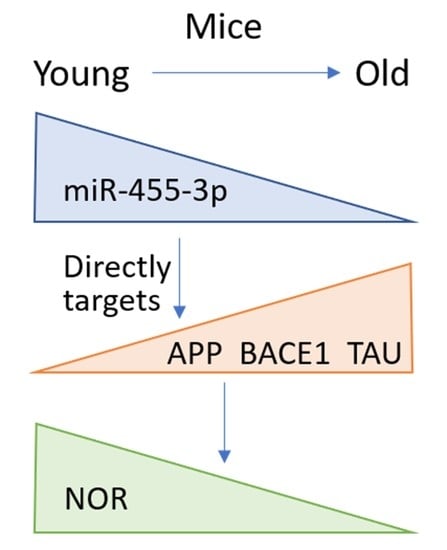
The microRNA-455 Null Mouse Has Memory Deficit and Increased Anxiety, Targeting Key Genes Involved in Alzheimer’s Disease
Abstract
:1. Introduction
2. Results
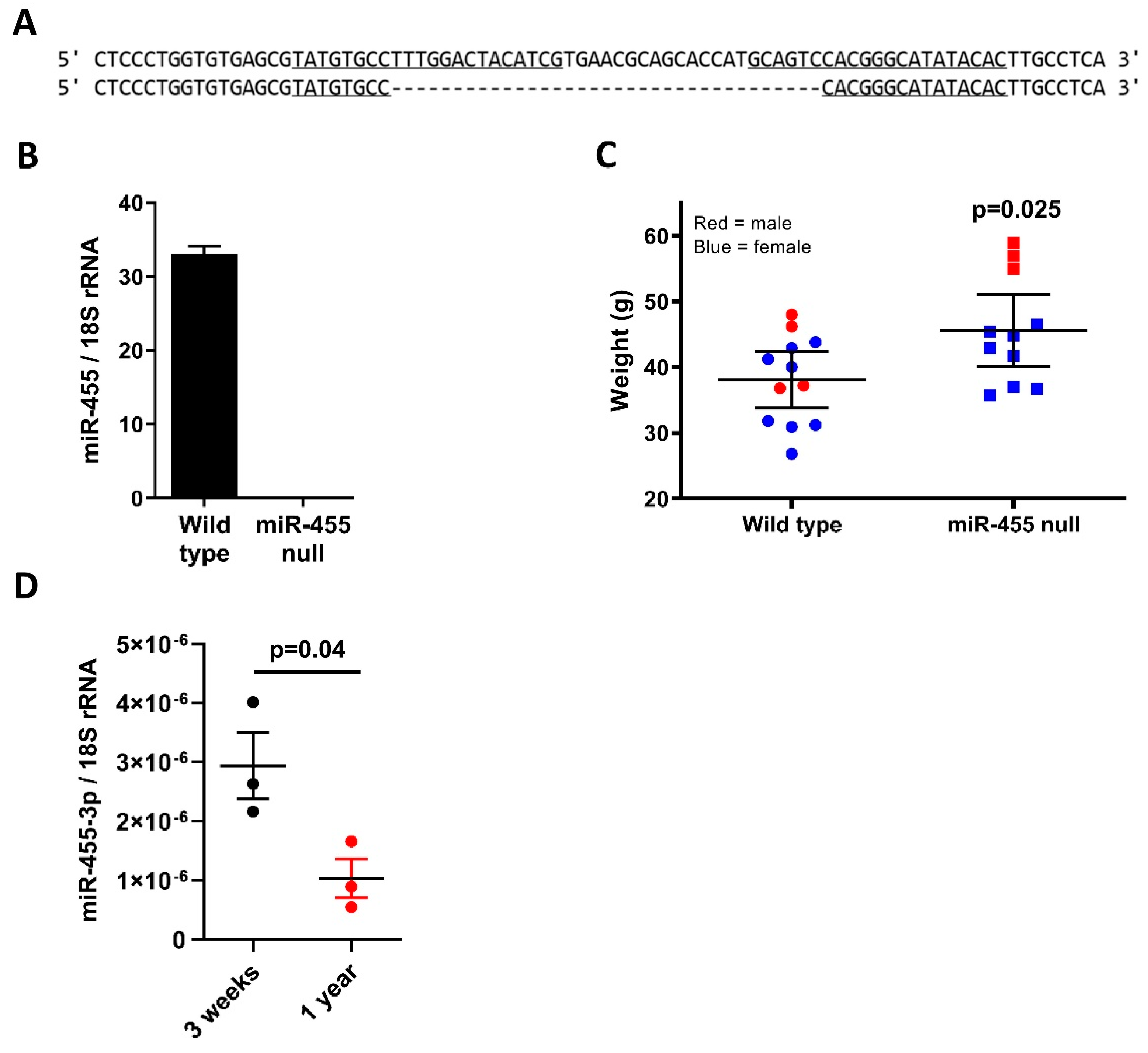
2.1. MicroRNA-455-3p Expression Decreases with Age and Is Absent in the Null Mouse Model
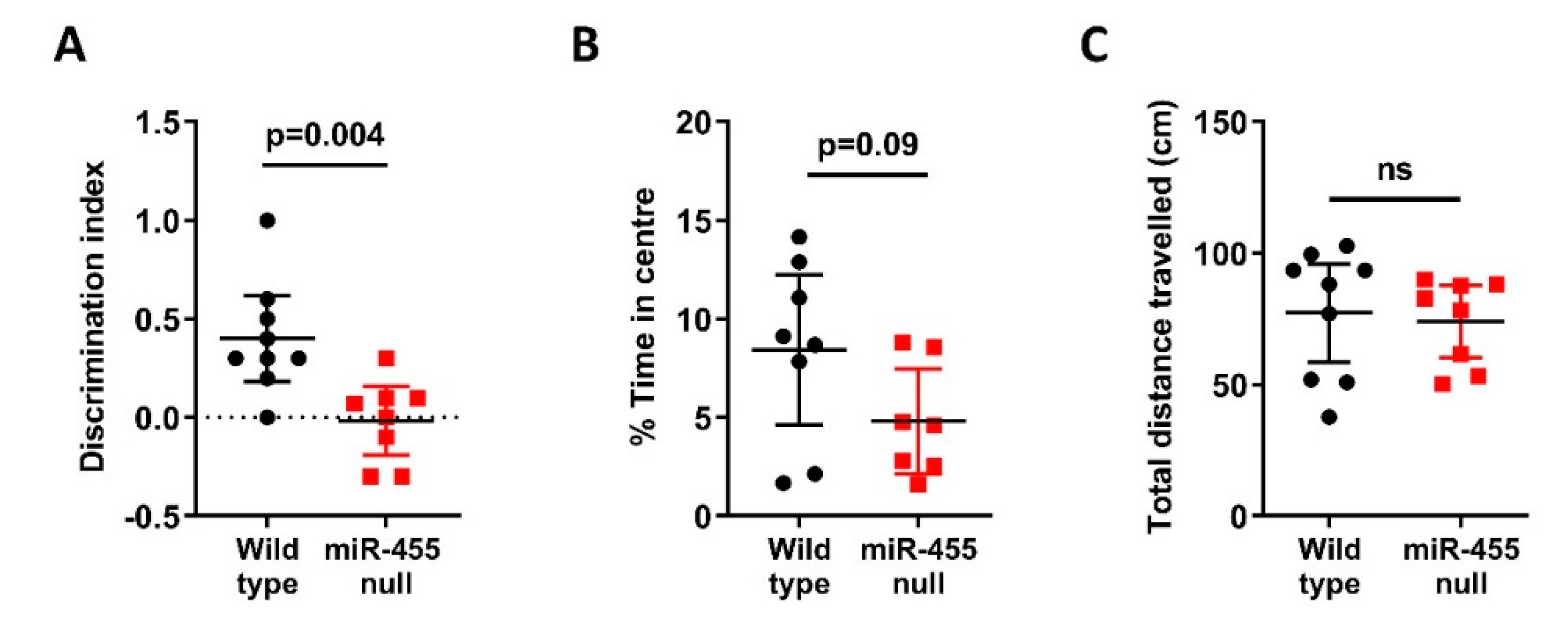
2.2. Recognition Memory Is Impaired in miR-455 Null Mice
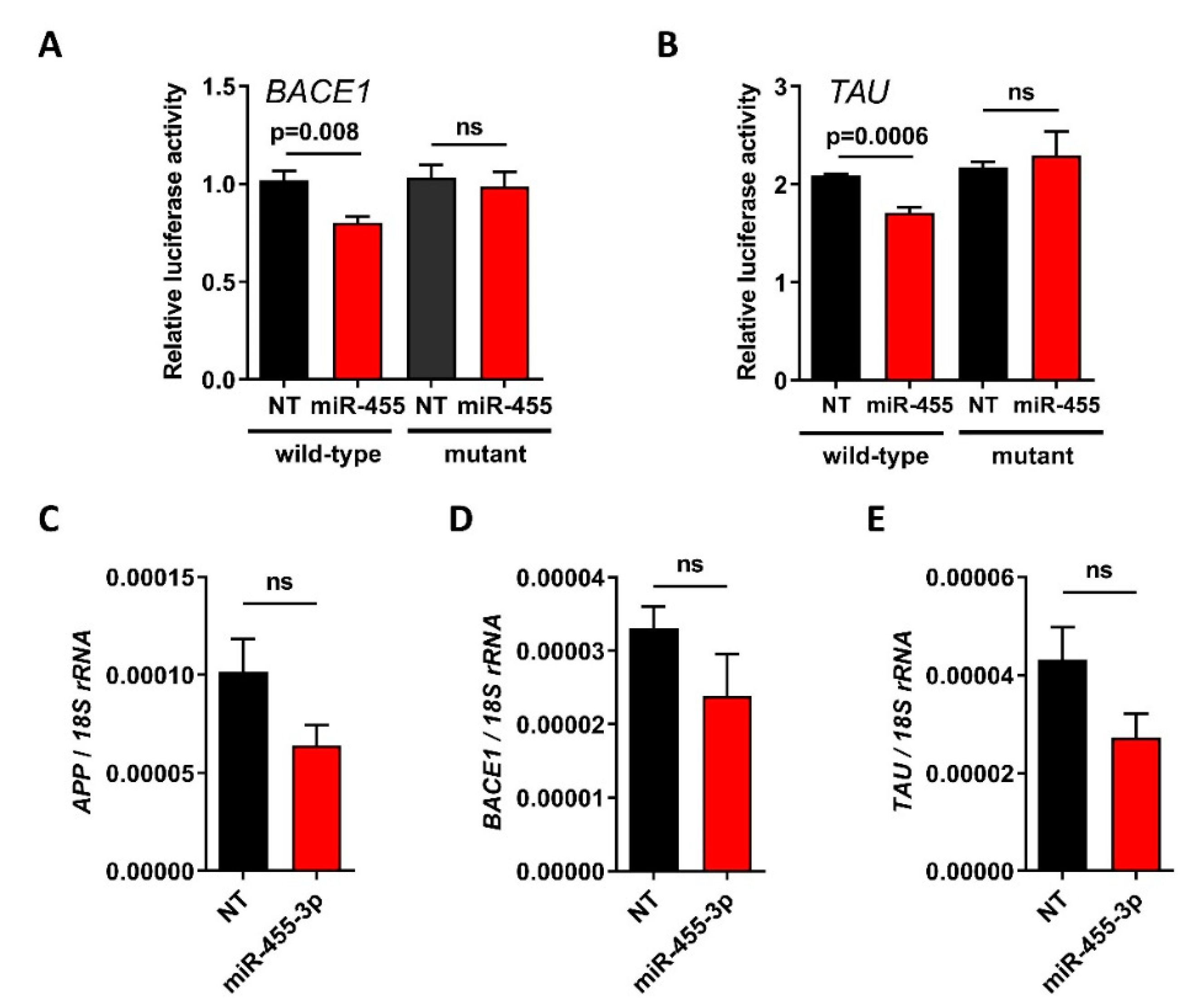
2.3. BACE1 and TAU Are Direct Targets of miR-455-3p
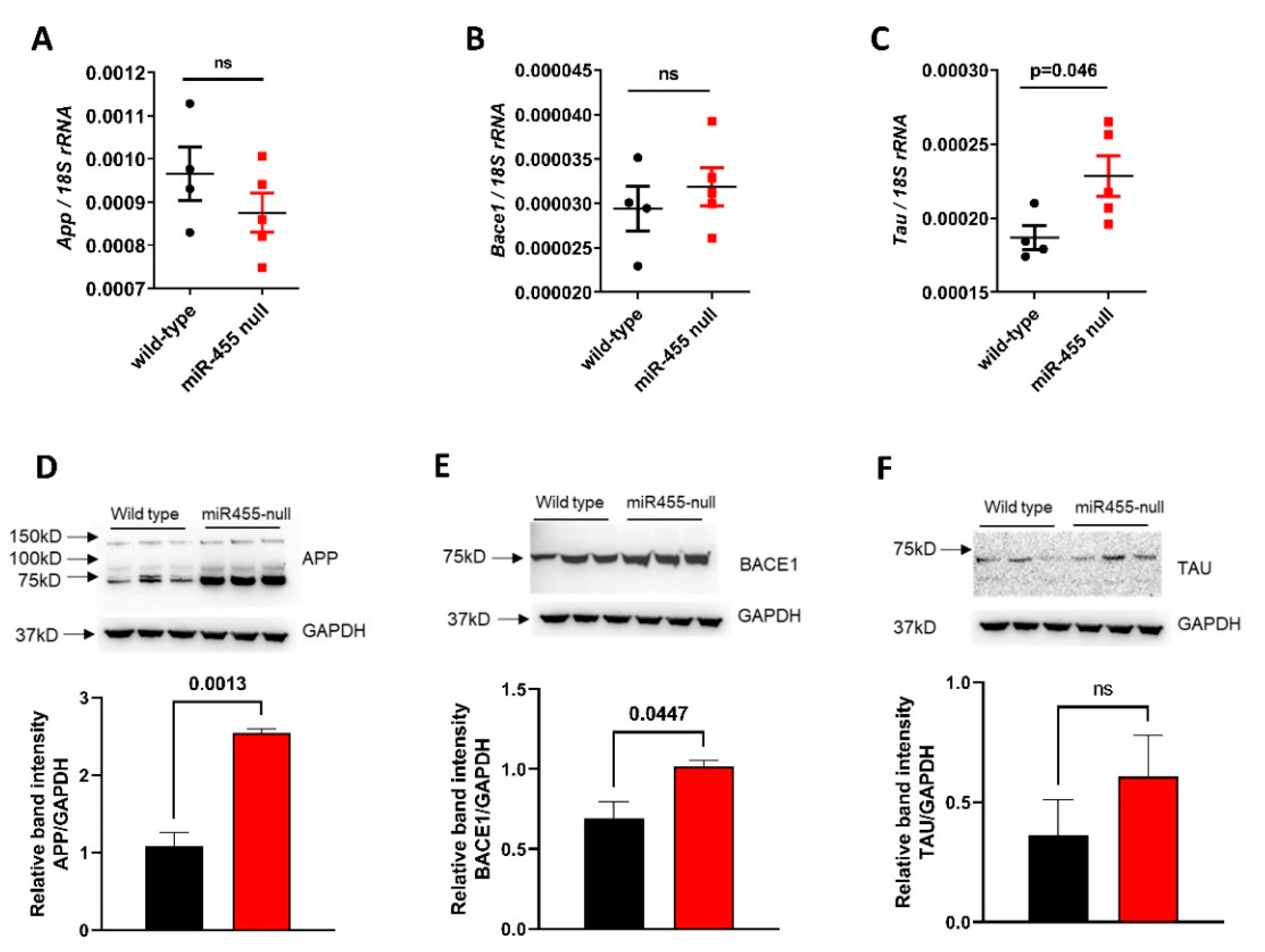
2.4. The Levels of APP, BACE1 and TAU Proteins Are Increased in miR-455 Null Mouse Hippocampus
3. Discussion
4. Materials and Methods
4.1. MiR-455 Null Mouse
4.2. Behavioural Assessment
4.3. Overexpression of miR-455-3p
4.4. RNA and Protein Extraction from Mouse Brains
4.5. Quantitative Real-Time PCR
4.6. Luciferase Assay
4.7. Western Blot
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 15 October 2021).
- Prince, M.; Bryce, R.; Albanese, E.; Wimo, A.; Ribeiro, W.; Ferri, C.P. The global prevalence of dementia: A systematic review and metaanalysis. Alzheimers Dement. 2013, 9, 63–75.e2. [Google Scholar] [CrossRef]
- Kumar, A.; Sidhu, J.; Goyal, A.; Tsao, J.W. Alzheimer Disease; StatPearls: Treasure Island, FL, USA, 2021. [Google Scholar]
- Picanco, L.C.D.S.; Ozela, P.F.; Brito, M.d.F.d.B.; Pinheiro, A.A.; Padilha, E.C.; Braga, F.S.; da Silva, C.H.T.d.P.; Dos Santos, C.B.R.; Rosa, J.M.C.; da Silva Hage-Melim, L.I. Alzheimer’s Disease: A Review from the Pathophysiology to Diagnosis, New Perspectives for Pharmacological Treatment. Curr. Med. Chem. 2018, 25, 3141–3159. [Google Scholar] [CrossRef] [PubMed]
