Insight into microRNAs-Mediated Communication between Liver and Brain: A Possible Approach for Understanding Acute Liver Failure?
Abstract
:1. Background
1.1. Liver Disease a Global Burden
1.2. Acute Liver Failure
1.3. Micro RNA
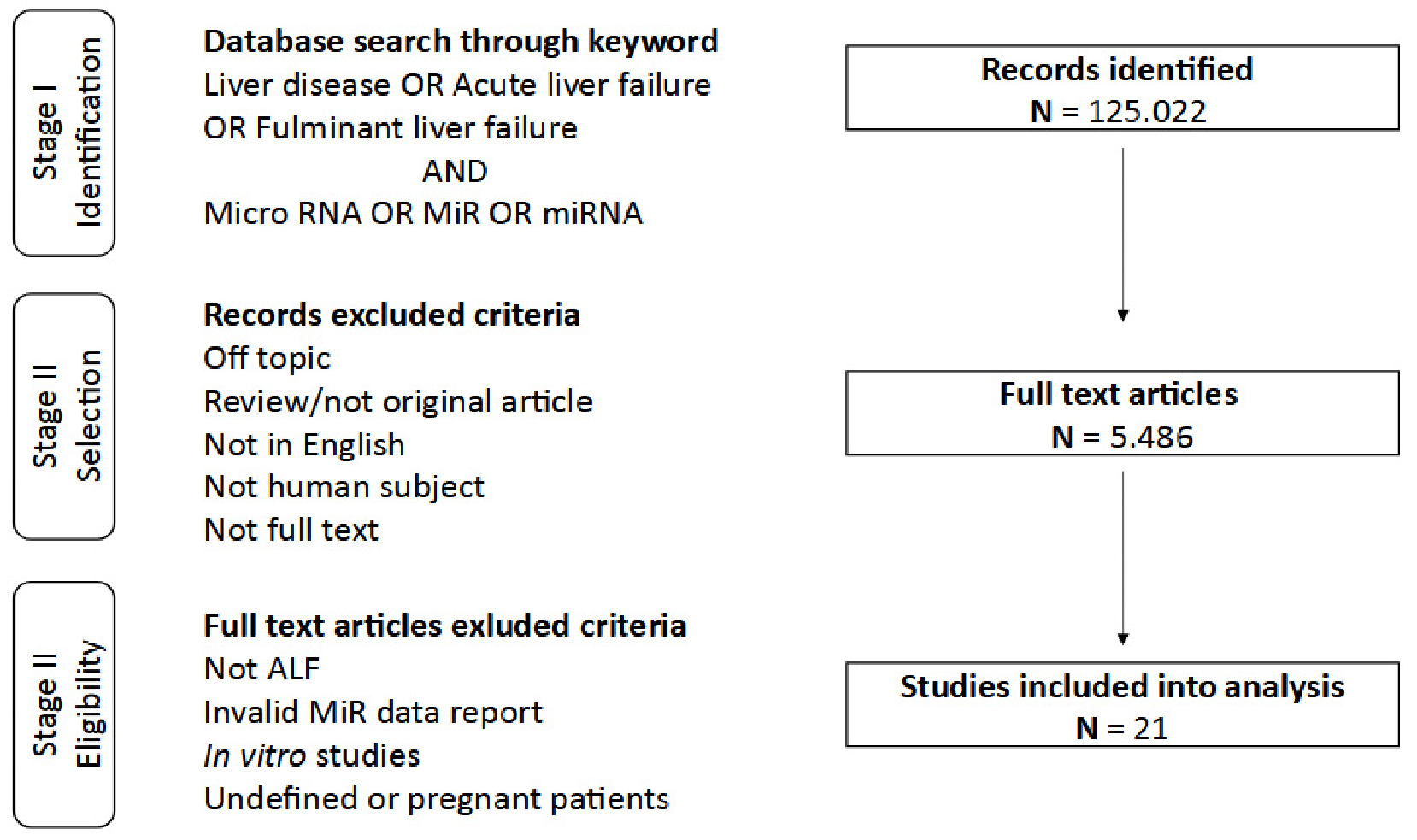
2. Study Concept and Data Analysis Strategy
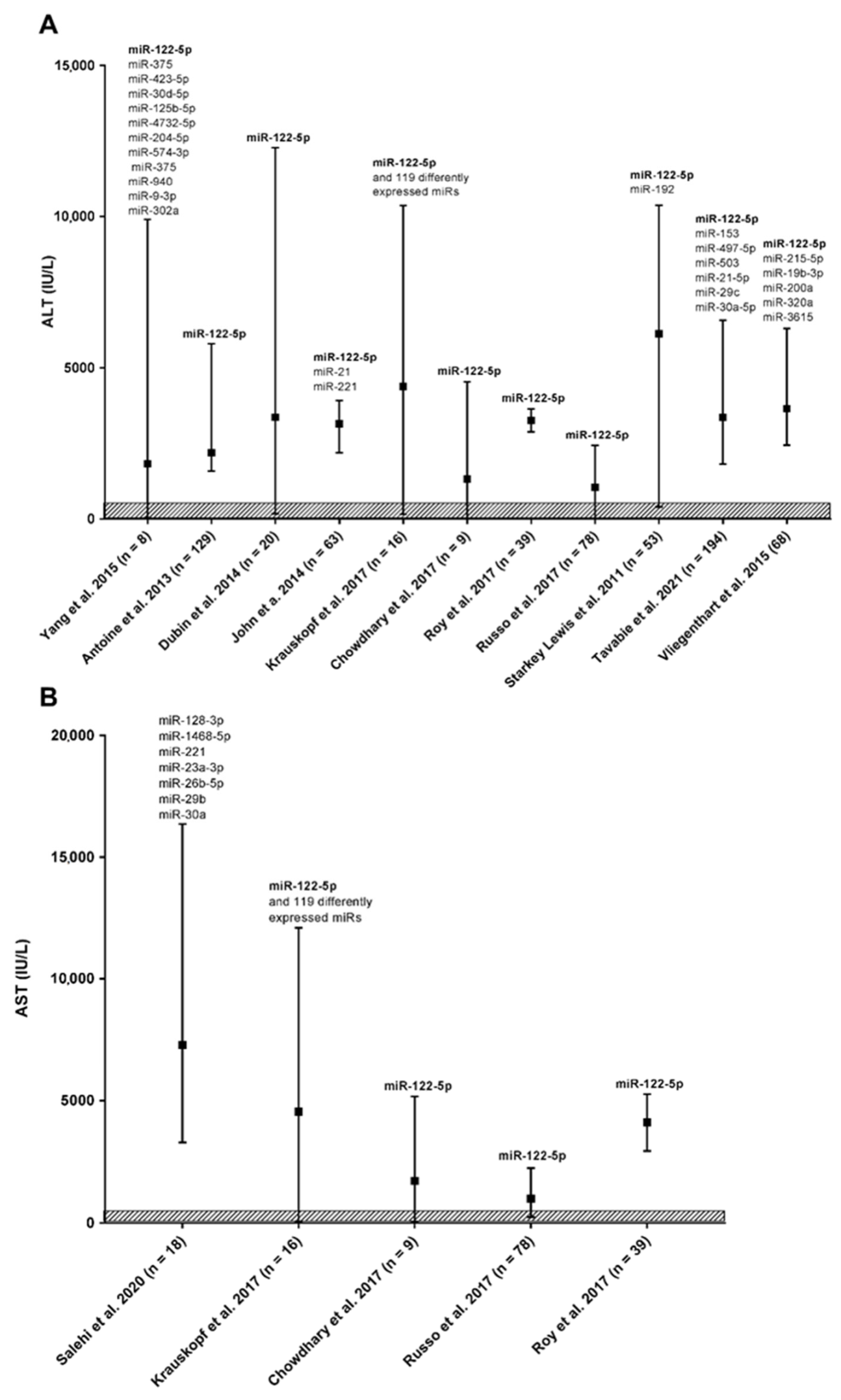
3. Research Summary
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mokdad, A.A.; Lopez, A.D.; Shahraz, S.; Lozano, R.; Mokdad, A.H.; Stanaway, J.; Murray, C.J.L.; Naghavi, M. Liver cirrhosis mortality in 187 countries between 1980 and 2010: A systematic analysis. BMC Med. 2014, 12, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Beste, L.A.; Leipertz, S.L.; Green, P.K.; Dominitz, J.A.; Ross, D.; Ioannou, G.N. Trends in Burden of Cirrhosis and Hepatocellular Carcinoma by Underlying Liver Disease in US Veterans, 2001–2013. Gastroenterology 2015, 149, 1471–1482.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO Global Health Estimates 2015: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2015; World Health Organization: Geneva, Switzerland, 2016.
