Orphan G-Protein Coupled Receptor GPRC5B Is Critical for Lymphatic Development
Abstract
1. Introduction
2. Results
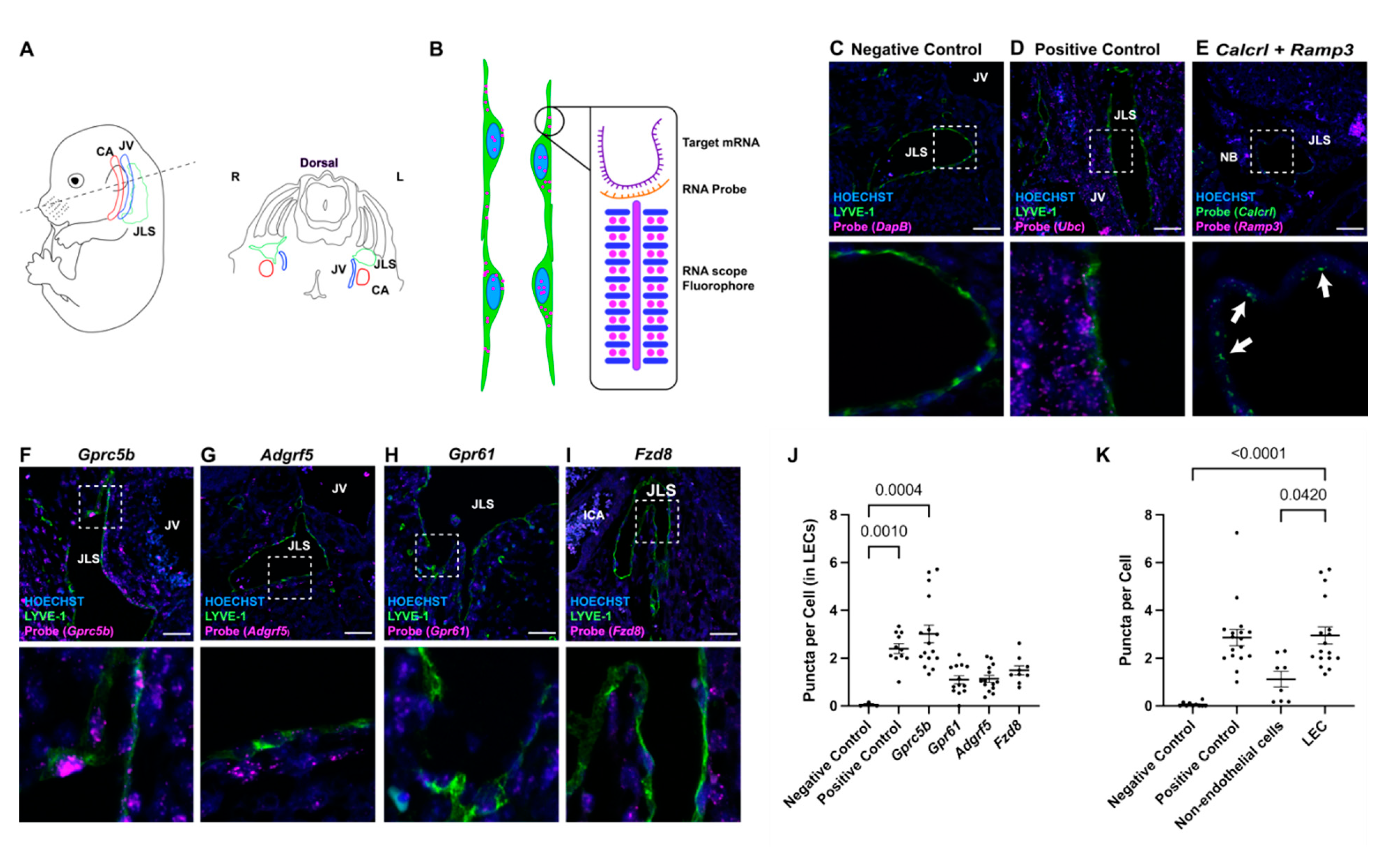
2.1. Lymphatic Endothelial Cell-Enriched GPCRs
2.2. In Situ Localization of Lymphatic Endothelial Cell-Enriched GPCR Expression
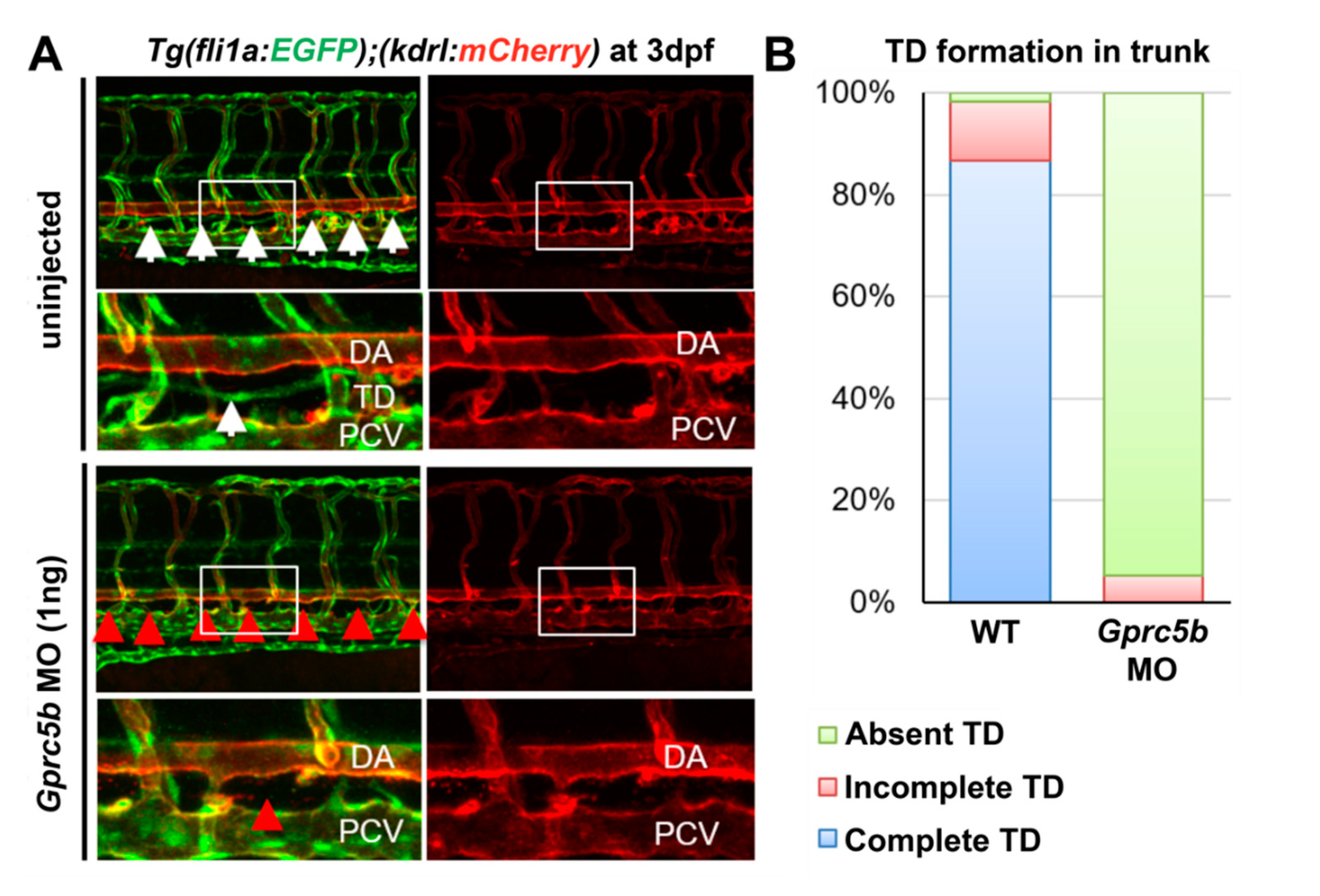
2.3. Gprc5b Is Required for Lymphatic Development in Zebrafish
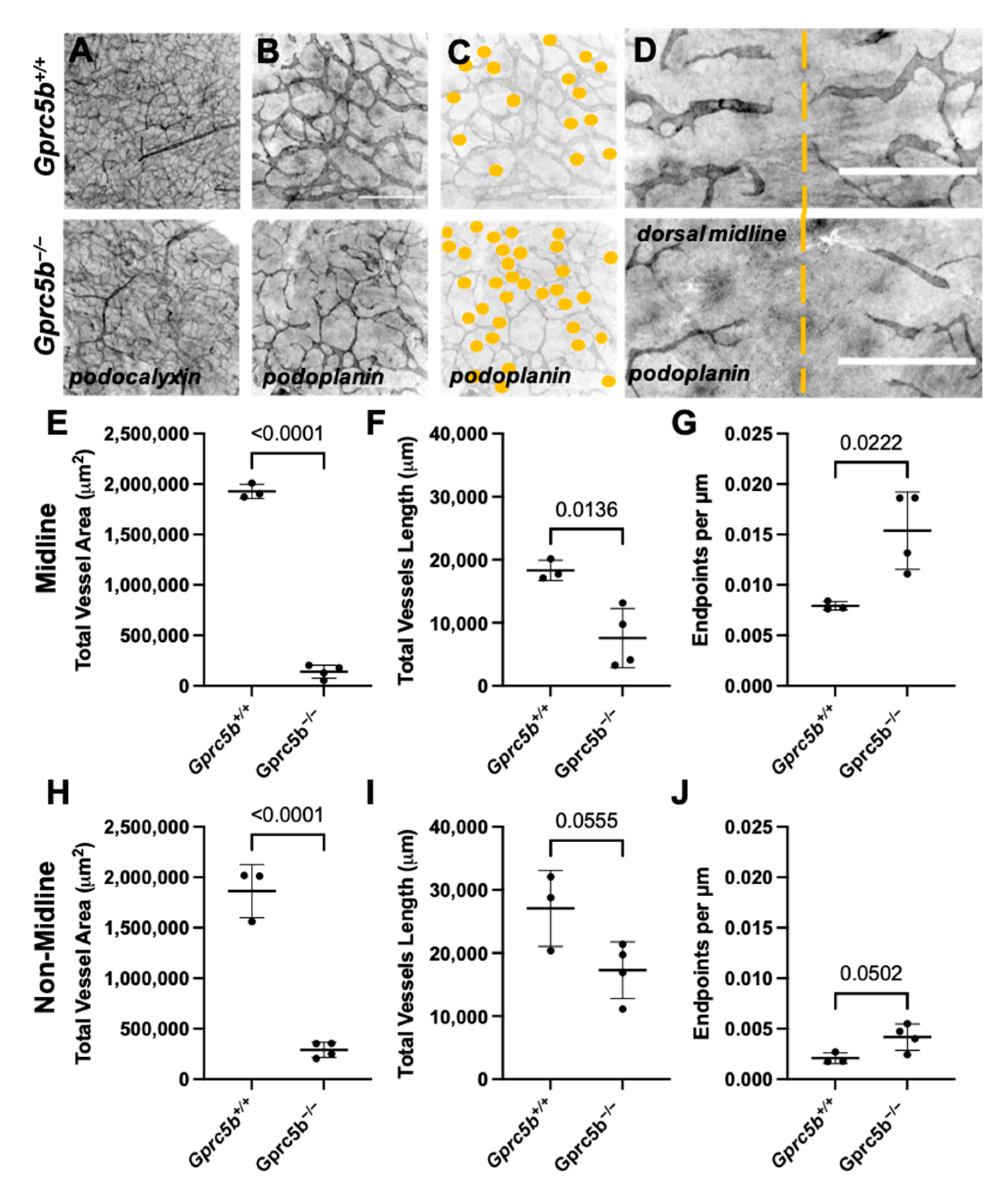
2.4. Lymphatic Development Is Attenuated in Murine Gprc5b−/− Embryos
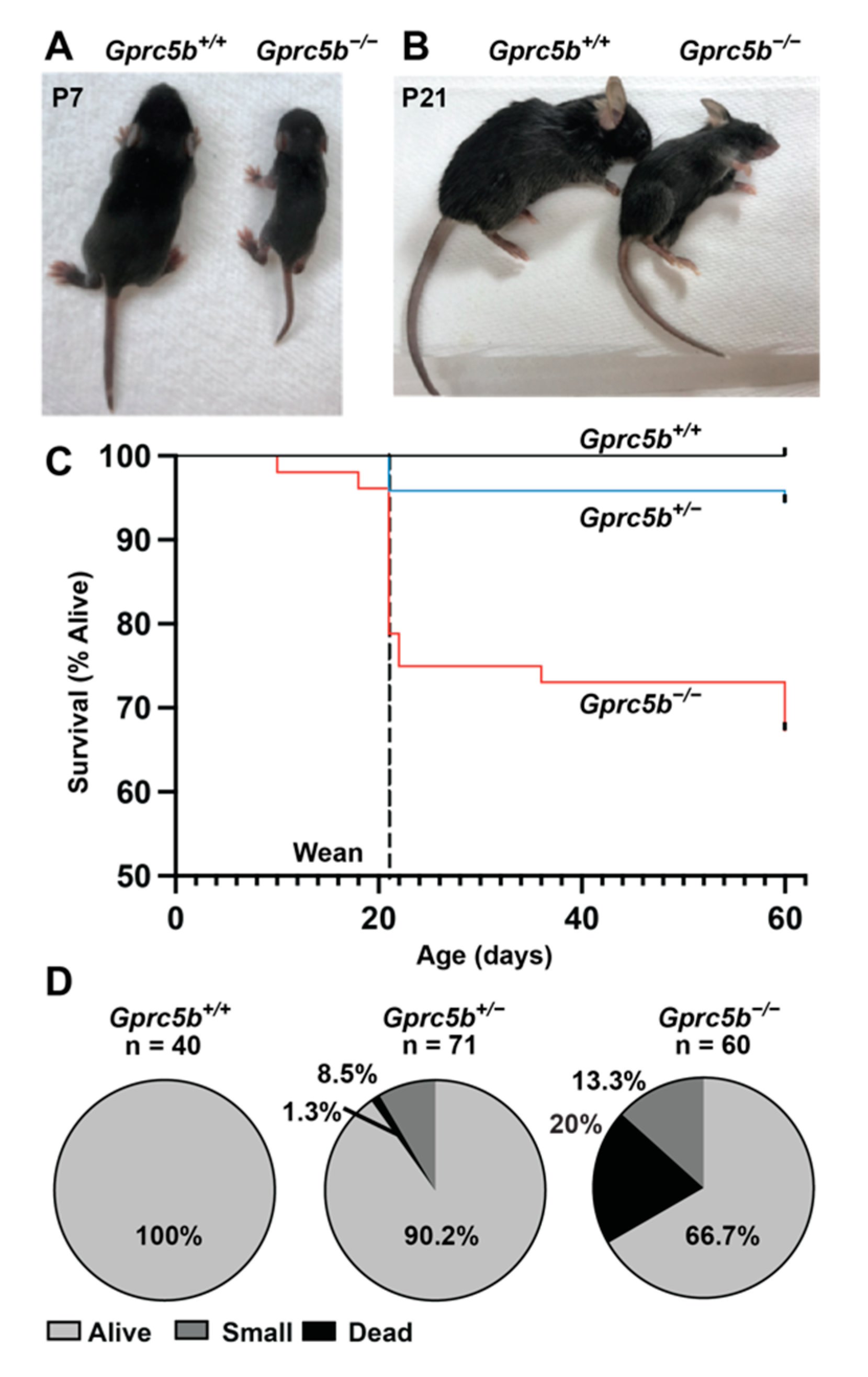
2.5. Some Gprc5b−/− Mice Exhibit Growth Restriction and Die Postnatally
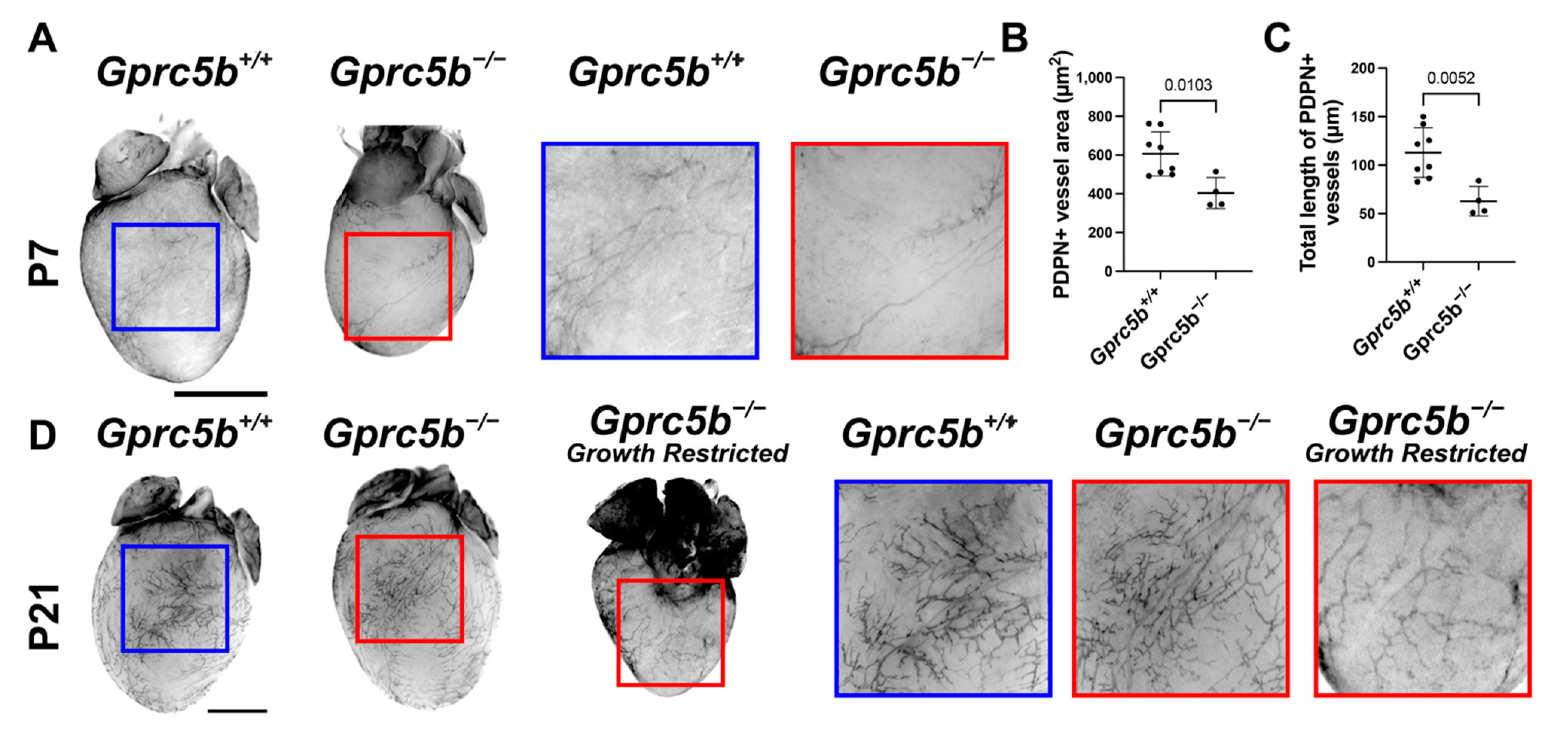
2.6. Gprc5b−/− Mice Exhibit Attenuated Postnatal Lymphatic Growth
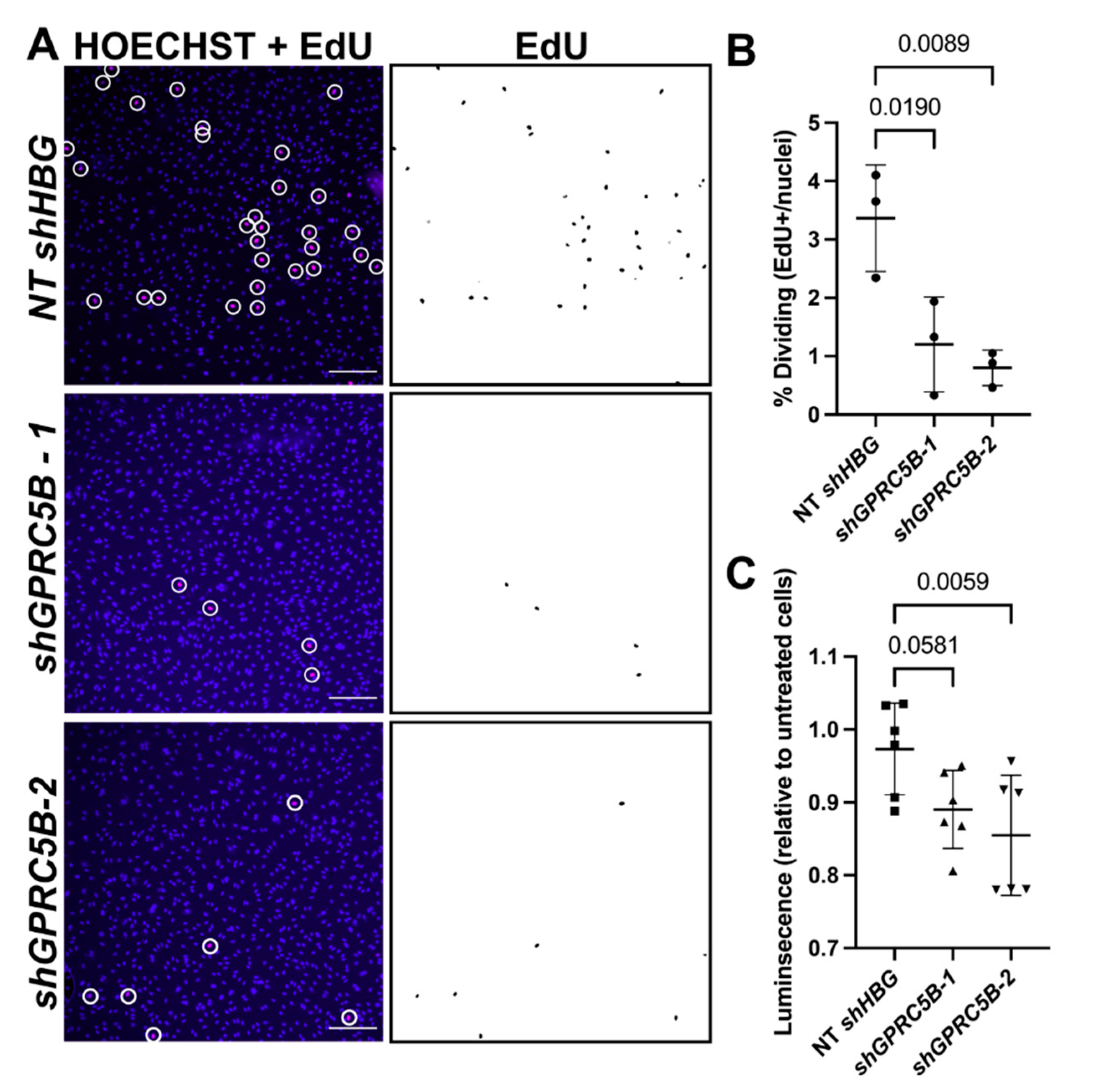
2.7. GPRC5B Deficiency Reduces Human LEC Proliferation and Viability In Vitro
3. Discussion
4. Materials and Methods
4.1. GPCR Pathway Focused Array
4.2. RNAscope and Immunohistochemistry
4.3. Animals
4.3.1. Zebrafish
4.3.2. Mice
4.4. Histology
4.5. Whole-Mount Immunostaining
4.6. Cell Culture
4.7. Lentivirus-Mediated Knockdown of GPRC5B in LECs
4.8. EdU Cell Proliferation Assay
4.9. Viability Quantification Using CellTiter-Glo
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, W.; Aspelund, A.; Alitalo, K. Lymphangiogenic Factors, Mechanisms, and Applications. J. Clin. Investig. 2014, 124, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Petrova, T.V.; Koh, G.Y. Biological functions of lymphatic vessels. Science 2020, 369, eaax4063. [Google Scholar] [CrossRef] [PubMed]
- Oliver, G.; Kipnis, J.; Randolph, G.J.; Harvey, N.L. The Lymphatic Vasculature in the 21st Century: Novel Functional Roles in Homeostasis and Disease. Cell 2020, 182, 270–296. [Google Scholar] [CrossRef] [PubMed]