- DeTure, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef]
- Dawkins, E.; Small, D.H. Insights into the physiological function of the β-amyloid precursor protein: Beyond Alzheimer’s disease. J. Neurochem. 2014, 129, 756–769. [Google Scholar] [CrossRef] [Green Version]
- Haass, C.; Kaether, C.; Thinakaran, G.; Sisodia, S. Trafficking and Proteolytic Processing of APP. Cold Spring Harb. Perspect. Med. 2012, 2, a006270. [Google Scholar] [CrossRef] [PubMed]
- Arnsten, A.F.T.; Datta, D.; Del Tredici, K.; Braak, H. Hypothesis: Tau pathology is an initiating factor in sporadic Alzheimer’s disease. Alzheimer’s Dement. 2020, 17, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar] [CrossRef]
- Kametani, F.; Hasegawa, M. Reconsideration of Amyloid Hypothesis and Tau Hypothesis in Alzheimer’s Disease. Front. Neurosci. 2018, 12, 25. [Google Scholar] [CrossRef] [Green Version]
- Calsolaro, V.; Edison, P. Neuroinflammation in Alzheimer’s disease: Current evidence and future directions. Alzheimers Dement. 2016, 12, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Toldi, J.; Vécsei, L. Exploring the Etiological Links behind Neurodegenerative Diseases: Inflammatory Cytokines and Bioactive Kynurenines. Int. J. Mol. Sci. 2020, 21, 2431. [Google Scholar] [CrossRef] [Green Version]