- Trey, C.; Davidson, C.S. The management of fulminant hepatic failure. Prog. Liver Dis. 1970, 3, 282–298. [Google Scholar]
- Williams, R.; Schalm, S.W.; O’Grady, J.G. Acute liver failure: Redefining the syndromes. Lancet 1993, 342, 273–275. [Google Scholar] [CrossRef]
- Kandiah, P.A.; Olson, J.C.; Subramanian, R.M. Emerging strategies for the treatment of patients with acute hepatic failure. Curr. Opin. Crit. Care 2016, 22, 142–151. [Google Scholar] [CrossRef]
- Bernal, W.; Auzinger, G.; Sizer, E.; Wendon, J. Intensive care management of acute liver failure. Semin. Liver Dis. 2008, 28, 188–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, R.J. Acute liver failure. Curr. Opin. Gastroenterol. 1999, 15, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Pimpin, L.; Cortez-Pinto, H.; Negro, F.; Corbould, E.; Lazarus, J.V.; Webber, L.; Sheron, N. Burden of liver disease in Europe: Epidemiology and analysis of risk factors to identify prevention policies. J. Hepatol. 2018, 69, 718–735. [Google Scholar] [CrossRef] [PubMed]
- Bernal, W.; Wendon, J. Acute liver failure. N. Engl. J. Med. 2013, 369, 2525–2534. [Google Scholar] [CrossRef]
- Andrade, R.J.; Aithal, G.P.; Björnsson, E.S.; Kaplowitz, N.; Kullak-Ublick, G.A.; Larrey, D.; Karlsen, T.H. EASL Clinical Practice Guidelines: Drug-induced liver injury. J. Hepatol. 2019, 70, 1222–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wai, C.T.; Tan, B.H.; Chan, C.L.; Sutedja, D.S.; Lee, Y.M.; Khor, C.; Lim, S.G. Drug-induced liver injury at an Asian center: A prospective study. Liver Int. 2007, 27, 465–474. [Google Scholar] [CrossRef]
- WHO. Recommendations and Guidance on Hepatitis C Virus Self-Testing; WHO: Geneva, Switzerland, 2021; ISBN 9789240031128. [Google Scholar]
- OMS, (Organização Mundial da Saúde). Global Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections, 2021; WHO: Geneva, Switzerland, 2021; Volume 53, ISBN 9788578110796. [Google Scholar]
- Zumla, A.; Chakaya, J.; Khan, M.; Fatima, R.; Wejse, C.; Al-Abri, S.; Fox, G.J.; Nachega, J.; Kapata, N.; Knipper, M.; et al. World Tuberculosis Day 2021 Theme—‘The Clock is Ticking’—And the world is running out of time to deliver the United Nations General Assembly commitments to End TB due to the COVID-19 pandemic. Int. J. Infect. Dis. 2021, 113 (Suppl. S1), S1–S6. [Google Scholar] [CrossRef]
- McKeating, C.; Cadden, I.; McDougall, N.; Jessop, L.; Quah, S.; Lavelle, M.; Griffths, A.; McCaughey, C. Progression from acute to chronic hepatitis B is more common in older adults. Ulster Med. J. 2018, 87, 177–180. [Google Scholar] [PubMed]
- Sahebjam, F.; Vierling, J.M. Autoimmune hepatitis. Front. Med. 2015, 9, 187–219. [Google Scholar] [CrossRef]
- Dong, V.; Nanchal, R.; Karvellas, C.J. Pathophysiology of Acute Liver Failure. Nutr. Clin. Pract. 2020, 35, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Aquilina, A.; Pirotta, T.; Aquilina, A. Acute liver failure and hepatic encephalopathy in exertional heat stroke. BMJ Case Rep. 2018, 2018, bcr-2018-224808. [Google Scholar] [CrossRef] [PubMed]
- Vilstrup, H.; Amodio, P.; Bajaj, J.; Cordoba, J.; Ferenci, P.; Mullen, K.D.; Weissenborn, K.; Wong, P. Hepatic encephalopathy in chronic liver disease: 2014 Practice Guideline by the American Association for the Study Of Liver Diseases and the European Association for the Study of the Liver. Hepatology 2014, 60, 715–735. [Google Scholar] [CrossRef] [PubMed]
- Bjerring, P.N.; Eefsen, M.; Hansen, B.A.; Larsen, F.S. The brain in acute liver failure. A tortuous path from hyperammonemia to cerebral edema. Metab. Brain Dis. 2009, 24, 5–14. [Google Scholar] [CrossRef]
- Wendon, J.; Cordoba, J.; Dhawan, A.; Larsen, F.S.; Manns, M.; Nevens, F.; Samuel, D.; Simpson, K.J.; Yaron, I.; Bernardi, M.; et al. EASL Clinical Practical Guidelines on the management of acute (fulminant) liver failure. J. Hepatol. 2017, 66, 1047–1081. [Google Scholar] [CrossRef]
- Weissenborn, K. Hepatic Encephalopathy: Definition, Clinical Grading and Diagnostic Principles. Drugs 2019, 79, 5–9. [Google Scholar] [CrossRef] [Green Version]
- Haj, M.; Rockey, D.C. Ammonia levels do not guide clinical management of patients with hepatic encephalopathy caused by cirrhosis. Am. J. Gastroenterol. 2020, 115, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Sheasgreen, C.; Lu, L.; Patel, A. Pathophysiology, diagnosis, and management of hepatic encephalopathy. Inflammopharmacology 2014, 22, 319–326. [Google Scholar] [CrossRef]
