Genome-Wide Identification of Gramineae Brassinosteroid-Related Genes and Their Roles in Plant Architecture and Salt Stress Adaptation
Abstract
:1. Introduction
2. Results
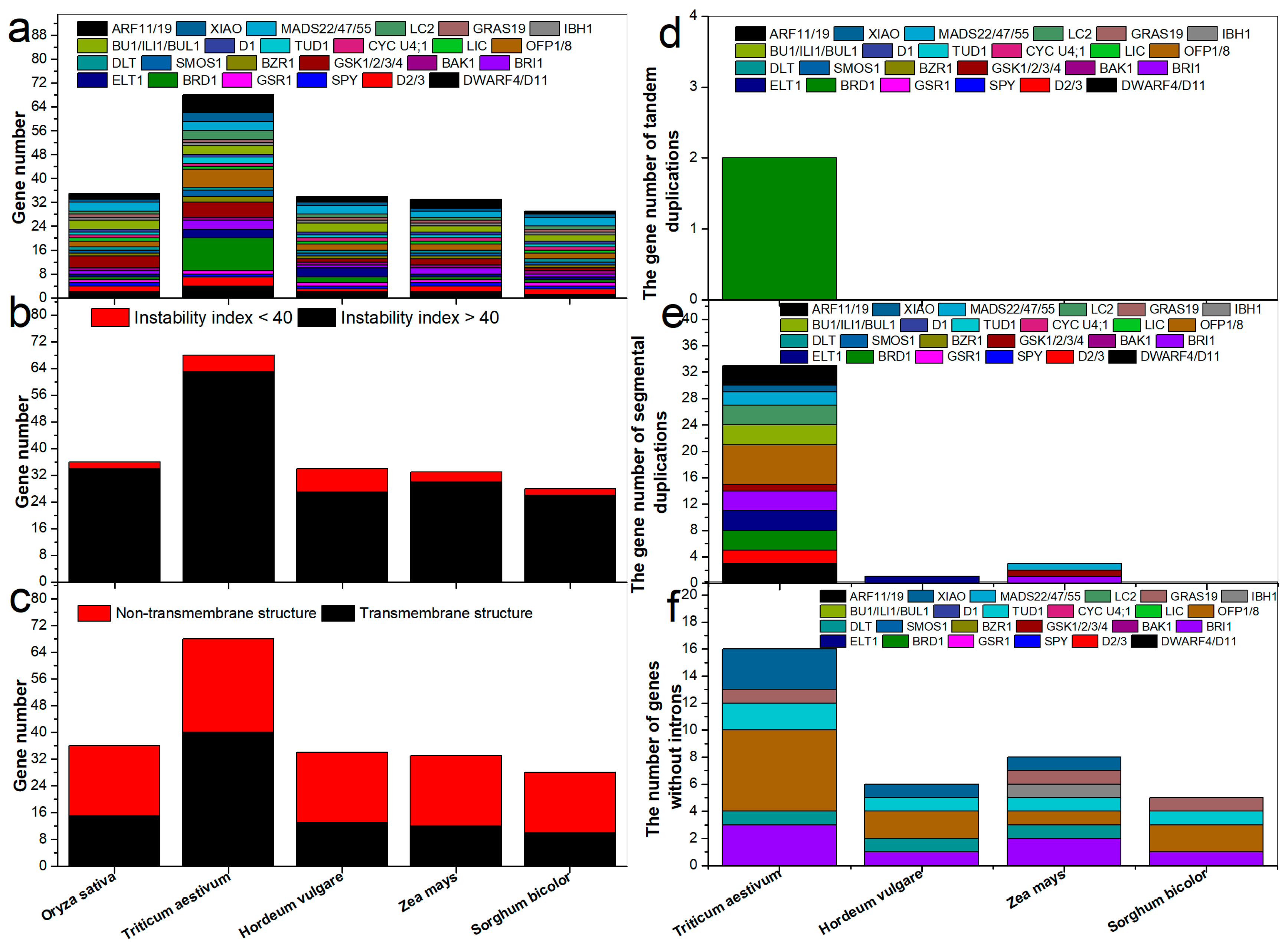
2.1. Identification of BR-Related Genes in Gramineae
2.2. Multiple Sequence Alignment, Secondary Structure, and Phylogenetic Analyses of BR-Related Proteins
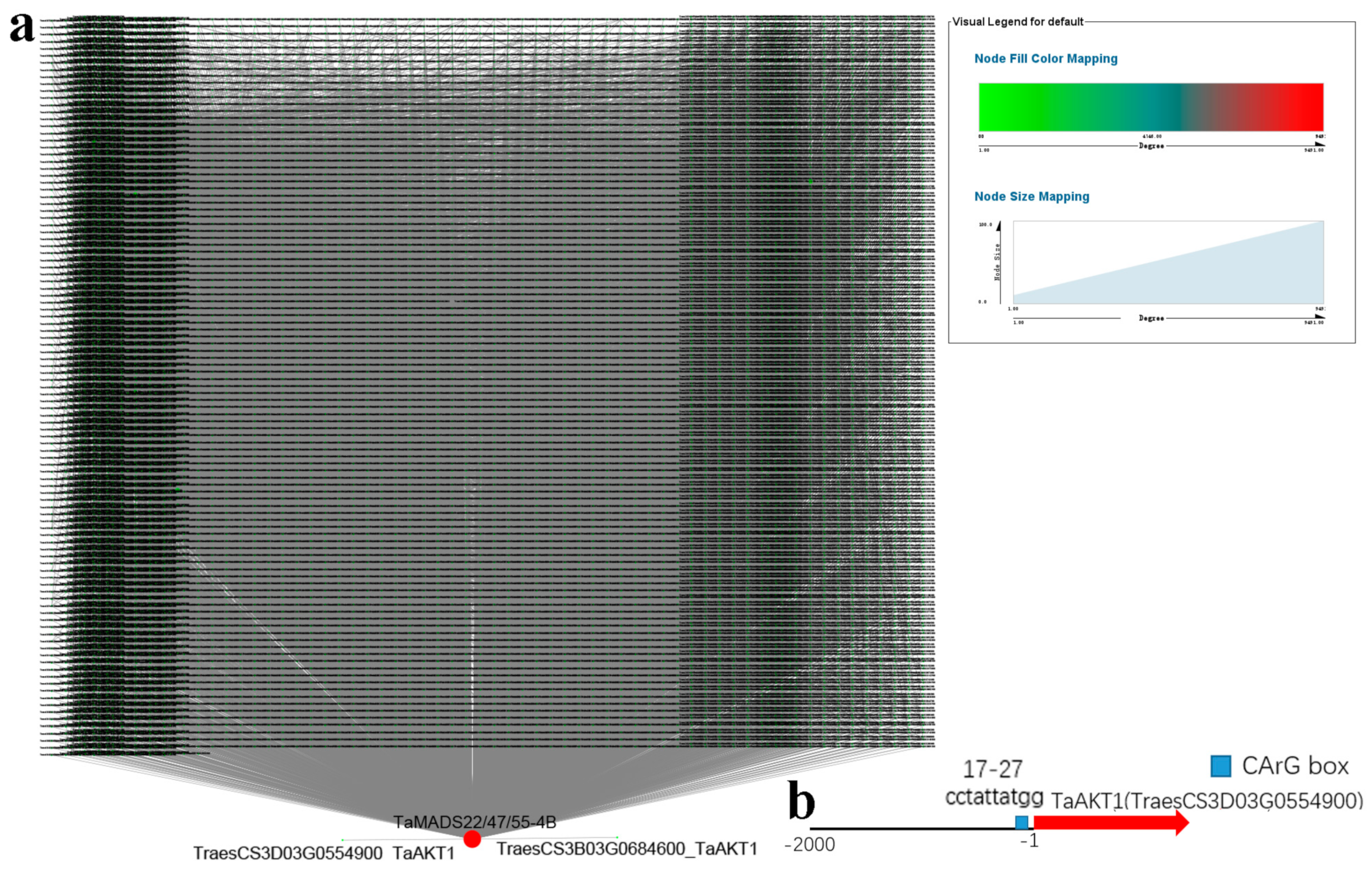
2.3. Transmembrane Structure, Synteny, Gene Structure, Promoter, and Protein-Protein Interaction Network Analyses
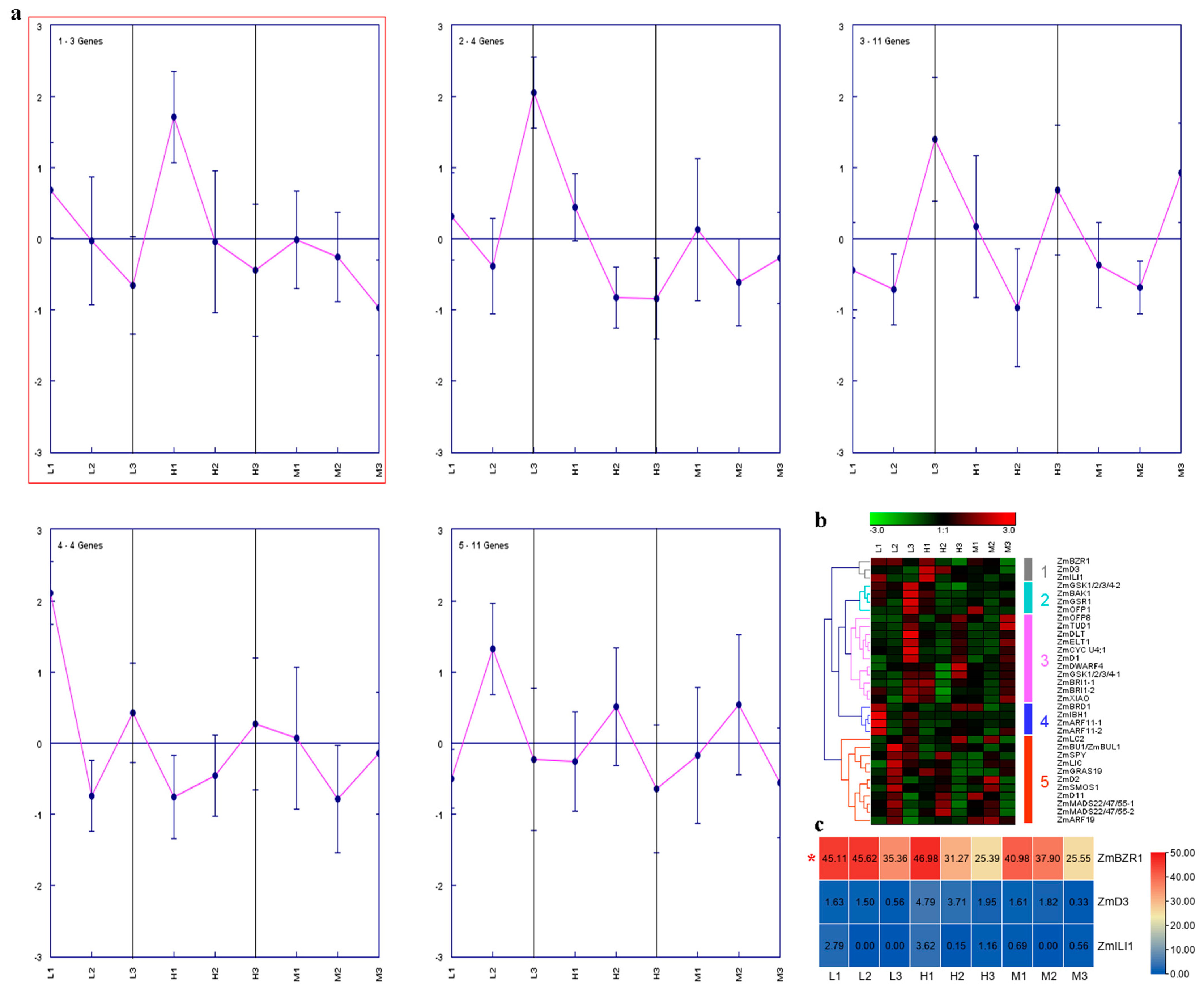
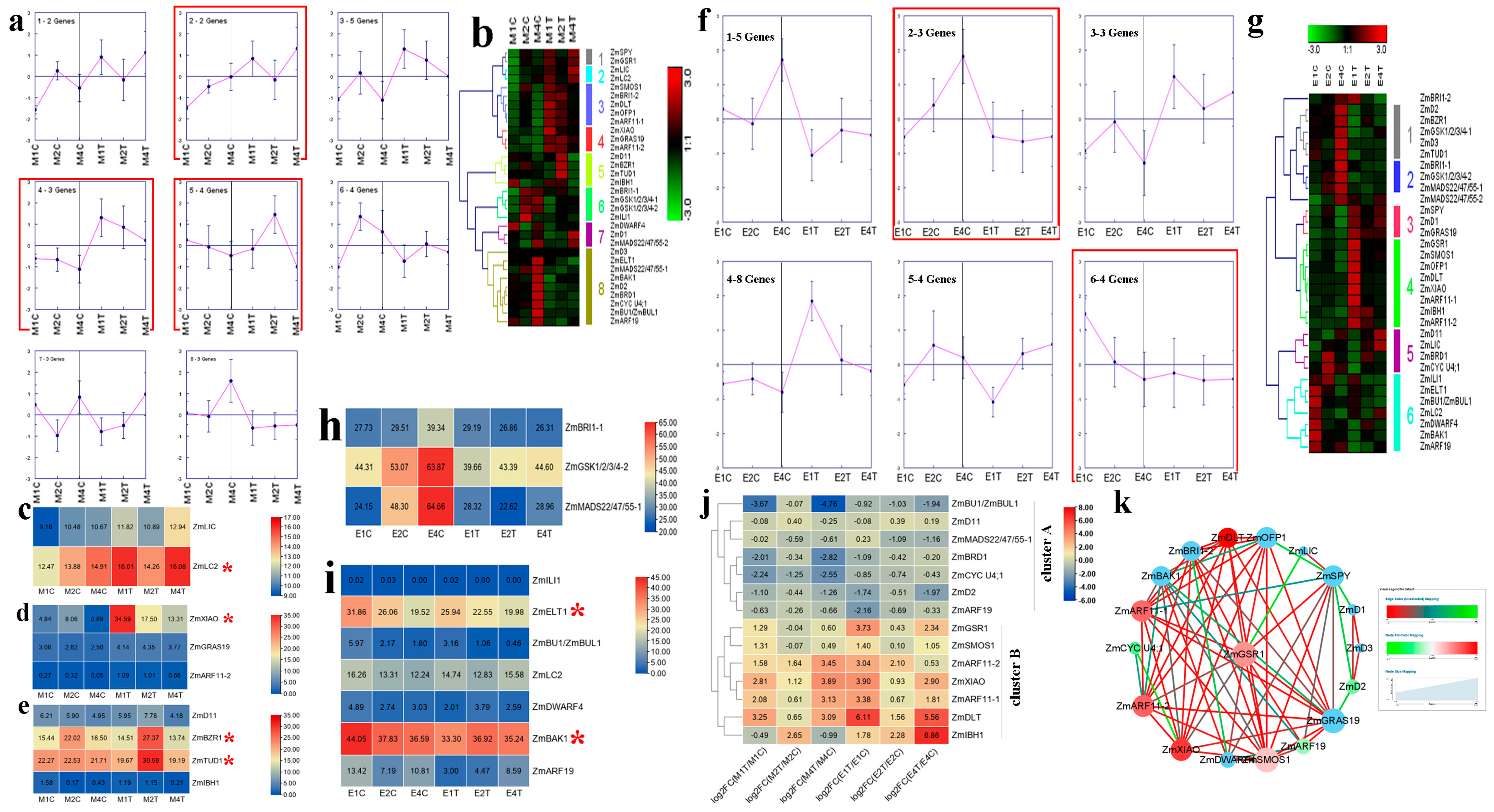
2.4. Expression Analysis of Maize BR-Related Genes in Regulating PH
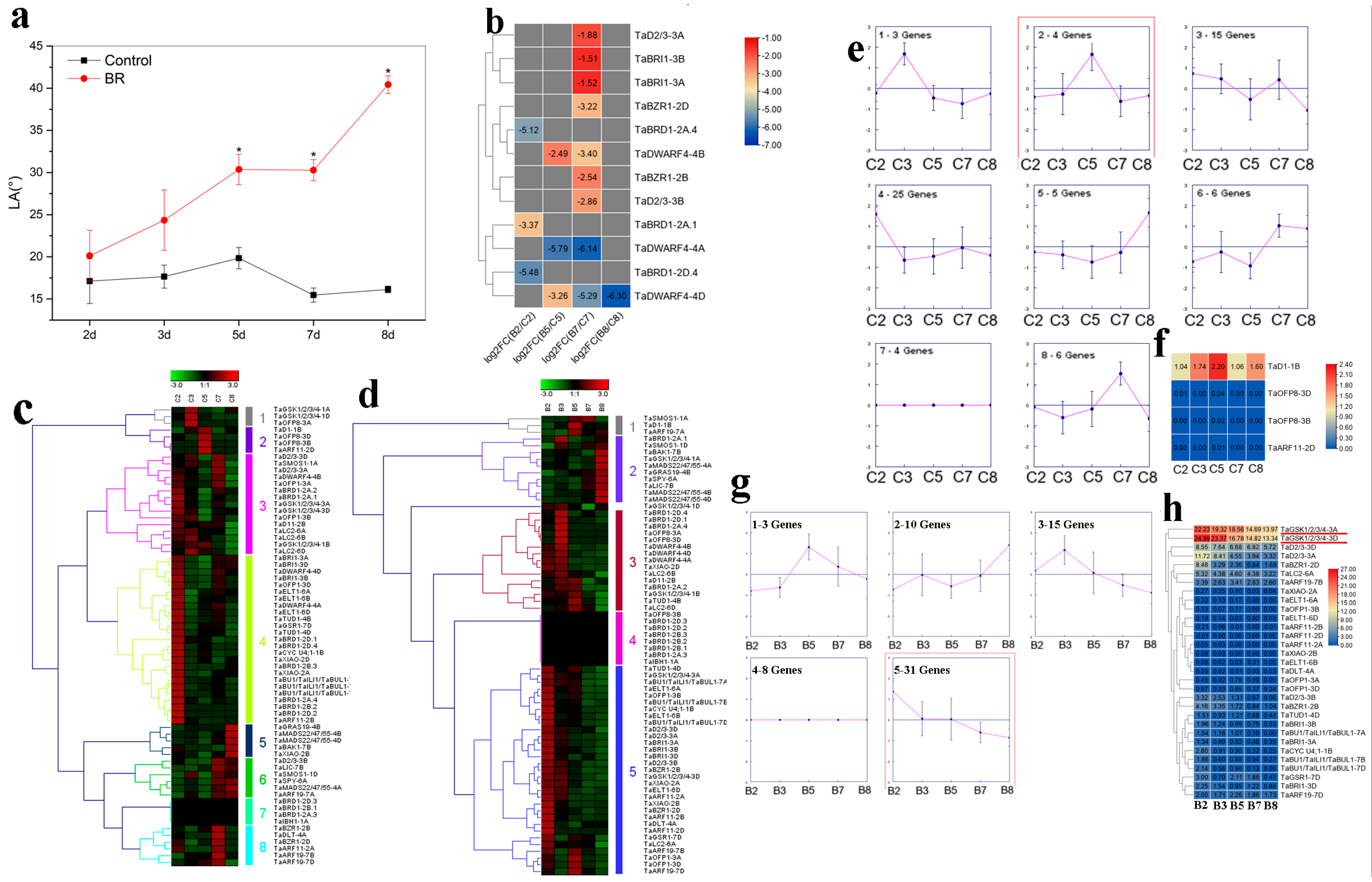
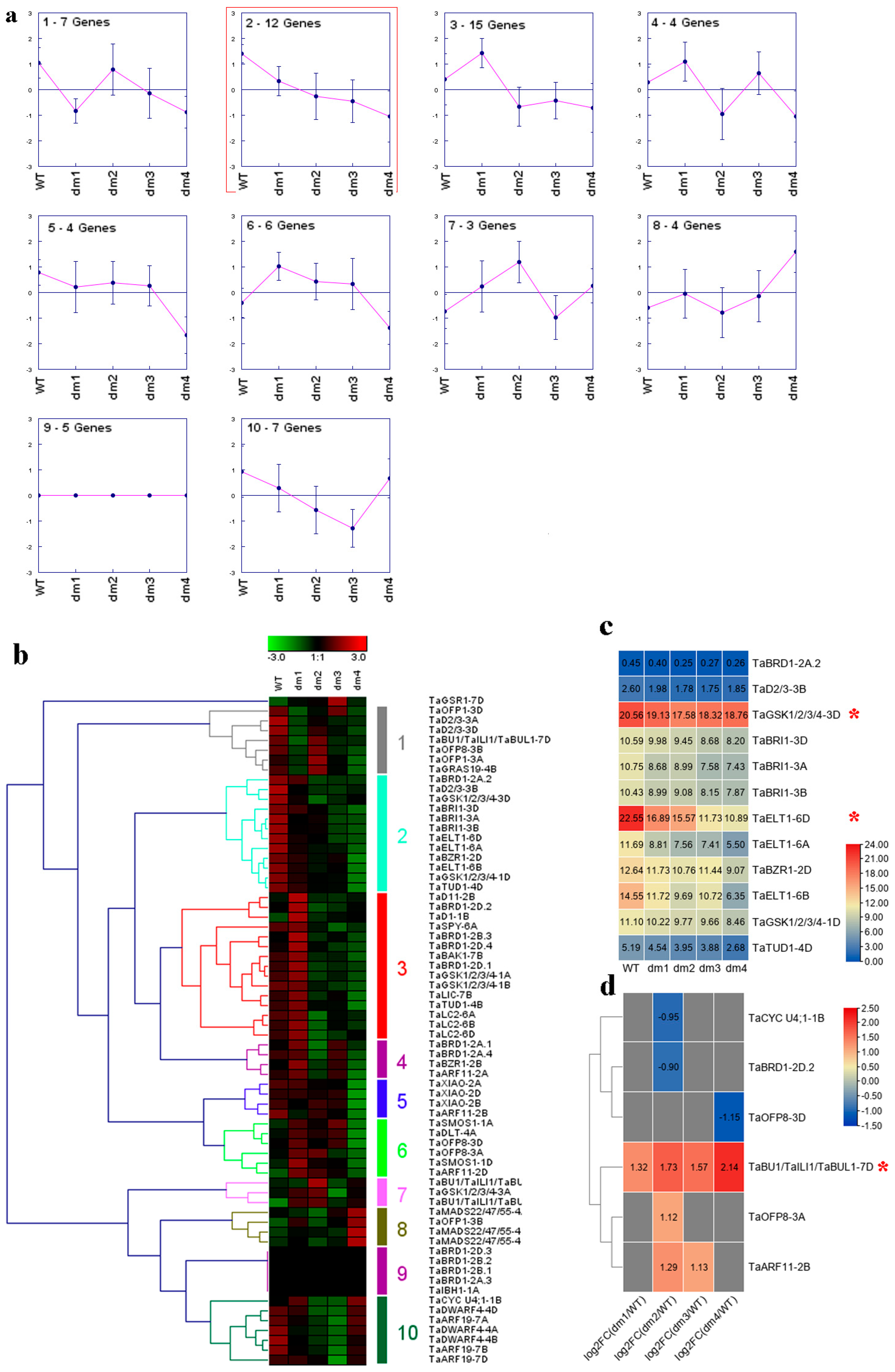
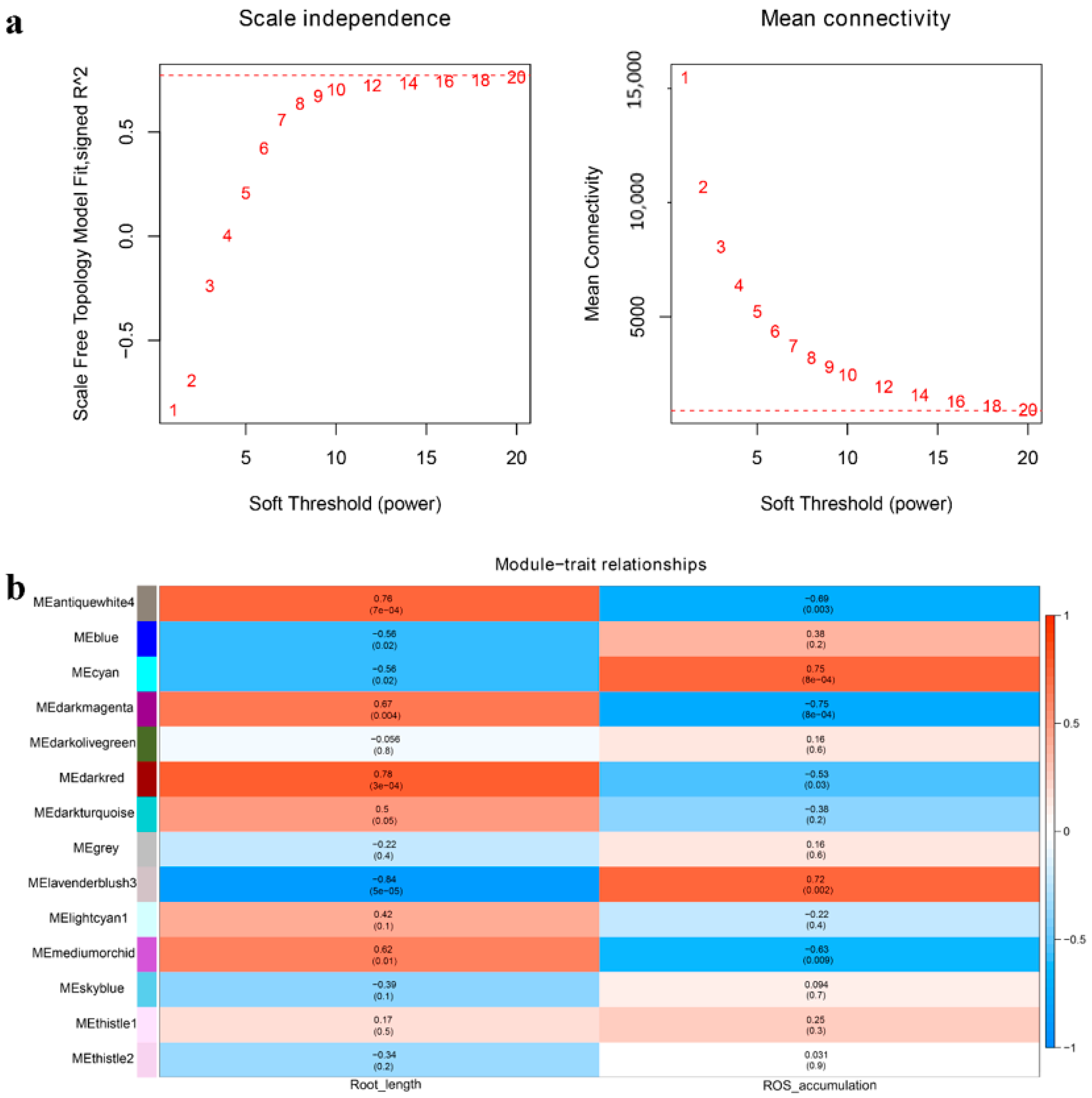
2.5. Transcriptome Analysis of Wheat BR-Related Genes in Regulating LA and PH
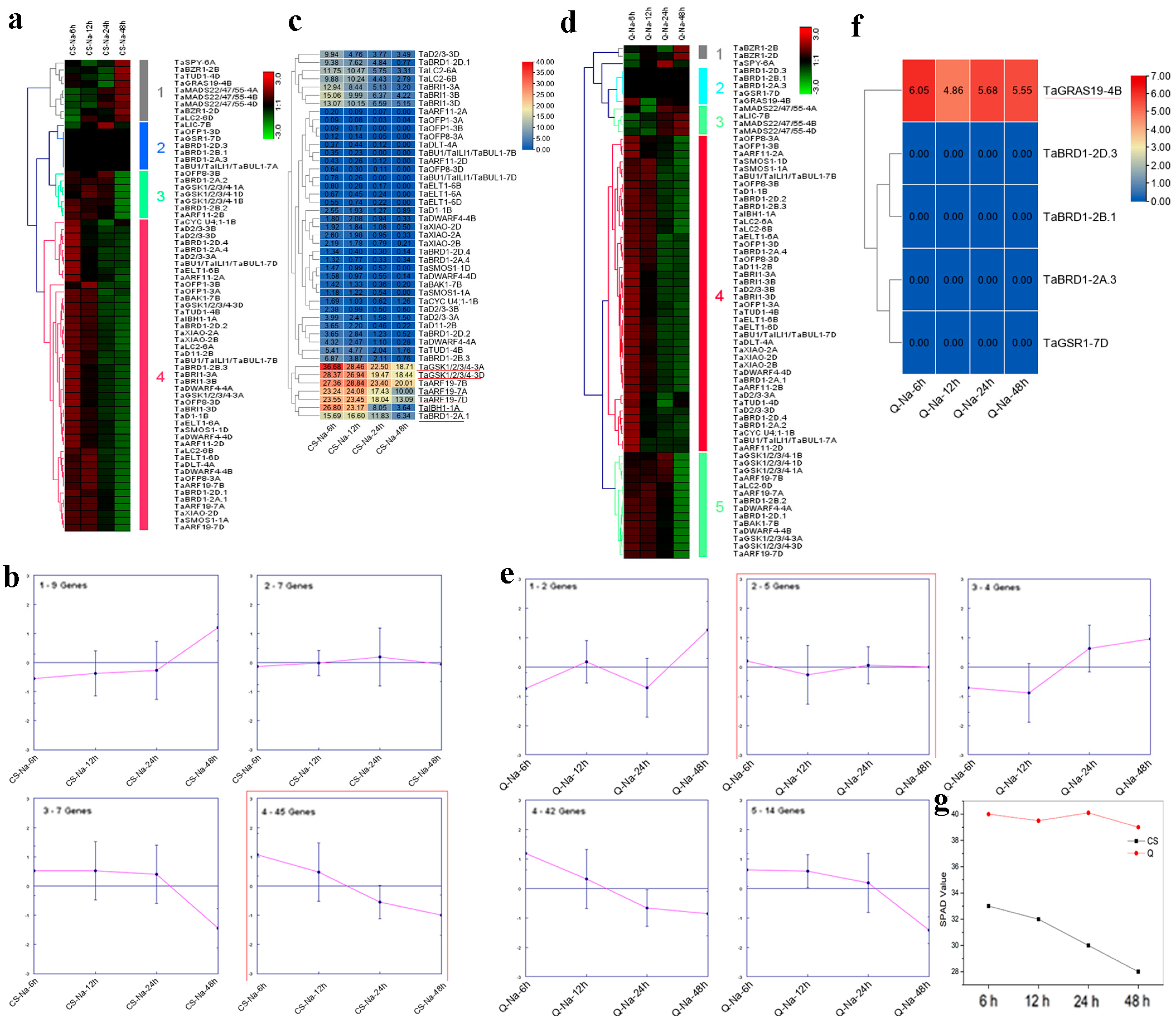
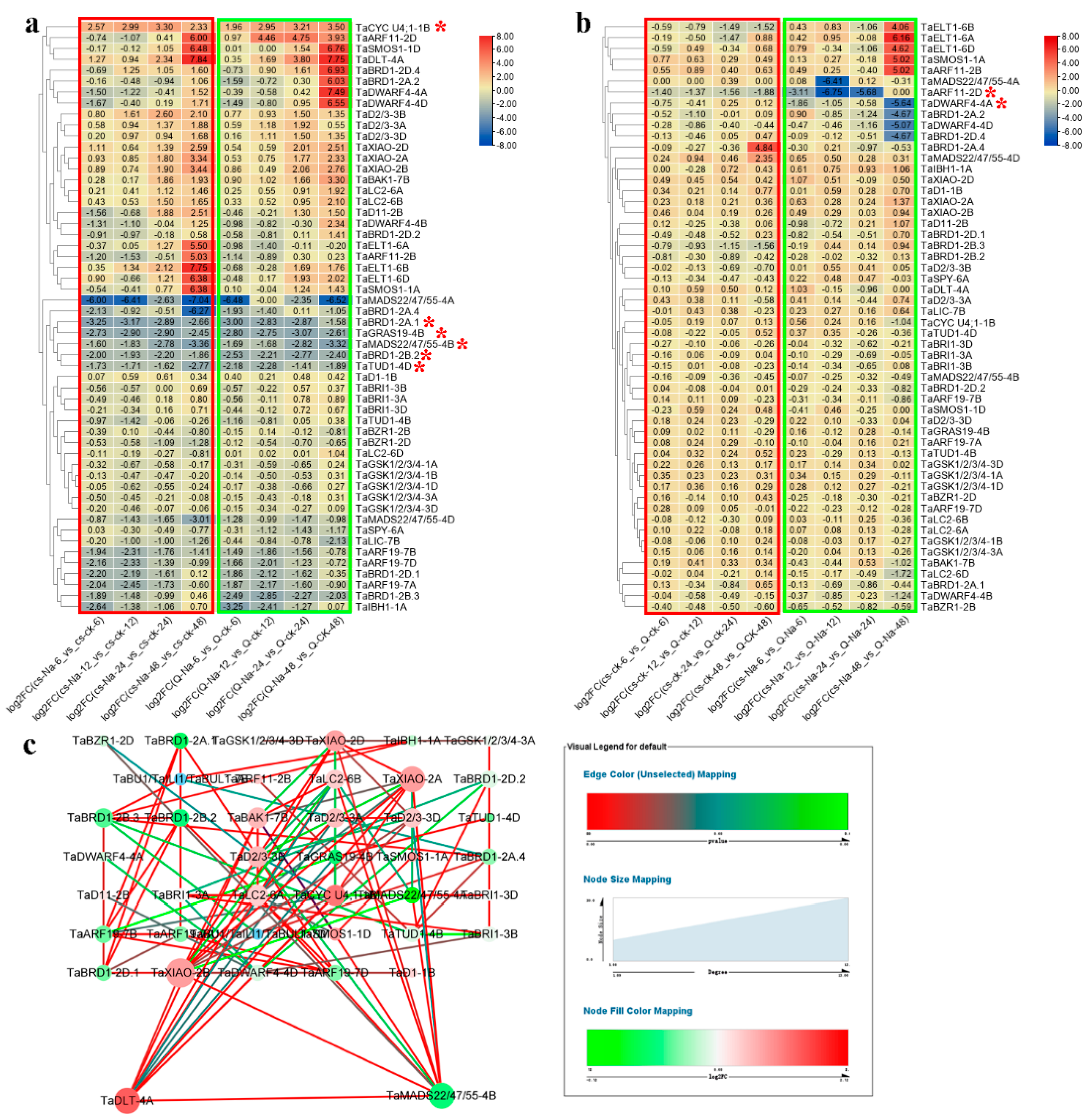
2.6. Expression Analyses of Wheat BR-Related Genes in Response to Salt Stress
3. Discussion
3.1. Comparison of BR-Related Genes among Gramineae Species
3.2. Putative Functions of Maize and Wheat BR-Related Genes in Plant Architecture and Salt Adaptation
4. Materials and Methods
4.1. Identification, Chromosomal Location, Chemical Characterization, Subcellular Localization, Multiple Sequence Alignment, and Protein Structure Analyses of BA-Related Genes