- Catanesi, M.; d’Angelo, M.; Tupone, M.G.; Benedetti, E.; Giordano, A.; Castelli, V.; Cimini, A. MicroRNAs Dysregulation and Mitochondrial Dysfunction in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 5986. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qin, L.; Tang, B. MicroRNAs in Alzheimer’s Disease. Front. Genet. 2019, 10, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelucci, F.; Cechova, K.; Valis, M.; Kuca, K.; Zhang, B.; Hort, J. MicroRNAs in Alzheimer’s Disease: Diagnostic Markers or Therapeutic Agents? Front. Pharmacol. 2019, 10, 665. [Google Scholar] [CrossRef]
- Brito, L.M.; Ribeiro-dos-Santos, Â.; Vidal, A.F.; de Araújo, G.S. Differential Expression and miRNA–Gene Interactions in Early and Late Mild Cognitive Impairment. Biology 2020, 9, 251. [Google Scholar] [CrossRef]
- Swingler, T.E.; Wheeler, G.; Carmont, V.; Elliott, H.R.; Barter, M.J.; Abu-Elmagd, M.; Donell, S.T.; Boot-Handford, R.P.; Hajihosseini, M.K.; Munsterberg, A.; et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012, 64, 1909–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Zhao, X.; Mao, G.; Zhang, Z.; Wen, X.; Zhang, C.; Liao, W.; Zhang, Z. MicroRNA-455-3p promotes TGF-β signaling and inhibits osteoarthritis development by directly targeting PAK2. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Reddy, P.H. A New Discovery of MicroRNA-455-3p in Alzheimer’s Disease. J. Alzheimers Dis. 2019, 72, S117–S130. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Vijayan, M.; Reddy, P.H. MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum. Mol. Genet. 2017, 26, 3808–3822. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Reddy, P.H. MicroRNA-455-3p as a Potential Biomarker for Alzheimer’s Disease: An Update. Front. Aging Neurosci. 2018, 10, 41. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Reddy, A.P.; Yin, X.; Reddy, P.H. Novel MicroRNA-455-3p and its protective effects against abnormal APP processing and amyloid beta toxicity in Alzheimer’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2428–2440. [Google Scholar] [CrossRef] [PubMed]
- Inukai, S.; de Lencastre, A.; Turner, M.; Slack, F. Novel microRNAs differentially expressed during aging in the mouse brain. PLoS ONE 2012, 7, e40028. [Google Scholar] [CrossRef] [Green Version]
- Mohammed, C.P.; Rhee, H.; Phee, B.K.; Kim, K.; Kim, H.J.; Lee, H.; Park, J.H.; Jung, J.H.; Kim, J.Y.; Kim, H.C.; et al. miR-204 downregulates EphB2 in aging mouse hippocampal neurons. Aging Cell 2016, 15, 380–388. [Google Scholar] [CrossRef]
- Mohammed, C.P.D.; Park, J.S.; Nam, H.G.; Kim, K. MicroRNAs in brain aging. Mech. Ageing Dev. 2017, 168, 3–9. [Google Scholar] [CrossRef]
- Cao, D.-D.; Li, L.; Chan, W.-Y. MicroRNAs: Key Regulators in the Central Nervous System and Their Implication in Neurological Diseases. Int. J. Mol. Sci. 2016, 17, 842. [Google Scholar] [CrossRef]
- Fitzpatrick, A.L.; Kuller, L.H.; Lopez, O.L.; Diehr, P.; O’Meara, E.S.; Longstreth, W.T., Jr.; Luchsinger, J.A. Midlife and late-life obesity and the risk of dementia: Cardiovascular health study. Arch. Neurol. 2009, 66, 336–342. [Google Scholar] [CrossRef] [Green Version]