- Gonzalez, J.J.; Tapper, E.B. A Prospective, Blinded Assessment of Ammonia Testing Demonstrates Low Utility Among Front-Line Clinicians. Clin. Gastroenterol. Hepatol. 2021, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Shalimar; Sheikh, M.F.; Mookerjee, R.P.; Agarwal, B.; Acharya, S.K.; Jalan, R. Prognostic Role of Ammonia in Patients With Cirrhosis. Hepatology 2019, 70, 982–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amodio, P.; Montagnese, S. Lights and shadows in hepatic encephalopathy diagnosis. J. Clin. Med. 2021, 10, 341. [Google Scholar] [CrossRef]
- Rahimi, R.S.; Rockey, D.C. Hepatic Encephalopathy: Pharmacological Therapies Targeting Ammonia. Semin. Liver Dis. 2016, 36, 48–55. [Google Scholar] [CrossRef] [Green Version]
- Abbott, N.J.; Patabendige, A.A.K.; Dolman, D.E.M.; Yusof, S.R.; Begley, D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef]
- Kato, M.; Hughes, R.D.; Keays, R.T.; Williams, R. Electron microscopic study of brain capillaries in cerebral edema from fulminant hepatic failure. Hepatology 1992, 15, 1060–1066. [Google Scholar] [CrossRef]
- Kong, L.Z.; Chandimali, N.; Han, Y.H.; Lee, D.H.; Kim, J.S.; Kim, S.U.; Kim, T.D.; Jeong, D.K.; Sun, H.N.; Lee, D.S.; et al. Pathogenesis, early diagnosis, and therapeutic management of alcoholic liver disease. Int. J. Mol. Sci. 2019, 20, 2712. [Google Scholar] [CrossRef] [Green Version]
- Torbenson, M.; Washington, K. Pathology of liver disease: Advances in the last 50 years. Hum. Pathol. 2020, 95, 78–98. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Duan, X.; Liu, C.; Li, Z. Digital quantitative analysis of microRNA in single cell based on ligation-depended polymerase colony (Polony). Biosens. Bioelectron. 2017, 95, 146–151. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.W.; Steuerwald, N.; Norton, H.J.; Anderson, W.E.; Foureau, D.; Chalasani, N.; Fontana, R.J.; Watkins, P.B.; Serrano, J.; Bonkovsky, H.L. Profiles of miRNAs in serum in severe acute drug induced liver injury and their prognostic significance. Liver Int. 2017, 37, 757–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Fan, J.; Belasco, J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 4034–4039. [Google Scholar] [CrossRef] [Green Version]
- Dragomir, M.P.; Knutsen, E.; Calin, G.A. SnapShot: Unconventional miRNA Functions. Cell 2018, 174, 1038. [Google Scholar] [CrossRef]
- Witwer, K.W. Circulating MicroRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef] [Green Version]
- Mar-Aguilar, F.; Mendoza-Ramírez, J.A.; Malagón-Santiago, I.; Espino-Silva, P.K.; Santuario-Facio, S.K.; Ruiz-Flores, P.; Rodríguez-Padilla, C.; Reséndez-Pérez, D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis. Markers 2013, 34, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Noferesti, S.S.; Sohel, M.M.H.; Hoelker, M.; Salilew-Wondim, D.; Tholen, E.; Looft, C.; Rings, F.; Neuhoff, C.; Schellander, K.; Tesfaye, D. Controlled ovarian hyperstimulation induced changes in the expression of circulatory miRNA in bovine follicular fluid and blood plasma. J. Ovarian Res. 2015, 8, 81. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antoine, D.J.; Dear, J.W.; Lewis, P.S.; Platt, V.; Coyle, J.; Masson, M.; Thanacoody, R.H.; Gray, A.J.; Webb, D.J.; Moggs, J.G.; et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology 2013, 58, 777–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhary, V.; Teng, K.Y.; Thakral, S.; Zhang, B.; Lin, C.H.; Wani, N.; Bruschweiler-Li, L.; Zhang, X.; James, L.; Yang, D.; et al. miRNA-122 Protects Mice and Human Hepatocytes from Acetaminophen Toxicity by Regulating Cytochrome P450 Family 1 Subfamily A Member 2 and Family 2 Subfamily E Member 1 Expression. Am. J. Pathol. 2017, 187, 2758–2774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, G.; Zamboni, F.; Tice, A.; Farci, P. Integrated ordination of miRNA and mRNA expression profiles. BMC Genom. 2015, 16, 767. [Google Scholar] [CrossRef] [Green Version]
- Dubin, P.H.; Yuan, H.; Devine, R.K.; Hynan, L.S.; Jain, M.K.; Lee, W.M.; Larson, A.M.; Liou, I.; Davern, T.; Fix, O.; et al. Micro-RNA-122 levels in acute liver failure and chronic hepatitis C. J. Med. Virol. 2014, 86, 1507–1514. [Google Scholar] [CrossRef] [Green Version]
- John, K.; Hadem, J.; Krech, T.; Wahl, K.; Manns, M.P.; Dooley, S.; Batkai, S.; Thum, T.; Schulze-Osthoff, K.; Bantel, H. MicroRNAs play a role in spontaneous recovery from acute liver failure. Hepatology 2014, 60, 1346–1355. [Google Scholar] [CrossRef]
- Krauskopf, J.; Caiment, F.; Claessen, S.M.; Johnson, K.J.; Warner, R.L.; Schomaker, S.J.; Burt, D.A.; Aubrecht, J.; Kleinjans, J.C. Application of high-throughput sequencing to circulating microRNAs reveals novel biomarkers for drug-induced liver injury. Toxicol. Sci. 2015, 143, 268–276. [Google Scholar] [CrossRef] [Green Version]