4.2. Phylogeny, Transmembrane Helices, Synteny, Gene Structure, Cis-Acting Elements in Promoters, Protein-Protein Interaction Networks, and Trend and Gene Co-Expression Network Analyses
4.3. Plant Materials, BR Treatment, and Transcriptome Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006, 24, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Morinaka, Y.; Sakamoto, T.; Inukai, Y.; Agetsuma, M.; Kitano, H.; Ashikari, M.; Matsuoka, M. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 2006, 141, 924–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Teng, W.; Yan, J.; Ma, X.; Meng, Y.; Dai, J.; Li, J. Genetic dissection of plant height by molecular markers using a population of recombinant inbred lines in maize. Euphytica 2007, 155, 117–124. [Google Scholar]
- Divi, U.K.; Krishna, P. Brassinosteroid: A biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol. 2009, 26, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Vriet, C.; Russinova, E.; Reuzeau, C. From squalene to brassinolide: The steroid metabolic and signaling pathways across the plant kingdom. Mol. Plant 2013, 6, 1738–1757. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Li, J. Regulation of brassinosteroid biosynthesis and inactivation. J. Integr. Plant Biol. 2012, 54, 746–759. [Google Scholar] [CrossRef]
- Tanabe, S.; Ashikari, M.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Yano, M.; Yoshimura, A.; Kitano, H. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 2005, 17, 776–790. [Google Scholar] [CrossRef] [Green Version]
- Hong, Z.; Ueguchi-Tanaka, M.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Hasegawa, Y.; Ashikari, M.; Kitano, H. The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 2005, 17, 2243–2254. [Google Scholar] [CrossRef] [Green Version]
- Mori, M.; Nomura, T.; Ooka, H.; Ishizaka, M.; Yokota, T.; Sugimoto, K.; Okabe, K.; Kajiwara, H. Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 2002, 130, 1152–1161. [Google Scholar] [CrossRef] [Green Version]
- Hong, Z.; Ueguchi-Tanaka, M.; Shimizu-Sato, S.; Inukai, Y.; Fujioka, S.; Shimada, Y.; Takatsuto, S.; Agetsuma, M. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002, 32, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Ihara, Y.; Wu, X.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Kitano, H. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000, 12, 1591–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Wang, L.; Wang, M.; Xu, Y.Y.; Luo, W.; Liu, Y.J.; Xu, Z.H.; Li, J. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 2009, 7, 791–806. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Shen, W.; He, Y.; Tian, Z.; Li, J. OVATE Family Protein 8 Positively Mediates Brassinosteroid Signaling through Interacting with the GSK3-like Kinase in Rice. PLoS Genet. 