- Alitalo, K. The lymphatic vasculature in disease. Nat. Med. 2011, 17, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Ambler, W.; Santambrogio, L.; Lu, T.T. Advances in understanding and examining lymphatic function: Relevance for understanding autoimmunity. Curr. Opin. Rheumatol. 2021, 34, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth, R.H.; Achen, M.G.; Stacker, S.A. The evolving role of lymphatics in cancer metastasis. Curr. Opin. Immunol. 2018, 53, 64–73. [Google Scholar] [CrossRef]
- Warren, A.G.; Brorson, H.; Borud, L.J.; Slavin, S.A. Lymphedema: A Comprehensive Review. Ann. Plast Surg. 2007, 59, 464–472. [Google Scholar] [CrossRef]
- Radhakrishnan, K.; Rockson, S.G. The Clinical Spectrum of Lymphatic Disease. Ann. N. Y. Acad. Sci. 2008, 1131, 155–184. [Google Scholar] [CrossRef]
- Rockson, S.G. Current concepts and future directions in the diagnosis and management of lymphatic vascular disease. Vasc. Med. 2010, 15, 223–231. [Google Scholar] [CrossRef]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in Gpcr Drug Discovery: New Agents, Targets and Indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Oprea, T.I.; Bologa, C.G.; Brunak, S.; Campbell, A.; Gan, G.N.; Gaulton, A.; Gomez, S.M.; Guha, R.; Hersey, A.; Holmes, J.; et al. Unexplored Therapeutic Opportunities in the Human Genome. Nat. Rev. Drug Discov. 2018, 17, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Drews, J. Genomic sciences and the medicine of tomorrow. Nat. Biotechnol. 1996, 14, 1516–1518. [Google Scholar] [CrossRef] [PubMed]
- Sheils, T.K.; Mathias, S.L.; Kelleher, K.J.; Siramshetty, V.B.; Nguyen, D.T.; Bologa, C.G.; Jensen, L.J.; Vidović, D.; Koleti, A.; Schürer, S.C.; et al. Tcrd and Pharos 2021: Mining the Human Proteome for Disease Biology. Nucleic. Acids Res. 2021, 49, D1334–D1346. [Google Scholar] [CrossRef]