- Krauskopf, J.; De Kok, T.M.; Schomaker, S.J.; Gosink, M.; Burt, D.A.; Chandler, P.; Warner, R.L.; Johnson, K.J.; Caiment, F.; Kleinjans, J.C.; et al. Serum microRNA signatures as “liquid biopsies” for interrogating hepatotoxic mechanisms and liver pathogenesis in human. PLoS ONE 2017, 12, e0177928. [Google Scholar] [CrossRef] [Green Version]
- Krauskopf, J.; Gosink, M.M.; Schomaker, S.; Caiment, F.; Warner, R.; Johnson, K.; Kleinjans, J.; Aubrecht, J. The microrna-based liquid biopsy improves early assessment of lethal acetaminophen poisoning: A case report. Am. J. Case Rep. 2020, 21, e919289. [Google Scholar] [CrossRef]
- Pan, K.; Wang, Y.; Pan, P.; Xu, G.; Mo, L.; Cao, L.; Wu, C.; Shen, X. The regulatory role of microRNA-mRNA co-expression in hepatitis B virus-associated acute liver failure. Ann. Hepatol. 2019, 18, 883–892. [Google Scholar] [CrossRef]
- Roy, S.; Bantel, H.; Wandrer, F.; Schneider, A.T.; Gautheron, J.; Vucur, M.; Tacke, F.; Trautwein, C.; Luedde, T.; Roderburg, C. miR-1224 inhibits cell proliferation in acute liver failure by targeting the antiapoptotic gene Nfib. J. Hepatol. 2017, 67, 966–978. [Google Scholar] [CrossRef] [PubMed]
- Salehi, S.; Tavabie, O.D.; Verma, S.; McPhail, M.J.W.; Farzaneh, F.; Bernal, W.; Menon, K.; Agarwal, K.; Aluvihare, V.R. Serum MicroRNA Signatures in Recovery From Acute and Chronic Liver Injury and Selection for Liver Transplantation. Liver Transplant. 2020, 26, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Starkey Lewis, P.J.; Dear, J.; Platt, V.; Simpson, K.J.; Craig, D.G.N.; Antoine, D.J.; French, N.S.; Dhaun, N.; Webb, D.J.; Costello, E.M.; et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 2011, 54, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Rooge, S.B.; Varshney, A.; Vasudevan, M.; Bhardwaj, A.; Venugopal, S.K.; Trehanpati, N.; Kumar, M.; Geffers, R.; Kumar, V.; et al. Global microRNA expression profiling in the liver biopsies of hepatitis B virus–infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatology 2018, 67, 1695–1709. [Google Scholar] [CrossRef] [Green Version]
- Tavabie, O.D.; Karvellas, C.J.; Salehi, S.; Speiser, J.L.; Rose, C.F.; Menon, K.; Prachalias, A.; Heneghan, M.A.; Agarwal, K.; Lee, W.M.; et al. A novel microRNA-based prognostic model outperforms standard prognostic models in patients with acetaminophen-induced acute liver failure. J. Hepatol. 2021, 75, 424–434. [Google Scholar] [CrossRef]
- Ward, J.; Kanchagar, C.; Veksler-Lublinsky, I.; Lee, R.C.; McGill, M.R.; Jaeschke, H.; Curry, S.C.; Ambros, V.R. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc. Natl. Acad. Sci. USA 2014, 111, 12169–12174. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.; Peng, S.F.; Fu, L.; Fu, X.Y.; Wu, D.X.; Liu, B.J.; Tan, D.M.; Ouyang, Y. Serum levels of miRNA in patients with hepatitis B virus-associated acute-on-chronic liver failure. Hepatobiliary Pancreat. Dis. Int. 2018, 17, 126–132. [Google Scholar] [CrossRef]
- Vliegenthart, A.D.B.; Shaffer, J.M.; Clarke, J.I.; Peeters, L.E.J.; Caporali, A.; Bateman, D.N.; Wood, D.M.; Dargan, P.I.; Craig, D.G.; Moore, J.K.; et al. Comprehensive microRNA profiling in acetaminophen toxicity identifies novel circulating biomarkers for human liver and kidney injury. Sci. Rep. 2015, 5, 15501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Salminen, W.F.; Shi, Q.; Greenhaw, J.; Gill, P.S.; Bhattacharyya, S.; Beger, R.D.; Mendrick, D.L.; Mattes, W.B.; James, L.P. Potential of extracellular microRNAs as biomarkers of acetaminophen toxicity in children. Toxicol. Appl. Pharmacol. 2015, 284, 180–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Zhang, X.; Dai, Z.; Feng, Y.; Li, Q.; Zhang, J.H.; Liu, X.; Chen, Y.; Feng, H. P2X7 Receptor Suppression Preserves Blood-Brain Barrier through Inhibiting RhoA Activation after Experimental Intracerebral Hemorrhage in Rats. Sci. Rep. 2016, 6, 23286. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Wu, L.; Gill, P.; Tolleson, W.H.; Chen, S.; Sun, J.; Knox, B.; Jin, Y.; Xiao, W.; Hong, H.; et al. Multiple microRNAs function as self-protective modules in acetaminophen-induced hepatotoxicity in humans. Arch. Toxicol. 2018, 92, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Stravitz, R.T.; Lee, W.M. Acute liver failure. Lancet 2019, 394, 869–881. [Google Scholar] [CrossRef]
- Rovegno, M.; Vera, M.; Ruiz, A.; Benítez, C. Current concepts in acute liver failure. Ann. Hepatol. 2019, 18, 543–552. [Google Scholar] [CrossRef]
- Jeong, S.H.; Lee, H.S. Hepatitis a: Clinical manifestations and management. Intervirology 2010, 53, 15–19. [Google Scholar] [CrossRef]