2016, 12, e1006118. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.Y.; Zhang, L.Y.; Gampala, S.S.; Zhu, S.W.; Song, W.Y.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, A.; Ueguchi-Tanaka, M.; Sakamoto, T.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Sazuka, T.; Ashikari, M. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 2006, 48, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Z.; Xu, Y.; Joo, S.H.; Kim, S.K.; Xue, Z.; Xu, Z.; Wang, Z. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 2009, 57, 498–510. [Google Scholar] [CrossRef]
- Yang, B.J.; Lin, W.H.; Fu, F.F.; Xu, Z.H.; Xue, H.W. Receptor-like protein ELT1 promotes brassinosteroid signaling through interacting with and suppressing the endocytosis-mediated degradation of receptor BRI1. Cell Res. 2017, 27, 1182–1185. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Sahni, S.; Prasad, B.D.; Liu, Q.; Grbic, V.; Sharpe, A.; Singh, S.P.; Krishna, P. Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 2016, 6, 28298. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gu, L.; Zhang, X.; Liu, M.; Jiang, H.; Cai, R.; Zhao, Y.; Cheng, B. Global transcriptome and weighted gene co-expression network analyses reveal hybrid-specific modules and candidate genes related to plant height development in maize. Plant Mol. Biol. 2018, 98, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Wang, X.; Tao, Q.; Guo, Q.; Zhou, Y.; Yi, F.; Huang, G.; Li, Y. Transcriptome dynamic landscape underlying the improvement of maize lodging resistance under coronatine treatment. BMC Plant Biol. 2021, 21, 202. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Zhou, C.; Guo, H.; Xie, Y.; Zhao, L.; Gu, J.; Zhao, S.; Ding, Y. Transcriptome sequencing reveals hotspot mutation regions and dwarfing mechanisms in wheat mutants induced by gamma-ray irradiation and EMS. J. Radiat. Res. 2020, 61, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Z.; Khan, A.A.; Lin, Q.; Han, Y.; Mu, P.; Liu, Y.; Zhang, H. Expression partitioning of homeologs and tandem duplications contribute to salt tolerance in wheat (Triticum aestivum L.). Sci. Rep. 2016, 6, 21476. [Google Scholar] [CrossRef]
- Sanchez-Barrena, M.J.; Chaves-Sanjuan, A.; Raddatz, N.; Mendoza, I.; Cortes, A.; Gago, F.; Gonzalez-Rubio, J.M.; Benavente, J.L. Recognition and Activation of the Plant AKT1 Potassium Channel by the Kinase CIPK23. Plant Physiol. 2020, 182, 2143–2153. [Google Scholar] [CrossRef] [Green Version]
- Parenicova, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S. Gene duplication and the uniqueness of vertebrate genomes circa 1970-1999. Semin. Cell Dev. Biol. 1999, 10, 517–522. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Stein, N. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Qiao, S.; Sun, S.; Wang, L.; Wu, Z.; Li, C.; Li, X.; Wang, T.; Leng, L. The RLA1/SMOS1 Transcription Factor Functions with OsBZR1 to Regulate Brassinosteroid Signaling and Rice Architecture. Plant Cell 2017, 29, 292–309. [Google Scholar] [CrossRef] [Green Version]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liu, D.; Zhang, G.; Tong, H.; Chu, C. Brassinosteroids Regulate OFP1, a DLT Interacting Protein, to Modulate Plant Architecture and Grain Morphology in Rice. Front. Plant Sci. 2017, 8, 1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.; Gendron, N.; Sun, C.Q.; Wang, Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, H.; Xiao, Y.; Liu, D.; Gao, S.; Liu, L.; Yin, Y.; Jin, Y.; Qian, Q. Brassinosteroid Regulates Cell Elongation by Modulating Gibberellin Metabolism in Rice. Plant Cell 2014, 26, 4376–4393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Liu, J.; Zhao, T.; Du, C.; Nie, S.; Zhang, Y.; Lv, S.; Huang, S. Modification of Threonine-1050 of SlBRI1 regulates BR Signalling and increases fruit yield of tomato. BMC Plant Biol. 2019, 19, 256. [Google Scholar] [CrossRef]
- Nakamura, A.; Fujioka, S.; Sunohara, H.; Kamiya, N.; Hong, Z.; Inukai, Y.; Miura, K.; Takatsuto, S. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 2006, 140, 580–590. [Google Scholar] [CrossRef] [Green Version]