- Kropiwnicki, E.; Binder, J.L.; Yang, J.J.; Holmes, J.; Lachmann, A.; Clarke, D.J.B.; Sheils, T.; Kelleher, K.J.; Metzger, V.T.; Bologa, C.G.; et al. Getting Started with the IDG KMC Datasets and Tools. Curr. Protoc. 2022, 2, e355. [Google Scholar] [CrossRef] [PubMed]
- Cannon, D.C.; Yang, J.J.; Mathias, S.L.; Ursu, O.; Mani, S.; Waller, A.; Schürer, S.C.; Jensen, L.J.; Sklar, L.A.; Bologa, C.G.; et al. TIN-X: Target importance and novelty explorer. Bioinformatics 2017, 33, 2601–2603. [Google Scholar] [CrossRef]
- Davis, R.B.; Kechele, D.O.; Blakeney, E.S.; Pawlak, J.B.; Caron, K.M. Lymphatic deletion of calcitonin receptor-like receptor exacerbates intestinal inflammation. JCI Insight 2017, 2, e92465. [Google Scholar] [CrossRef] [PubMed]
- Fritz-Six, K.L.; Dunworth, W.P.; Li, M.; Caron, K.M. Adrenomedullin signaling is necessary for murine lymphatic vascular development. J. Clin. Investig. 2008, 118, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Mackie, D.I.; Al Mutairi, F.; Davis, R.B.; Kechele, D.O.; Nielsen, N.; Snyder, J.; Caron, M.G.; Kliman, H.J.; Berg, J.S.; Simms, J.; et al. hCALCRL mutation causes autosomal recessive nonimmune hydrops fetalis with lymphatic dysplasia. J. Exp. Med. 2018, 215, 2339–2353. [Google Scholar] [CrossRef]
- Gonzalez-Garay, M.; Aldrich, M.B.; Rasmussen, J.; Guilliod, R.; Lapinski, P.E.; King, P.D.; Sevick-Muraca, E.M. A novel mutation in CELSR1 is associated with hereditary lymphedema. Vasc. Cell 2016, 8, 1. [Google Scholar] [CrossRef]
- Hisano, Y.; Kono, M.; Cartier, A.; Engelbrecht, E.; Kano, K.; Kawakami, K.; Xiong, Y.; Piao, W.; Galvani, S.; Yanagida, K.; et al. Lysolipid receptor cross-talk regulates lymphatic endothelial junctions in lymph nodes. J. Exp. Med. 2019, 216, 1582–1598. [Google Scholar] [CrossRef]