- Kwo, P.Y.; Cohen, S.M.; Lim, J.K. ACG Clinical Guideline: Evaluation of Abnormal Liver Chemistries. Am. J. Gastroenterol. 2017, 112, 18–35. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yuan, Q.; Balakrishnan, A.; Bantel, H.; Klusmann, J.H.; Manns, M.P.; Ott, M.; Cantz, T.; Sharma, A.D. MicroRNA-125b-5p mimic inhibits acute liver failure. Nat. Commun. 2016, 7, 11916. [Google Scholar] [CrossRef] [Green Version]
- Bandiera, S.; Pfeffer, S.; Baumert, T.F.; Zeisel, M.B. MiR-122—A key factor and therapeutic target in liver disease. J. Hepatol. 2015, 62, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Jopling, C.L. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012, 9, 137–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howell, L.S.; Ireland, L.; Park, B.K.; Goldring, C.E. MiR-122 and other microRNAs as potential circulating biomarkers of drug-induced liver injury. Expert Rev. Mol. Diagn. 2018, 18, 47–54. [Google Scholar] [CrossRef]
- Fu, S.; Wu, D.; Jiang, W.; Li, J.; Long, J.; Jia, C.; Zhou, T. Molecular biomarkers in drug-induced liver injury: Challenges and future perspectives. Front. Pharmacol. 2020, 10, 1667. [Google Scholar] [CrossRef] [Green Version]
- Tornavaca, O.; Chia, M.; Dufton, N.; Almagro, L.O.; Conway, D.E.; Randi, A.M.; Schwartz, M.A.; Matter, K.; Balda, M.S. ZO-1 controls endothelial adherens junctions, cell-cell tension, angiogenesis, and barrier formation. J. Cell Biol. 2015, 208, 821–838. [Google Scholar] [CrossRef] [Green Version]
- Martínez, C.; Rodinõ-Janeiro, B.K.; Lobo, B.; Stanifer, M.L.; Klaus, B.; Granzow, M.; González-Castro, A.M.; Salvo-Romero, E.; Alonso-Cotoner, C.; Pigrau, M.; et al. MiR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut 2017, 66, 1597–1610. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.C.; Lin, X.L.; Li, J.; Zhang, T.T.; Wang, H.Y.; Shi, J.W.; Yang, S.; Zhao, W.T.; Xie, R.Y.; Wei, F.; et al. MicroRNA-122 triggers mesenchymal-epithelial transition and suppresses hepatocellular carcinoma cell motility and invasion by targeting RhoA. PLoS ONE 2014, 9, e101330. [Google Scholar] [CrossRef] [PubMed]
- Osmak, G.; Kiselev, I.; Baulina, N.; Favorova, O. From mirna target gene network to mirna function: mir-375 might regulate apoptosis and actin dynamics in the heart muscle via rho-gtpases-dependent pathways. Int. J. Mol. Sci. 2020, 21, 9670. [Google Scholar] [CrossRef]
- Zhang, J.; Ying, Z.Z.; Tang, Z.L.; Long, L.Q.; Li, K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 2012, 287, 21093–21101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jingushi, K.; Kashiwagi, Y.; Ueda, Y.; Kitae, K.; Hase, H.; Nakata, W.; Fujita, K.; Uemura, M.; Nonomura, N.; Tsujikawa, K. High miR-122 expression promotes malignant phenotypes in ccRCC by targeting occludin. Int. J. Oncol. 2017, 51, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Xu, H.W.; Lu, X.R.; Xu, Q.F.; Tao, M.H.; Dai, Y.M. Overexpression of miR-122 Impairs Intestinal Barrier Function and Aggravates Acute Pancreatitis by Downregulating Occludin Expression. Biochem. Genet. 2021. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, M.; Liu, X.; Fei, L.; Shen, J.; Chen, D. miR-122-5p regulates the tight junction of the blood-testis barrier of mice via occludin: miR-122-5p can regulate the tight junction. Basic Clin. Androl. 2021, 31, 7. [Google Scholar] [CrossRef]
- Shojaie, L.; Iorga, A.; Dara, L. Cell death in liver diseases: A review. Int. J. Mol. Sci. 2020, 21, 9682. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Huo, Y.; Yin, S.; Hu, H. Mechanisms of acetaminophen-induced liver injury and its implications for therapeutic interventions. Redox Biol. 2018, 17, 274–283. [Google Scholar] [CrossRef]
- Iwarson, S.; Lundin, P.; Hermodsson, S. Liver morphology in acute viral hepatitis related to the hepatitis B antigen. J. Clin. Pathol. 1972, 25, 850–855. [Google Scholar] [CrossRef] [Green Version]
- Sanphui, P.; Biswas, S.C. FoxO3a is activated and executes neuron death via Bim in response to β-amyloid. Cell Death Dis. 2013, 4, e625. [Google Scholar] [CrossRef] [PubMed]
- Winkler, E.A.; Bell, R.D.; Zlokovic, B.V. Central nervous system pericytes in health and disease. Nat. Neurosci. 2011, 14, 1398–1405. [Google Scholar] [CrossRef] [Green Version]
- Malek, N.; Mrówczyńska, E.; Michrowska, A.; Mazurkiewicz, E.; Pavlyk, I.; Mazur, A.J. Knockout of ACTB and ACTG1 with CRISPR/Cas9(D10A) technique shows that non-muscle β and γ actin are not equal in relation to human melanoma cells’ motility and focal adhesion formation. Int. J. Mol. Sci. 2020, 21, 2746. [Google Scholar] [CrossRef] [Green Version]
- Von Bernhardi, R.; Cornejo, F.; Parada, G.E.; Eugenín, J. Role of TGFβ signaling in the pathogenesis of Alzheimer’s disease. Front. Cell. Neurosci. 2015, 9, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Zhao, S.; Liu, B.; Zhang, Q.; Li, Y.; Liu, J.; Shen, Y.; Ding, X.; Lin, J.; Wu, Y.; et al. Perturbations of BMP/TGF-β and VEGF/VEGFR signalling pathways in non-syndromic sporadic brain arteriovenous malformations (BAVM). J. Med. Genet. 2018, 55, 675–684. [Google Scholar] [CrossRef]
