and precision were evaluated to ensure the quality of the analytical method. Linearity was determined by injecting tribenuron-methyl working standard solutions at concentrations of 0.001, 0.005, 0.01, 0.05, 0.1 and 0.2 mg/L followed by linear regression analysis. Precision was calculated as relative standard deviation (RSD) from recovery studies with standard spiked samples (n = 4) at levels of 0.01, 0.1 and 0.2 mg/L (for the elution matrix) and 0.016, 0.16 and 0.32 mg/L (for the extract matrix).
Data of tribenuron-methyl absorption and metabolism were subjected to ANOVA with Duncan’s test by IBM SPSS Statistics 21. Four replicates were conducted for each line.
5. Conclusions
In summary, CYP77B34 from D. sophia is a typical P450 protein localized in the ER. The constitutive and induced expression levels of the CYP77B34 gene in TR D. sophia was significantly higher than that in TS plants. Arabidopsis expressing CYP77B34 exhibited obvious resistance to tribenuron-methyl, carfentrazone-ethyl and oxyfluorfen. The P450 inhibitor malathion could reverse the transgenic Arabidopsis resistance to three herbicides. Compared to that of CK-GFP, three 77B34 lines possessed stronger metabolic ability for tribenuron-methyl. In addition, transgenic Arabidopsis expressing CYP77B34 also displayed tolerance of bromoxynil and pretilachlor. The results provided direct evidence on CYP77B34 endowing D. sophia resistance to tribenuron-methyl. At the same time, it also proved that cross-resistance patterns caused by P450 could be unpredictable.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23105812/s1.
Author Contributions
M.Z. and J.S. conceived and designed the research and coordinated the project. J.S. and M.Z. wrote and revised the manuscript. J.S. performed the main experiments and analyzed the data. Q.Y., L.H., L.Z. and X.L. provided advice on the experimental design. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31672047).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interests.
References
- Baucom, R.S. The remarkable repeated evolution of herbicide resistance. Am. J. Bot. 2016, 103, 181–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powles, S.B.; Yu, Q. Evolution in Action: Plants Resistant to Herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Délye, C.; Jasieniuk, M.; Corre, V.L. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Suzukawa, A.K.; Bobadilla, L.K.; Mallory-Smith, C.; Brunharo, C.A.C.G. Non-target-Site Resistance in Lolium spp. Globally: A Review. Front. Plant Sci. 2021, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.S.; Tranel, P.J.; Stewart, N.C., Jr. Non-target-site herbicide resistance: A family business. Trends Plant Sci. 2007, 12, 6–13. [Google Scholar] [CrossRef]
- Carey, V.F.; Hoagland, R.E.; Talbert, R.E. Resistance mechanism of propanil-resistant barnyardgrass. In-vivo metabolism of the propanil molecule. Pestic. Sci. 1997, 49, 333–338. [Google Scholar] [CrossRef]
- Yu, Q.; Powles, S. Metabolism-Based Herbicide Resistance and Cross-Resistance in Crop Weeds: A Threat to Herbicide Sustainability and Global Crop Production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef] [Green Version]
- Werck-Reichhart, D.; Hehn, A.; Didierjean, L. Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 2000, 5, 116–123. [Google Scholar] [CrossRef]
- Takahashi, N.; Nakazawa, M.; Shibata, K.; Yokota, T.; Matsui, M. shk1-D, a dwarf Arabidopsis mutant caused by activation of the CYP72C1 gene, has altered brassinosteroid levels. Plant J. 2005, 42, 13–22. [Google Scholar] [CrossRef]