- Tatin, F.; Renaud-Gabardos, E.; Godet, A.-C.; Hantelys, F.; Pujol, F.; Morfoisse, F.; Calise, D.; Viars, F.; Valet, P.; Masri, B.; et al. Apelin modulates pathological remodeling of lymphatic endothelium after myocardial infarction. JCI Insight 2017, 2, e93887. [Google Scholar] [CrossRef] [PubMed]
- Dunworth, W.P.; Caron, K.M. G Protein–Coupled Receptors as Potential Drug Targets for Lymphangiogenesis and Lymphatic Vascular Diseases. Arter. Thromb. Vasc. Biol. 2009, 29, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, R.; Vaziri, H.; Gautam, A.; Ballesteros, E.; Karimeddini, D.; Wu, G.Y. Chylous Ascites: A Review of Pathogenesis, Diagnosis and Treatment. J. Clin. Transl. Hepatol. 2018, 6, 105–113. [Google Scholar] [CrossRef]
- Xiong, Y.; Piao, W.; Brinkman, C.C.; Li, L.; Kulinski, J.M.; Olivera, A.; Cartier, A.; Hla, T.; Hippen, K.L.; Blazar, B.R.; et al. CD4 T cell sphingosine 1-phosphate receptor (S1PR)1 and S1PR4 and endothelial S1PR2 regulate afferent lymphatic migration. Sci. Immunol. 2019, 4, eaav1263. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Yanagida, K.; Akwii, R.G.; Choi, D.; Chen, L.; Ho, Y.; Cha, B.; Mahamud, R.; de Ruiz, K.B.; Ichise, H.; et al. S1PR1 regulates the quiescence of lymphatic vessels by inhibiting laminar shear stress–dependent VEGF-C signaling. JCI Insight 2020, 5, e137652. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Kim, Y.-J.; Oshima, E.; Shimizu, C.; Kiyonari, H.; Abe, T.; Higashi, H.; Yamada, K.; Hirabayashi, Y. Comparative characterization of GPRC5B and GPRC5CLacZ knockin mice; behavioral abnormalities in GPRC5B-deficient mice. Biochem. Biophys. Res. Commun. 2011, 412, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Klotz, L.; Norman, S.; Vieira, J.M.; Masters, M.; Rohling, M.; Dubé, K.N.; Bollini, S.; Matsuzaki, F.; Carr, C.A.; Riley, P.R. Cardiac lymphatics are heterogeneous in origin and respond to injury. Nature 2015, 522, 62–67. [Google Scholar] [CrossRef]