- Chung, J.; Marini, S.; Pera, J.; Norrving, B.; Jimenez-Conde, J.; Roquer, J.; Fernandez-Cadenas, I.; Tirschwell, D.L.; Selim, M.; Brown, D.L.; et al. Genome-wide association study of cerebral small vessel disease reveals established and novel loci. Brain 2019, 142, 3176–3189. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, X. The Roles of TGF-β Signaling in Cerebrovascular Diseases. Front. Cell Dev. Biol. 2020, 8, 567682. [Google Scholar] [CrossRef] [PubMed]
- Armulik, A.; Genové, G.; Mäe, M.; Nisancioglu, M.H.; Wallgard, E.; Niaudet, C.; He, L.; Norlin, J.; Lindblom, P.; Strittmatter, K.; et al. Pericytes regulate the blood-brain barrier. Nature 2010, 468, 557–561. [Google Scholar] [CrossRef] [Green Version]
- Bell, R.D.; Winkler, E.A.; Sagare, A.P.; Singh, I.; LaRue, B.; Deane, R.; Zlokovic, B.V. Pericytes Control Key Neurovascular Functions and Neuronal Phenotype in the Adult Brain and during Brain Aging. Neuron 2010, 68, 409–427. [Google Scholar] [CrossRef] [Green Version]
- Siu, E.R.; Wong, E.W.P.; Mruk, D.D.; Sze, K.L.; Porto, C.S.; Cheng, C.Y. An occludin-focal adhesion kinase protein complex at the blood-testis barrier: A study using the cadmium model. Endocrinology 2009, 150, 3336–3344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatovic, S.M.; Dimitrijevic, O.B.; Keep, R.F.; Andjelkovic, A.V. Protein kinase Cα-RhoA cross-talk in CCL2-induced alterations in brain endothelial permeability. J. Biol. Chem. 2006, 281, 8379–8388. [Google Scholar] [CrossRef] [Green Version]
- Mushimiyimana, I.; Tomas Bosch, V.; Niskanen, H.; Downes, N.L.; Moreau, P.R.; Hartigan, K.; Ylä-Herttuala, S.; Laham-Karam, N.; Kaikkonen, M.U. Genomic Landscapes of Noncoding RNAs Regulating VEGFA and VEGFC Expression in Endothelial Cells. Mol. Cell. Biol. 2021, 41, MCB-00594. [Google Scholar] [CrossRef]
- Gui, P.; Wu, X.; Ling, S.; Stotz, S.C.; Winkfein, R.J.; Wilson, E.; Davis, G.E.; Braun, A.P.; Zamponi, G.W.; Davis, M.J. Integrin receptor activation triggers converging regulation of Cav1.2 calcium channels by c-Src and protein kinase A pathways. J. Biol. Chem. 2006, 281, 14015–14025. [Google Scholar] [CrossRef] [Green Version]
- Morawski, M.; Filippov, M.; Tzinia, A.; Tsilibary, E.; Vargova, L. ECM in Brain Aging and Dementia, 1st ed.; Elsevier B.V.: Amsterdam, The Netherlands, 2014; Volume 214, ISBN 9780444634863. [Google Scholar]
- Jhunjhunwala, S.; Jiang, Z.; Stawiski, E.W.; Gnad, F.; Liu, J.; Mayba, O.; Du, P.; Diao, J.; Johnson, S.; Wong, K.F.; et al. Diverse modes of genomic alteration in hepatocellular carcinoma. Genome Biol. 2014, 15, 436. [Google Scholar] [CrossRef] [Green Version]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, 1–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonneh-Barkay, D.; Wiley, C.A. Brain extracellular matrix in neurodegeneration. Brain Pathol. 2009, 19, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Van Horssen, J.; Dijkstra, C.D.; De Vries, H.E. The extracellular matrix in multiple sclerosis pathology. J. Neurochem. 2007, 103, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Bamaga, A.; Al-Lozi, M.; Weihl, C. A novel mutation in AGRN gene causing congenital myasthenic syndrome with distal myopathy. Neuromuscul. Disord. 2017, 27, S222. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Li, M.; Yin, L.; Fu, G.; Liu, Z. Role of thrombospondin–1 and thrombospondin–2 in cardiovascular diseases (Review). Int. J. Mol. Med. 2020, 45, 1275–1293. [Google Scholar] [CrossRef] [Green Version]
- Hammond, J.W.; Cai, D.; Verhey, K.J. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 2008, 20, 71–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto-Komatsu, A.; Hirase, T.; Asaka, M.; Node, K. Angiotensin II induces microtubule reorganization mediated by a deacetylase SIRT2 in endothelial cells. Hypertens. Res. 2011, 34, 949–956. [Google Scholar] [CrossRef] [Green Version]
- Pierce, K.L.; Premont, R.T.; Lefkowitz, R.J.; Hughes, T.H. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 2002, 3, 639–650. [Google Scholar] [CrossRef]
- Rai, V.; Nath, K.; Saraswat, V.A.; Purwar, A.; Rathore, R.K.S.; Gupta, R.K. Measurement of cytotoxic and interstitial components of cerebral edema in acute hepatic failure by diffusion tensor imaging. J. Magn. Reson. Imaging 2008, 28, 334–341. [Google Scholar] [CrossRef]
- Kale, R.A.; Gupta, R.K.; Saraswat, V.A.; Hasan, K.M.; Trivedi, R.; Mishra, A.M.; Ranjan, P.; Pandey, C.M.; Narayana, P.A. Demonstration of interstitial cerebral edema with diffusion tensor MR imaging in type C hepatic encephalopathy. Hepatology 2006, 43, 698–706. [Google Scholar] [CrossRef]