- Profenno, L.A.; Porsteinsson, A.P.; Faraone, S.V. Meta-analysis of Alzheimer’s disease risk with obesity, diabetes, and related disorders. Biol. Psychiatry 2010, 67, 505–512. [Google Scholar] [CrossRef]
- Atti, A.R.; Valente, S.; Iodice, A.; Caramella, I.; Ferrari, B.; Albert, U.; Mandelli, L.; De Ronchi, D. Metabolic Syndrome, Mild Cognitive Impairment, and Dementia: A Meta-Analysis of Longitudinal Studies. Am. J. Geriatr. Psychiatry 2019, 27, 625–637. [Google Scholar] [CrossRef]
- Borshchev, Y.Y.; Uspensky, Y.P.; Galagudza, M.M. Pathogenetic pathways of cognitive dysfunction and dementia in metabolic syndrome. Life Sci. 2019, 237, 116932. [Google Scholar] [CrossRef] [PubMed]
- Tolppanen, A.M.; Solomon, A.; Soininen, H.; Kivipelto, M. Midlife vascular risk factors and Alzheimer’s disease: Evidence from epidemiological studies. J. Alzheimers Dis. 2012, 32, 531–540. [Google Scholar] [CrossRef]
- Cunnane, S.C.; Trushina, E.; Morland, C.; Prigione, A.; Casadesus, G.; Andrews, Z.B.; Beal, M.F.; Bergersen, L.H.; Brinton, R.D.; de la Monte, S.; et al. Brain energy rescue: An emerging therapeutic concept for neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2020, 19, 609–633. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.E.; Lim, C.S.; Kim, J.I.; Seo, D.; Chun, H.; Yu, N.K.; Lee, J.; Kang, S.J.; Ko, H.G.; Choi, J.H.; et al. The Brain-Enriched MicroRNA miR-9-3p Regulates Synaptic Plasticity and Memory. J. Neurosci. 2016, 36, 8641–8652. [Google Scholar] [CrossRef] [PubMed]
- Michely, J.; Kraft, S.; Muller, U. miR-12 and miR-124 contribute to defined early phases of long-lasting and transient memory. Sci. Rep. 2017, 7, 7910. [Google Scholar] [CrossRef] [Green Version]