- Valtcheva, N.; Primorac, A.; Jurisic, G.; Hollmén, M.; Detmar, M. The Orphan Adhesion G Protein-coupled Receptor GPR97 Regulates Migration of Lymphatic Endothelial Cells via the Small GTPases RhoA and Cdc42. J. Biol. Chem. 2013, 288, 35736–35748. [Google Scholar] [CrossRef]
- Robbins, M.; Michalovich, D.; Hill, J.; Calver, A.; Medhurst, A.; Gloger, I.; Sims, M.; Middlemiss, D.; Pangalos, M. Molecular Cloning and Characterization of Two Novel Retinoic Acid-Inducible Orphan G-Protein-Coupled Receptors (GPRC5B and GPRC5C). Genomics 2000, 67, 8–18. [Google Scholar] [CrossRef]
- Robbins, M.J.; Charles, K.J.; Harrison, D.C.; Pangalos, M.N. Localisation of the GPRC5B receptor in the rat brain and spinal cord. Mol. Brain Res. 2002, 106, 136–144. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Sano, T.; Nabetani, T.; Asano, Y.; Hirabayashi, Y. GPRC5B Activates Obesity-Associated Inflammatory Signaling in Adipocytes. Sci. Signal. 2012, 5, ra85. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, Y.; Kim, Y.-J. Roles of GPRC5 family proteins: Focusing on GPRC5B and lipid-mediated signalling. J. Biochem. 2020, 167, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, S.; Moller-Hackbarth, K.; Li, X.; Rodriguez, P.Q.; Charrin, E.; Schwarz, A.; Nystrom, J.; Wernerson, A.O.; Lal, M.; Patrakka, J. Gprc5b Modulates Inflammatory Response in Glomerular Diseases Via Nf-Kappab Pathway. J. Am. Soc. Nephrol. 2019, 30, 1573–1586. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Kohyama-Koganeya, A.; Kinoshita, M.O.; Tatsukawa, T.; Shimizu, C.; Oshima, E.; Yamada, K.; Le, T.D.; Akagi, T.; Tohyama, K.; et al. Loss of GPRC5B impairs synapse formation of Purkinje cells with cerebellar nuclear neurons and disrupts cerebellar synaptic plasticity and motor learning. Neurosci. Res. 2018, 136, 33–47. [Google Scholar] [CrossRef]
- von Samson-Himmelstjerna, F.A.; Freundt, G.; Nitz, J.-T.; Stelter, F.; Luedde, M.; Wieland, T.; Frey, N.; Hippe, H.-J. The orphan receptor GPRC5B modulates inflammatory and fibrotic pathways in cardiac fibroblasts and mice hearts. Biochem. Biophys. Res. Commun. 2019, 514, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.G.; Sanderson, E.; Elsworth, B.; Tilling, K.; Smith, G.D. Use of genetic variation to separate the effects of early and later life adiposity on disease risk: Mendelian randomisation study. BMJ 2020, 369, m1203. [Google Scholar] [CrossRef]
- Carvalho, J.; Chennupati, R.; Li, R.; Günther, S.; Kaur, H.; Zhao, W.; Tonack, S.; Kurz, M.; Mößlein, N.; Bünemann, M.; et al. Orphan G Protein–Coupled Receptor GPRC5B Controls Smooth Muscle Contractility and Differentiation by Inhibiting Prostacyclin Receptor Signaling. Circulation 2020, 141, 1168–1183. [Google Scholar] [CrossRef]
- Atanes, P.; Ruz-Maldonado, I.; Hawkes, R.; Liu, B.; Persaud, S.J.; Amisten, S. Identifying Signalling Pathways Regulated by Gprc5b in Beta-Cells by Crispr-Cas9-Mediated Genome Editing. Cell Physiol. Biochem. 2018, 45, 656–666. [Google Scholar] [CrossRef]
- Kurabayashi, N.; Nguyen, M.D.; Sanada, K. The G protein-coupled receptor GPRC5B contributes to neurogenesis in the developing mouse neocortex. Development 2013, 140, 4335–4346. [Google Scholar] [CrossRef][Green Version]