- Cui, W.; Sun, C.; Liu, P. Alterations of Blood-Brain Barrier and Associated Factors in Acute Liver Failure. Gastroenterol. Res. Pract. 2013, 2013, 841707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, L.A.; Lee, K.C.L.; Jimenez, C.P.; Alibhai, H.; Chang, Y.; Leckie, P.J.; Mookerjee, R.P.; Davies, N.A.; Andreola, F.; Jalan, R. Circulating microRNAs reveal time course of organ injury in a porcine model of acetaminophen-induced acute liver failure. PLoS ONE 2015, 10, e0128076. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407. [Google Scholar] [CrossRef] [Green Version]
- Cogswell, J.; Ward, J.; Taylor, I.; Waters, M.; Shi, Y.; Cannon, B.; Kelnar, K.; Kemppainen, J.; Brown, D.; Chen, C.; et al. Identification of miRNA Changes in Alzheimer’s.pdf. J. Alzheimer’s Dis. 2008, 14, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J.; Alexandrov, P.N.; Zhao, Y.; Hill, J.M.; Bhattacharjee, S. Spreading of Alzheimer’s disease inflammatory signaling through soluble micro-RNA. Neuroreport 2012, 23, 621–626. [Google Scholar] [CrossRef] [Green Version]
- Sethi, P.; Lukiw, W.J. Micro-RNA abundance and stability in human brain: Specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci. Lett. 2009, 459, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. Micro RNA -125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Cause | Causative Factor | Therapy | Frequencyin Adults * |
|---|---|---|---|
| Intoxication | Acetaminophen | N-acetyl cysteine | 58% |
| Isoniazid | Hydration | ||
| Other drug-driven liver damage | |||
| Mushrooms poisoning | Penicillin | ||
| Viral | Hepatitis B/C/A/E/ ** | Antiviral therapy | 10% |
| Metabolic | Wilson disease | Copper chelation | 7% |
| Autoimmune hepatitis | Steroids | ||
| Acute fatty liver of pregnancy | Delivery of the fetus | ||
| Other | Budd-Chiari syndrome | Surgery | 5% |
| Ischemic/sepsis shock | Hemodynamic stabilization | ||
| Heat stroke | Hydration | ||
| Unknown | Undetermined etiology | 14% |
| Ref. | Year | Disease | Specimen | Region | N ** | Analysis Method | Changed miRNAs | Notes |
|---|---|---|---|---|---|---|---|---|
| [48] | 2013 | Acetaminophen overdose | Plasma | UK | 129 | RT-qPCR | 1 | |
| [49] | 2017 | ALF * | Plasma/liver | USA | 9/4 | RT-qPCR | 1 | |
| [50] | 2015 | HBV | Liver | Italy | 4/10 | RT-qPCR | 17 | |
| [51] | 2014 | ALF * | Serum | USA | 35/12 | RT-qPCR | 1 | |
| [52] | 2014 | ALF * | Serum | Germany | 63/15 | RT-qPCR | 3 | |
| [53] | 2015 | Acetaminophen overdose | Serum | Netherlands | 6/6 | RT-qPCR, HiSeq 2000 | 3 | |
| [54] | 2017 | Acetaminophen/HBV | Serum | USA | 16/22 | Illumina HiSeq 2000 | 132 | |
| [55] | 2020 | Acetaminophen overdose | Serum | Netherlands | 1 | RT-qPCR, NGS | 57 | Case Study |
| [56] | 2019 | HBV | Liver | China | 4/10 | Microarray | 38 | |
| [57] | 2017 | ALF * | Serum/liver | USA | 39/5 | RT-qPCR | 1 | |
| [38] | 2017 | Drug induced ALF | Serum | USA | 78/40 | GeneChip® 3.0 miRNA microarrays | 1 | |
| [58] | 2020 | Acetaminophen overdose | Serum | UK | 18/undefined | RT-qPCR | 7 | HE grade diagnosed |
| [59] | 2011 | Acetaminophen overdose | Serum | UK | 53/25 | RT-qPCR | 2 | Controlled Clinical Trial/HE grade |
| [60] | 2018 | HBV | Liver | India | 30/6 | Microarray | 1 | |
| [61] | 2021 | Acetaminophen overdose | Serum | USA | 194 | RT-qPCR | 7 | HE grade diagnosed |
| [62] | 2014 | Acetaminophen overdose | Serum/plasma | USA | 42/12 | RT-qPCR | 3 | |
| [63] | 2018 | HBV | Serum | China | 55 | RT-qPCR | 3 | |
| [64] | 2015 | Acetaminophen overdose | Serum | UK | 68 | RT-qPCR (miScript System) | 6 | |
| [65] | 2015 | Acetaminophen overdose | Serum/urine | USA | 8/10 | Whole genome PCR array | 12 | |
| [66] | 2016 | Acetaminophen overdose | Serum/liver | Germany | 9/4 | RT-qPCR | 1 | same cohort as Chowdhary et al., 2017 [49] |
| [67] | 2018 | Acetaminophen overdose | Serum | USA | 8/10 | RT-qPCR | 4 | same cohort as Yang X et al., 2015 [65] |
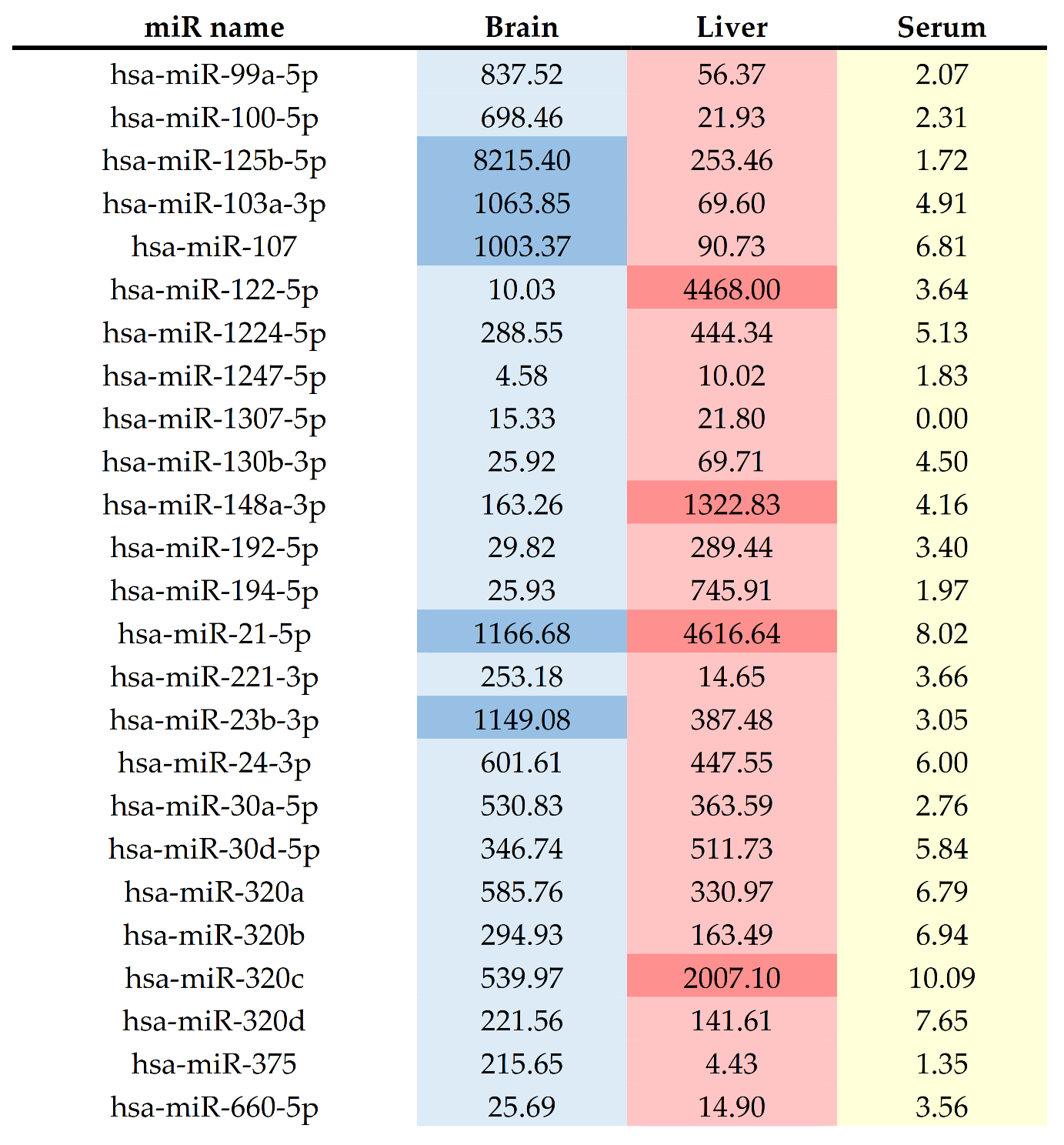
| Family | MiRNA | Localization | Cluster Members | References |
|---|---|---|---|---|
| miR-10 | miR-99a-5p | chr21 | [54,55] | |
| miR-10 | miR-100-5p | chr11 | [54,55] | |
| miR-10 | miR-125b-5p | chr11 | [54,55,65,72] | |
| miR-103 | miR-103a-3p | chr20 | miR-103b-2 | [54,55] |
| miR-103 | miR-107 | chr10 | [54,55] | |
| miR0122 | miR-122-5p | chr18 | miR-122b | [49,53,54,65] |
| miR-1224 | miR-1224-5p | chr3 | [38,48,51,52,59,61,62,63,64] | |
| miR-1247 | miR-1247-5p | chr14 | [54,55] | |
| miR-1307 | miR-1307-5p | chr10 | [54,55] | |
| miR-130 | miR-130b-3p | chr22 | miR-301b | [54,55] |