- Regnault, T.; Daviere, J.M.; Heintz, D.; Lange, T.; Achard, P. The gibberellin biosynthetic genes AtKAO1 and AtKAO2 have overlapping roles throughout Arabidopsis development. Plant J. 2014, 80, 462–474. [Google Scholar] [CrossRef]
- Yang, S.H.; Zeevaart, J.A.D. Expression of ABA 8’-hydroxylases in relation to leaf water relations and seed development in bean. Plant J. 2006, 47, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.L.; Zhang, R.Z.; Li, D.L.; Wang, F. Integrating Transcriptome and Coexpression Network Analyses to Characterize Salicylic Acid- and Jasmonic Acid-Related Genes in Tolerant Poplars Infected with Rust. Int. J. Mol. Sci. 2021, 22, 5001. [Google Scholar] [CrossRef] [PubMed]
- Schoenbohm, C.; Martens, S.; Eder, C.; Forkmann, G.; Weisshaar, B. Identification of the Arabidopsis thaliana flavonoid 3'-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol. Chem. 2000, 381, 749–753. [Google Scholar] [CrossRef]
- Shiono, K.; Yamauchi, T.; Yamazaki, S.; Mohanty, B.; Malik, A.I.; Nagamura, Y.; Nishizawa, N.K.; Tsutsumi, N.; Colmer, T.D.; Nakazono, M. Microarray analysis of laser-microdissected tissues indicates the biosynthesis of suberin in the outer part of roots during formation of a barrier to radial oxygen loss in rice (Oryza sativa). J. Exp. Bot. 2014, 65, 4795–4806. [Google Scholar] [CrossRef] [Green Version]
- Park, H.; Park, G.; Jeon, W.; Ahn, J.-O.; Yang, Y.-H.; Choi, K.-Y. Whole-cell biocatalysis using cytochrome P450 monooxygenases for biotransformation of sustainable bioresources (fatty acids, fatty alkanes, and aromatic amino acids). Biotechnol. Adv. 2020, 40, 107504. [Google Scholar] [CrossRef]
- Mao, H.J.; Liu, J.; Ren, F.; Peters, R.J.; Wang, Q. Characterization of CYP71Z18 indicates a role in maize zealexin biosynthesis. Phytochemistry 2016, 121, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Gang, D.R.; Beuerle, T.; Ullmann, P.; Werck-Reichhart, D.; Pichersky, E. Differential production of meta hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases. Plant Physiol. 2002, 130, 1536–1544. [Google Scholar] [CrossRef] [Green Version]
- Hamberger, B.; Bak, S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos. Trans. R. Soc. B-Biol. Sci. 2013, 368, 1612. [Google Scholar] [CrossRef] [Green Version]
- Schlezinger, J.J.; Struntz, W.D.J.; Goldstone, J.V.; Stegeman, J.J. Uncoupling of cytochrome P450 1A and stimulation of reactive oxygen species production by co-planar polychlorinated biphenyl congeners. Aquat. Toxicol. 2006, 77, 422–432. [Google Scholar] [CrossRef]
- Dai, D.; Cao, Y.; Falls, G.; Levi, P.E.; Hodgson, E.; Rose, R.L. Modulation of mouse P450 isoforms CYP1A2, CYP2B10, CYP2E1, and CYP3A by the environmental chemicals mirex, 2,2-bis(p-chlorophenyl)-1,1-dichloroethylene, vinclozolin, and flutamide. Pest. Biochem. Physiol. 2001, 70, 127–141. [Google Scholar] [CrossRef]
- Deng, W.; Cao, Y.; Yang, Q.; Liu, M.J.; Mei, Y.; Zheng, M.Q. Different cross-resistance patterns to AHAS herbicides of two tribenuron-methyl resistant flixweed ( Descurainia sophia L.) biotypes in China. Pestic. Biochem. Physiol. 2014, 112, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Liu, M.J.; Yang, Q.; Mei, Y.; Li, X.F.; Zheng, M.Q. Tribenuron-methyl resistance and mutation diversity of Pro197 in flixweed (Descurainia sophia L.) accessions from China. Pestic. Biochem. Physiol. 2015, 117, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Yang, Q.; Zhang, Y.; Jiao, H.; Mei, Y.; Li, X.; Zheng, M. Cross-resistance patterns to acetolactate synthase (ALS)-inhibiting herbicides of flixweed (Descurainia sophia L.) conferred by different combinations of ALS isozymes with a Pro-197-Thr mutation or a novel Trp-574-Leu mutation. Pestic. Biochem. Physiol. 2017, 136, 41. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, J.Y.; Shen, J.; Xu, Y.F.; Liu, H.J.; Deng, W.; Li, X.F.; Zheng, M.Q. Metabolic Resistance to Acetolactate Synthase Inhibiting Herbicide Tribenuron-Methyl in Descurainia sophia L. Mediated by Cytochrome P450 Enzymes. J. Agric. Food Chem. 2018, 66, 4319–4327. [Google Scholar] [CrossRef]