- Lawson, N.; Weinstein, B.M. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Dev. Biol. 2002, 248, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Chi, N.C.; Shaw, R.M.; De Val, S.; Kang, G.; Jan, L.Y.; Black, B.L.; Stainier, D.Y. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008, 22, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Klein, K.R.; Karpinich, N.O.; Espenschied, S.; Willcockson, H.H.; Dunworth, W.P.; Hoopes, S.L.; Kushner, E.J.; Bautch, V.L.; Caron, K.M. Decoy Receptor CXCR7 Modulates Adrenomedullin-Mediated Cardiac and Lymphatic Vascular Development. Dev. Cell 2014, 30, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Zudaire, E.; Gambardella, L.; Kurcz, C.; Vermeren, S. A computational tool for quantitative analysis of vascular networks. PLoS ONE 2011, 6, e27385. [Google Scholar] [CrossRef] [PubMed]

| Target ID | Gene Symbol | Receptor Protein Name | p-Value | Log2 Fold Change |
|---|---|---|---|---|
| GPRC5B-Hs00212116_m1 | GPRC5B | GPCR Receptor Class C Group 5 Member B (Orphan) | 0.002 | 11.83 |
| GPR116-Hs00391810_m1 | ADGRF5 | Adhesion G Protein-Coupled Receptor F5 (Orphan) | 0.010 | 6.75 |
| FZD8-Hs00259040_s1 | FZD8 | Frizzled Class Receptor 8 (Orphan) | 0.019 | 3.10 |
| EDG1-Hs00173499_m1 | S1PR1 | Sphingosine-1-Phosphate Receptor 1 | 0.033 | 2.87 |
| SSTR5-Hs00265647_s1 | SSTR5 | Somatostatin Receptor 5 | 0.036 | 2.10 |
| FKSG83-Hs00259293_s1 | VN1R10P | Vomeronasal 1 Receptor 10 Pseudogene | 0.041 | 1.85 |
| GPR10-Hs00244685_s1 | PRLHR | Prolactin-releasing peptide receptor | 0.049 | 1.82 |
| PPYR1-Hs00275980_s1 | NPY4R | Neuropeptide Y Receptor Y4; Pancreatic Polypeptide Y | 0.008 | 1.71 |
| GPR55-Hs00271662_s1 | GPR55 | G Protein-Coupled Receptor 55 | 0.010 | 1.64 |
| CHRM2-Hs00265208_s1 | CHRM2 | Cholinergic Receptor Muscarinic 2 | 0.011 | 1.46 |
| MC3R-Hs00252036_s1 | MC3R | Melanocortin 3 Receptor | 0.043 | 1.37 |