| miR-148 | miR-148a-3p | chr7 | [54,55] | |
| miRR-192 | miR-192-5p | chr11 | miR-194-2; miR-6750; miR-6749 | [54,55,64] |
| miR-194 | miR-194-5p | chr1 | miR-215 | [54,55] |
| miR-21 | miR-21-5p | chr17 | [54,61,62] | |
| miR-221 | miR-221-3p | chrX | [52,54] | |
| miR-23 | miR-23b-3p | chr9 | miR-27b; miR-3074; miR-24-1 | [54,55] |
| miR-24 | miR-24-3p | chr9 | miR-27b; miR-3074; miR-23b | [54,55] |
| miR-30 | miR-30a-5p | chr6 | [54,61] | |
| miR-30 | miR-30d-5p | chr8 | [55,65] | |
| miR-320 | miR-320a | chr8 | [54,64,67] | |
| miR-320 | miR-320b | chr1 | [54,67] | |
| miR-320 | miR-320c | chr18 | [54,67] | |
| miR-320 | miR-320d | chr13 | [54,67] | |
| miR-375 | miR-375 | chr2 | [55,65] | |
| miR-188 | miR-660-5p | chrX | [54,55] |
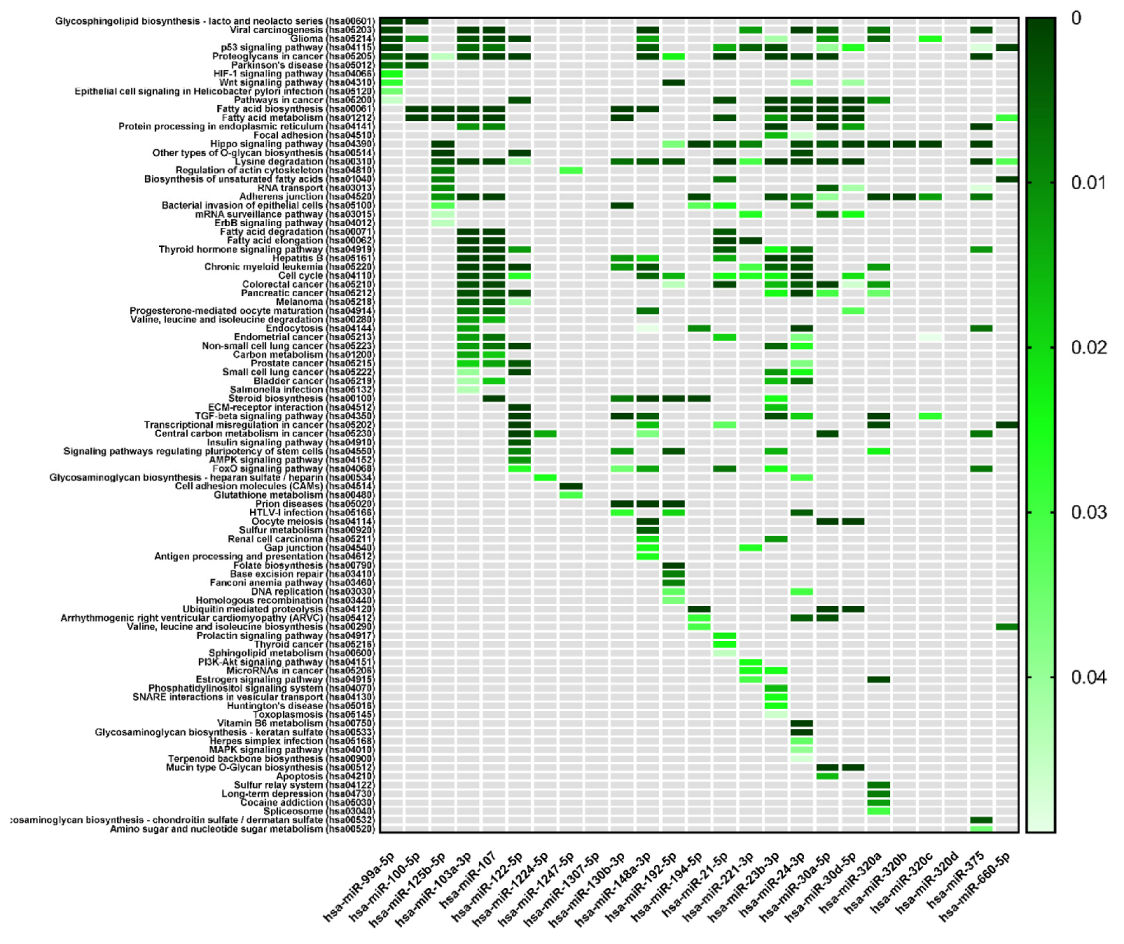
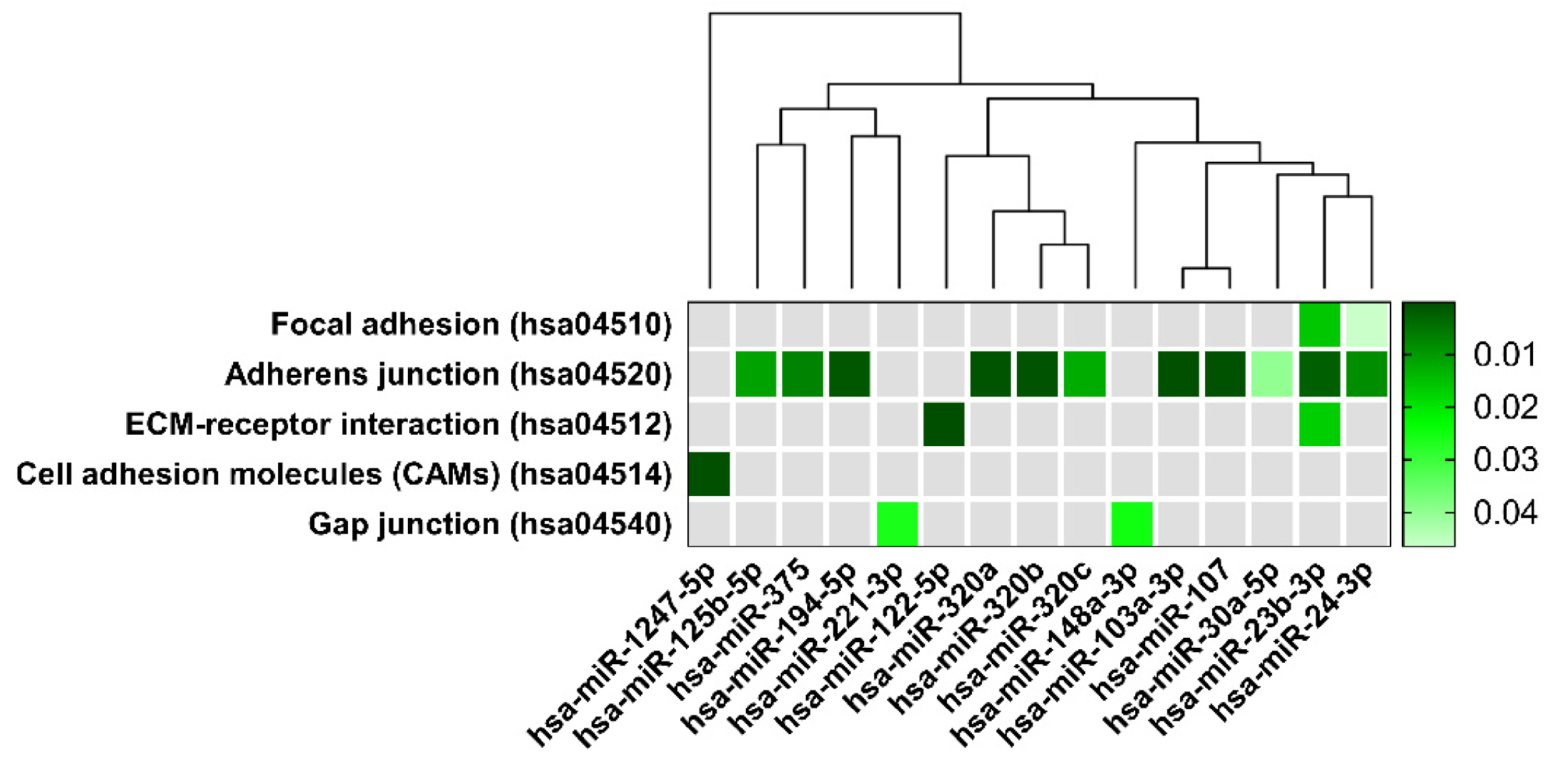
| KEGG Pathways | miRNA Name | Gene Name | p-Value |
|---|---|---|---|
| Focal adhesion | miR-125b-2-3p | BRAF, GSK3B, ITGB1, LAMB1, THBS1, PAK2, VCL, RHOA, PAK3, FYN, PPP1R12A, JUN, DIAPH1, VEGFC, PTEN, BIRC2 | 4.7 × 10−2 |
| miR-23b-3p | TLN2, PRKCA, ACTN2, MYLK4, MET, ITGB1, FLNC, CRKL, THBS1, MYL12B, PAK2, TNXB, PP1CC, BCL2, EGFR, FYN, COL6A1, AKT2, ARHGAP35, PPP1R12A, MAPK9, PIK3R3, CCND1, CTNNB1, COL4A2, PAK6, PRKCB, LAMC1, PDK‘, LAMC2, ITGA6, VEGFA, PTEN, MAPK1, KDR, MYL12A, XIAP, COL4A1, PPP1CB | 1.5 × 10−2 | |
| miR-24-3p | ACTB, GSK3B, ITGB1, ITGB8, FLNC, LAMA5, PIK3CB, CAV1, PXN, RAF1, BCAR1, EGFR, MYLK2, CAV2, ITGB5, ARHGAP35, PTK2, ITGA11, JUN, PIK3R3, CTNNB1, DIAPH1, FLNB, FLT1, FLNA, RAC1, LAMC1, VASP, FN1, TNC, BIRC3, PDPK1, VEGFA, TLN1, KDR, LAMA4 | 4.6 × 10−2 | |
| Adherens junction | miR-125b-5p | ERBB2, ACTG1, IQGAP1, MLLT4, CTNNB1, CTNNA1, WASF2, INSR, FGFR1 | 10−2 |
| miR-103a-3p | ACTB, CTNND1, ACTG1, IQGAP1, IGF1R, MLLT4, SMAD4, CTNNB1, CTNNA1, ACTN1, WASF2, CSNK2A1, FARP2, PTPRJ, CDC42, PTPRB, MAP3K7, CREBBP, TGFBR2, PVRL1 | 1.2 × 10−3 | |
| miR-107 | ACTB1, CTNND1, ACTG1, IQGAP1, IGF1R, PTPRF, MLLT4, DMAD4, CTNNB1, CTNNA1, ACTN1, WASF2, CSNK2A1, FARP2, PTPRJ, MAPK3, CDC42, PVRL3, PTPRB, MAP3K7, CREBBP, TGFBR2, PVRL1 | 3 × 10−4 | |
| miR-194-5p | TGFBR1, WASL, ACTG1, LMO7, IGF1R, PVRL4, RAC1 | 9.6 × 10−4 | |
| miR-23b-3p | ACTN2, CSNK2A2, MET, CTNND1, LMO7, IQGAP1, SMAD3, EGFR, TJP1, FYN, SMAD4, CTNNB1, WASF2, CSNK2A1, FARP2, PTPRJ, YES1, MAPK1, TGFBR2 | 1.9 × 10−3 | |
| miR-24-3p | ACTB, CSNK2A2, SMAD2, PVRL2, SMAD3, EGFR, PTPRF, NLK, CTNNB1, CTNNA1, WASF2, RAC1, EP300, YES1, CREBBP, TGFBR2, PVRL1 | 8.4 × 10−3 | |
| miR-30a-5p | CSNK2A2, MET, WASL, CTNND1, SMAD2, PTPN1, IGFR1, EGFR, NLK, RAC1, PVRL3, MAPK1, MAP3K7, TGFBR2 | 3.9 × 10−2 | |
| miR-320a | SMAD2, SMAD3, IGF1R, VCL, TJP1, CDH1, CTNNB1, RAC1, INSR, SSX2IP, MAPK1, TGFBR2 | 5.2 × 10−4 | |
| miR-320b | SMAD3, TJP1, CDH1, CTNNB1, RAC1, INSR, SSX2IP, MAPK1 | 4 × 10−4 | |
| miR-320c | SMAD3, TJP1, CDH1, CTNNB1, INSR, SSX2IP, MAPK1 | 1.2 × 10−3 | |
| miR-375 | ERBB2, IQGAP1, PTPN1, IGF1R, PTPRF, RHOA, CTNNA1, WASF2, CDC42, PARD3 | 6.9 × 10−4 | |
| ECM-receptor interaction | miR-122-5p | ITGB1, ITGB8, LAMB1, LAMA5, COL27A1, AGRN, LAMC3, COL4A2, COL5A1, COL4A3, CD44 | 8.3 × 10−8 |
| miR-23b-3p | ITGB1, THBS1, TNXB, COL6A1, COL4A2, LAMC1, LAMC2, ITGA6, HMMR, COL4A1 | 1.6 × 10−2 | |
| Cell adhesion molecules | miR-1247-5p | CDH2 | 9.6 × 10−7 |
| Gap junction | miR-148a-3p | ADCY1, ADCY8, GNAS, MAP2K5, RAF1, TUBB, TUBB6, CDK1, KRAS, TUBA8, GNA11, TUBB2B, SOS1, GNAI2, HTR2C, TUBB4B, MAPK1, GRB2, GNAI1 | 2.4 × 10−2 |
| miR-221-3p | RAF1, CDK1, PDGFD, GNAI2, GJA1 | 2.6 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orzeł-Gajowik, K.; Milewski, K.; Zielińska, M. Insight into microRNAs-Mediated Communication between Liver and Brain: A Possible Approach for Understanding Acute Liver Failure? Int. J. Mol. Sci. 2022, 23, 224. https://doi.org/10.3390/ijms23010224
Orzeł-Gajowik K, Milewski K, Zielińska M. Insight into microRNAs-Mediated Communication between Liver and Brain: A Possible Approach for Understanding Acute Liver Failure? International Journal of Molecular Sciences. 2022; 23(1):224. https://doi.org/10.3390/ijms23010224
Chicago/Turabian StyleOrzeł-Gajowik, Karolina, Krzysztof Milewski, and Magdalena Zielińska. 2022. "Insight into microRNAs-Mediated Communication between Liver and Brain: A Possible Approach for Understanding Acute Liver Failure?" International Journal of Molecular Sciences 23, no. 1: 224. https://doi.org/10.3390/ijms23010224
APA StyleOrzeł-Gajowik, K., Milewski, K., & Zielińska, M. (2022). Insight into microRNAs-Mediated Communication between Liver and Brain: A Possible Approach for Understanding Acute Liver Failure? International Journal of Molecular Sciences, 23(1), 224. https://doi.org/10.3390/ijms23010224