- Lippi, G.; Fernandes, C.C.; Ewell, L.A.; John, D.; Romoli, B.; Curia, G.; Taylor, S.R.; Frady, E.P.; Jensen, A.B.; Liu, J.C.; et al. MicroRNA-101 Regulates Multiple Developmental Programs to Constrain Excitation in Adult Neural Networks. Neuron 2016, 92, 1337–1351. [Google Scholar] [CrossRef] [Green Version]
- Mathew, R.S.; Tatarakis, A.; Rudenko, A.; Johnson-Venkatesh, E.M.; Yang, Y.J.; Murphy, E.A.; Todd, T.P.; Schepers, S.T.; Siuti, N.; Martorell, A.J.; et al. A microRNA negative feedback loop downregulates vesicle transport and inhibits fear memory. Elife 2016, 5, 463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, K.F.; Sakamoto, K.; Aten, S.; Snider, K.H.; Loeser, J.; Hesse, A.M.; Page, C.E.; Pelz, C.; Arthur, J.S.; Impey, S.; et al. Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learn. Mem. 2016, 23, 61–71. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, B.B.; Ouyang, Y.B.; Xu, L.; Sun, X.; Giffard, R.G.; Stary, C.M. Postinjury Inhibition of miR-181a Promotes Restoration of Hippocampal CA1 Neurons after Transient Forebrain Ischemia in Rats. eNeuro 2019, 6. [Google Scholar] [CrossRef]
- Zhang, S.F.; Chen, J.C.; Zhang, J.; Xu, J.G. miR-181a involves in the hippocampus-dependent memory formation via targeting PRKAA1. Sci. Rep. 2017, 7, 8480. [Google Scholar] [CrossRef] [Green Version]
- Biundo, F.; Del Prete, D.; Zhang, H.; Arancio, O.; D’Adamio, L. A role for tau in learning, memory and synaptic plasticity. Sci. Rep. 2018, 8, 3184. [Google Scholar] [CrossRef] [Green Version]