| HTR1F-Hs00265296_s1 | HTR1F | Serotonin Receptor 1F | 0041 | 1.35 |
| GPR61-Hs00259108_s1 | GPR61 | G Protein-Coupled Receptor 61 (Orphan) | 0.046 | 1.34 |
| VN1R2-Hs00545195_s1 | VN1R2 | Vomeronasal Type-1 Receptor 2, GPCR25 | 0.036 | 1.33 |
| ADRB2-Hs00240532_s1 | ADRB2 | Beta-2 Adrenergic Receptor | 0.047 | 1.31 |
| GPR120-Hs00664157_s1 | FFAR4 | Free Fatty Acid Receptor 4 | 0.032 | 1.25 |
| OR2C3-Hs00812303_s1 | OR2C3 | Olfactory Receptor Family 2 Subfamily C Member 3 | 0.040 | 1.24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Nelson-Maney, N.P.; Bálint, L.; Kwon, H.-B.; Davis, R.B.; Dy, D.C.M.; Dunleavey, J.M.; St. Croix, B.; Caron, K.M. Orphan G-Protein Coupled Receptor GPRC5B Is Critical for Lymphatic Development. Int. J. Mol. Sci. 2022, 23, 5712. https://doi.org/10.3390/ijms23105712
Xu W, Nelson-Maney NP, Bálint L, Kwon H-B, Davis RB, Dy DCM, Dunleavey JM, St. Croix B, Caron KM. Orphan G-Protein Coupled Receptor GPRC5B Is Critical for Lymphatic Development. International Journal of Molecular Sciences. 2022; 23(10):5712. https://doi.org/10.3390/ijms23105712
Chicago/Turabian StyleXu, Wenjing, Nathan P. Nelson-Maney, László Bálint, Hyouk-Bum Kwon, Reema B. Davis, Danielle C. M. Dy, James M. Dunleavey, Brad St. Croix, and Kathleen M. Caron. 2022. "Orphan G-Protein Coupled Receptor GPRC5B Is Critical for Lymphatic Development" International Journal of Molecular Sciences 23, no. 10: 5712. https://doi.org/10.3390/ijms23105712
APA StyleXu, W., Nelson-Maney, N. P., Bálint, L., Kwon, H.-B., Davis, R. B., Dy, D. C. M., Dunleavey, J. M., St. Croix, B., & Caron, K. M. (2022). Orphan G-Protein Coupled Receptor GPRC5B Is Critical for Lymphatic Development. International Journal of Molecular Sciences, 23(10), 5712. https://doi.org/10.3390/ijms23105712