- Kir, G.; Ye, H.; Nelissen, H.; Neelakandan, A.K.; Kusnandar, A.S.; Luo, A.; Inze, D.; Sylvester, A.W. RNA Interference Knockdown of BRASSINOSTEROID INSENSITIVE1 in Maize Reveals Novel Functions for Brassinosteroid Signaling in Controlling Plant Architecture. Plant Physiol. 2015, 169, 826–839. [Google Scholar] [CrossRef] [Green Version]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Zhao, Z.; Zhou, S.; Liu, L.; Kong, W.; Chen, H.; Long, W.; Feng, Z. Top Bending Panicle1 is involved in brassinosteroid signaling and regulates the plant architecture in rice. Plant Physiol. Biochem. 2017, 121, 1–13. [Google Scholar] [CrossRef]
- Sun, S.; Chen, D.; Li, X.; Qiao, S.; Shi, C.; Li, C.; Shen, H.; Wang, X. Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev. Cell 2015, 34, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Tong, H.; Liu, L.; Jin, Y.; Du, L.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 2012, 24, 2562–2577. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Lu, S.J.; Wang, M.J.; He, H.; Sun, L.; Wang, H.; Liu, X.H.; Jiang, L. A Novel QTL qTGW3 Encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that Interacts with OsARF4 to Negatively Regulate Grain Size and Weight in Rice. Mol. Plant 2018, 11, 736–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.; Zhu, J.Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Feng, Y.; Zhou, T.; Xu, F. Genome-scale mRNA transcriptomic insights into the responses of oilseed rape (Brassica napus L.) to varying boron availabilities. Plant Soil 2017, 416, 205–225. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnere, G. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Ma, S.; Zhang, K.; Liu, X.; Hu, L.; Wang, W.; Zheng, L. Genome-Wide Identification of Gramineae Brassinosteroid-Related Genes and Their Roles in Plant Architecture and Salt Stress Adaptation. Int. J. Mol. Sci. 2022, 23, 5551. https://doi.org/10.3390/ijms23105551
Huang J, Ma S, Zhang K, Liu X, Hu L, Wang W, Zheng L. Genome-Wide Identification of Gramineae Brassinosteroid-Related Genes and Their Roles in Plant Architecture and Salt Stress Adaptation. International Journal of Molecular Sciences. 2022; 23(10):5551. https://doi.org/10.3390/ijms23105551
Chicago/Turabian StyleHuang, Jinyong, Shengjie Ma, Kaiyan Zhang, Xiaohan Liu, Linlin Hu, Wenming Wang, and Liwei Zheng. 2022. "Genome-Wide Identification of Gramineae Brassinosteroid-Related Genes and Their Roles in Plant Architecture and Salt Stress Adaptation" International Journal of Molecular Sciences 23, no. 10: 5551. https://doi.org/10.3390/ijms23105551
APA StyleHuang, J., Ma, S., Zhang, K., Liu, X., Hu, L., Wang, W., & Zheng, L. (2022). Genome-Wide Identification of Gramineae Brassinosteroid-Related Genes and Their Roles in Plant Architecture and Salt Stress Adaptation. International Journal of Molecular Sciences, 23(10), 5551. https://doi.org/10.3390/ijms23105551