- Evans, C.E.; Thomas, R.S.; Freeman, T.J.; Hvoslef-Eide, M.; Good, M.A.; Kidd, E.J. Selective reduction of APP-BACE1 activity improves memory via NMDA-NR2B receptor-mediated mechanisms in aged PDAPP mice. Neurobiol. Aging 2019, 75, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, H.; Takahashi, H.; Tarui, N.; Matsui, J.; Tomita, T.; Hirode, M.; Sagayama, M.; Maeda, R.; Kawamoto, M.; Hirai, K.; et al. A noncompetitive BACE1 inhibitor TAK-070 ameliorates Abeta pathology and behavioral deficits in a mouse model of Alzheimer’s disease. J. Neurosci. 2010, 30, 11157–11166. [Google Scholar] [CrossRef]
- Webster, S.J.; Bachstetter, A.D.; Van Eldik, L.J. Comprehensive behavioral characterization of an APP/PS-1 double knock-in mouse model of Alzheimer’s disease. Alzheimers Res. Ther. 2013, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- Homolak, J.; Mudrovcic, M.; Vukic, B.; Toljan, K. Circadian Rhythm and Alzheimer’s Disease. Med. Sci. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyles, L.; Pontifex, M.G.; Rodriguez-Ramiro, I.; Anis-Alavi, M.A.; Jelane, K.S.; Snelling, T.; Solito, E.; Fonseca, S.; Carvalho, A.L.; Carding, S.R.; et al. Regulation of blood–brain barrier integrity by microbiome-associated methylamines and cognition by trimethylamine N-oxide. Microbiome 2021, 9, 235. [Google Scholar] [CrossRef]
- Pontifex, M.G.; Martinsen, A.; Saleh, R.N.M.; Harden, G.; Tejera, N.; Müller, M.; Fox, C.; Vauzour, D.; Minihane, A.M. APOE4 genotype exacerbates the impact of menopause on cognition and synaptic plasticity in APOE-TR mice. FASEB J. 2021, 35, e21583. [Google Scholar] [CrossRef] [PubMed]