- Yang, Q.; Deng, W.; Li, X.; Yu, Q.; Zheng, M. Target-site and non-target-site based resistance to the herbicide tribenuron-methyl in flixweed (Descurainia sophia L.). BMC Genom. 2016, 17, 551. [Google Scholar] [CrossRef] [Green Version]
- Siminszky, B. Plant cytochrome P450-mediated herbicide metabolism. Phytochem. Rev. 2006, 5, 445–458. [Google Scholar] [CrossRef]
- Busi, R.; Gaines, T.A.; Powles, S. Phorate can reverse P450 metabolism-based herbicide resistance in Lolium rigidum. Pest Manag. Sci. 2016, 73, 410. [Google Scholar] [CrossRef]
- Gaines, T.A.; Lorentz, L.; Figge, A.; Herrmann, J.; Maiwald, F.; Ott, M.-C.; Han, H.; Busi, R.; Yu, Q.; Powles, S.B.; et al. RNA-Seq transcriptome analysis to identify genes involved in metabolism-based diclofop resistance in Lolium rigidum. Plant J. 2014, 78, 865–876. [Google Scholar] [CrossRef]
- Duhoux, A.; Carrère, S.; Duhoux, A.; Délye, C. Transcriptional markers enable identification of rye-grass (Lolium sp.) plants with non-target -site-based resistance to herbicides inhibiting acetolactate-synthase. Plant Sci. 2017, 257, 22–36. [Google Scholar] [CrossRef]
- Dimaano, N.G.; Yamaguchi, T.; Fukunishi, K.; Tominaga, T.; Iwakami, S. Functional characterization of cytochrome P450 CYP81A subfamily to disclose the pattern of cross-resistance in Echinochloa phyllopogon. Plant Mol. Biol. 2020, 102, 403–416. [Google Scholar] [CrossRef]
- Iwakami, S.; Endo, M.; Saika, H.; Okuno, J.; Nakamura, N.; Yokoyama, M.; Watanabe, H.; Toki, S.; Uchino, A.; Inamura, T. Cytochrome P450 CYP81A12 and CYP81A21 Are Associated with Resistance to Two Acetolactate Synthase Inhibitors in Echinochloa phyllopogon. Plant Physiol. 2014, 165, 618–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.; Iwakami, S.; Yamaguchi, T.; Uchino, A.; Sunohara, Y.; Matsumoto, H. Role of CYP81A cytochrome P450s in clomazone metabolism in Echinochloa phyllopogon. Plant Sci. 2019, 283, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Kamidate, Y.; Yamaguchi, T.; Ishizaka, M.; Endo, M.; Suda, H.; Nagai, K.; Sunohara, Y.; Toki, S.; Uchino, A.; et al. CYP81A P450s are involved in concomitant cross-resistance to ALS and ACCase herbicides in Echinochloa phyllopogon. New Phytol. 2018, 221, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Guo, Q.; Wang, J.; Shi, L.; Yang, X.; Zhou, Y.; Yu, Q.; Bai, L. CYP81A68 confers metabolic resistance to ALS and ACCase-inhibiting herbicides and its epigenetic regulation in Echinochloa crus-galli. J. Hazard. Mater. 2022, 428, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Yu, Q.; Beffa, R.; González, S.; Maiwald, F.; Wang, J.; Powles, S.B. Cytochrome P450 CYP81A10v7 in Lolium rigidum confers metabolic resistance to herbicides across at least five modes of action. Plant J. 2020, 105, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Reycaballero, J.; Menéndez, J.; Osuna, M.D.; Salas, M.; Torra, J. Target-site and non-target-site resistance mechanisms to ALS inhibiting herbicides in Papaver rhoeas. Pestic. Biochem. Physiol. 2017, 138, 57–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakka, S.; Godar, A.S.; Thompson, C.R.; Peterson, D.E.; Jugulam, M. Rapid detoxification via glutathione S-transferase (GST) conjugation confers a high level of atrazine resistance in Palmer amaranth (Amaranthus palmeri). Pest Manag. Sci. 2017, 73, 2236–2243. [Google Scholar] [CrossRef]