- Dere, E.; Huston, J.P.; De Souza Silva, M.A. The pharmacology, neuroanatomy and neurogenetics of one-trial object recognition in rodents. Neurosci. Biobehav. Rev. 2007, 31, 673–704. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swingler, T.E.; Niu, L.; Pontifex, M.G.; Vauzour, D.; Clark, I.M. The microRNA-455 Null Mouse Has Memory Deficit and Increased Anxiety, Targeting Key Genes Involved in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 554. https://doi.org/10.3390/ijms23010554
Swingler TE, Niu L, Pontifex MG, Vauzour D, Clark IM. The microRNA-455 Null Mouse Has Memory Deficit and Increased Anxiety, Targeting Key Genes Involved in Alzheimer’s Disease. International Journal of Molecular Sciences. 2022; 23(1):554. https://doi.org/10.3390/ijms23010554
Chicago/Turabian StyleSwingler, Tracey E., Lingzi Niu, Matthew G. Pontifex, David Vauzour, and Ian M. Clark. 2022. "The microRNA-455 Null Mouse Has Memory Deficit and Increased Anxiety, Targeting Key Genes Involved in Alzheimer’s Disease" International Journal of Molecular Sciences 23, no. 1: 554. https://doi.org/10.3390/ijms23010554
APA StyleSwingler, T. E., Niu, L., Pontifex, M. G., Vauzour, D., & Clark, I. M. (2022). The microRNA-455 Null Mouse Has Memory Deficit and Increased Anxiety, Targeting Key Genes Involved in Alzheimer’s Disease. International Journal of Molecular Sciences, 23(1), 554. https://doi.org/10.3390/ijms23010554