- Veldhuis, L.J.; Hall, L.M.; O'Donovan, J.T.; Dyer, W.; Hall, J.C. Metabolism-based resistance of a wild mustard (Sinapis arvensis L.) biotype to ethametsulfuron-methyl. J. Agric. Food Chem. 2000, 48, 2986–2990. [Google Scholar] [CrossRef]
- Zhang, X.L.; Wang, H.Z.; Bei, F.; Wu, C.X.; Zhang, L.L.; Jia, S.S.; Wang, J.X.; Liu, W.T. Investigating the Mechanism of Metabolic Resistance to Tribenuron-Methyl in Capsella bursa-pastoris (L.) Medik. by Full-Length Transcriptome Assembly Combined with RNA-Seq. J. Agric. Food Chem. 2021, 69, 3692–3701. [Google Scholar] [CrossRef]
- Délye, C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: A major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 2013, 69, 176–187. [Google Scholar] [CrossRef]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef] [PubMed]
- Pineau, E.; Sauveplane, V.; Grienenberger, E.; Bassard, J.-E.; Beisson, F.; Pinot, F. CYP77B1 a fatty acid epoxygenase specific to flowering plants. Plant Sci. 2021, 307, 110905. [Google Scholar] [CrossRef] [PubMed]
- Philippe, G.; Sorensen, I.; Jiao, C.; Sun, X.P.; Fei, Z.J.; Domozych, D.S.; Rose, J.K.C. Cutin and suberin: Assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 2020, 55, 11–20. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Pollard, M.; Sauveplane, V.; Pinot, F.; Ohlrogge, J.; Beisson, F. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc. Natl. Acad. Sci. USA 2009, 106, 22008–22013. [Google Scholar] [CrossRef] [Green Version]
- Sauveplane, V.; Kandel, S.; Kastner, P.E.; Ehlting, J.; Compagnon, V.; Werck-Reichhart, D.; Pinot, F. Arabidopsis thaliana CYP77A4 is the first cytochrome P450 able to catalyze the epoxidation of free fatty acids in plants. FEBS J. 2009, 276, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Kawade, K.; Li, Y.; Koga, H.; Sawada, Y.; Okamoto, M.; Kuwahara, A.; Tsukaya, H.; Hirai, M.Y. The cytochrome P450 CYP77A4 is involved in auxin-mediated patterning of the Arabidopsis thaliana embryo. Development 2018, 145, dev168369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, Y.; Peng, H.; Sun, J.; Yang, Z.; Yang, H.; Liu, G.; Hu, H. Characterization of two CYP77 gene family members related to development of ornamental organs in petunia. J. Genet. 2016, 95, 177–181. [Google Scholar] [CrossRef]
- Dimaano, N.G.; Iwakami, S. CytochromeP450-mediated herbicide metabolism in plants: Current understanding and prospects. Pest Manag. Sci. 2021, 77, 22–32. [Google Scholar] [CrossRef]
- Chevet, E.; Cameron, P.H.; Pelletier, M.F.; Thomas, D.Y.; Bergeron, J.J.M. The endoplasmic reticulum: Integration of protein folding, quality control, signaling and degradation. Curr. Opin. Struct. Biol. 2001, 11, 120–124. [Google Scholar] [CrossRef]
- Yang, Q.; Deng, W.; Wang, S.; Liu, H.; Li, X.; Zheng, M. Effects of resistance mutations of Pro197, Asp376 and Trp574 on the characterization of acetohydroxyacid synthase (AHAS) isozymes. Pest Manag. Sci. 2018, 74, 1870–1879. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for parip growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, L.; Ma, Z.; Zhang, X.; Huang, Y.; Li, R.; Miao, Y.; Li, R. The Small GTPase RABA2a Recruits SNARE Proteins to Regulate Secretory Pathway in Parallel with the Exocyst Complex in Arabidopsis. Mol. Plant 2021, 15, 398–418. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).