Granzymes—Their Role in Colorectal Cancer
Abstract
1. Colorectal Cancer—An Urgent Need for Novel Biomarkers
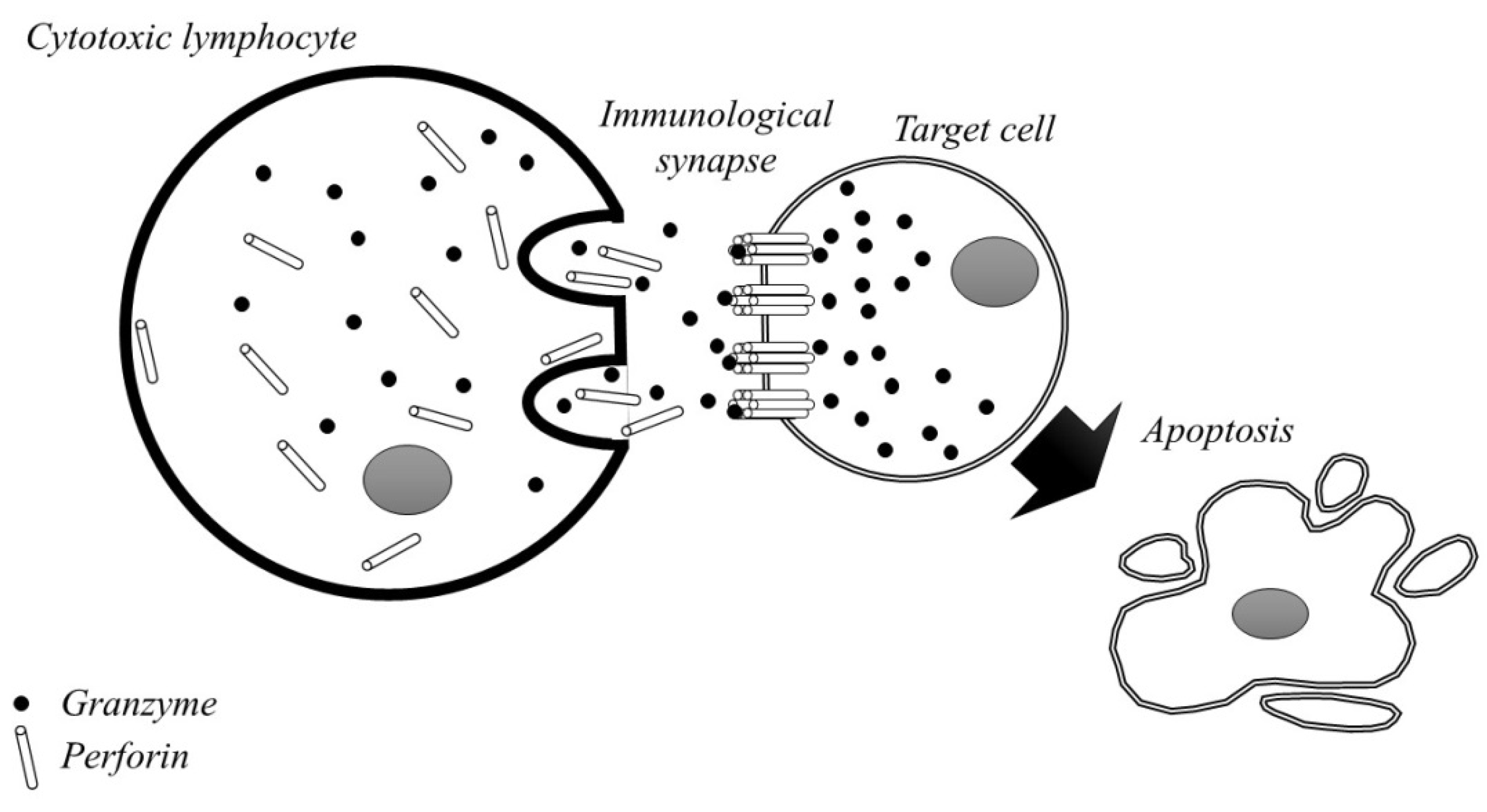
2. Granzymes—Structure, Function and Apoptosis
3. GZMs in Inflammation
4. Inflammation and Carcinogenesis
5. GZMs in CRC
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stoffel, E.M.; Murphy, C.C. Epidemiology and Mechanisms of the Increasing Incidence of Colon and Rectal Cancers in Young Adults. Gastroenterology 2020, 158, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Han, Z.; Li, X.; Huang, A.; Shi, J.; Gu, J. Epidemiology and risk factors of colorectal cancer in China. Chin. J. Cancer Res. 2020, 32, 729–741. [Google Scholar] [CrossRef]
- Yu, H.; Hemminki, K. Genetic epidemiology of colorectal cancer and associated cancers. Mutagenesis 2019, 35, 207–219. [Google Scholar] [CrossRef]
- Onyoh, E.F.; Hsu, W.-F.; Chang, L.-C.; Lee, Y.-C.; Wu, M.-S.; Chiu, H.-M. The Rise of Colorectal Cancer in Asia: Epidemiology, Screening, and Management. Curr. Gastroenterol. Rep. 2019, 21, 36. [Google Scholar] [CrossRef]
- Weitz, J.; Koch, M.; Debus, J.; Höhler, T.; Galle, P.R.; Büchler, M.W. Colorectal cancer. Lancet 2005, 365, 153–165. [Google Scholar] [CrossRef]
- Dariya, B.; Aliya, S.; Merchant, N.; Alam, A.; Nagaraju, G.P. Colorectal Cancer Biology, Diagnosis, and Therapeutic Approaches. Crit. Rev. Oncog. 2020, 25, 71–94. [Google Scholar] [CrossRef]
- Carethers, J.M.; Jung, B.H. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 2015, 149, 1177–1190.e3. [Google Scholar] [CrossRef]
- Edin, S.; Wikberg, M.L.; Dahlin, A.M.; Rutegård, J.; Öberg, Å.; Oldenborg, P.A.; Palmqvist, R. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS ONE 2012, 7, e47045. [Google Scholar] [CrossRef]
- Nguyen, L.H.; Goel, A.; Chung, D.C. Pathways of Colorectal Carcinogenesis. Gastroenterology 2020, 158, 291–302. [Google Scholar] [CrossRef]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Guo, Y.; Bao, Y.; Ma, M.; Yang, W. Identification of Key Candidate Genes and Pathways in Colorectal Cancer by Integrated Bioinformatical Analysis. Int. J. Mol. Sci. 2017, 18, 722. [Google Scholar] [CrossRef] [PubMed]
- Sakai, E.; Fukuyo, M.; Ohata, K.; Matsusaka, K.; Doi, N.; Mano, Y.; Takane, K.; Abe, H.; Yagi, K.; Matsuhashi, N.; et al. Genetic and epigenetic aberrations occurring in colorectal tumors associated with serrated pathway. Int. J. Cancer 2015, 138, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, G.L.; Bottarelli, L.; Azzoni, C.; De Angelis, N.; Leandro, G.; Di Mario, F.; Gaiani, F.; Negri, F. Microsatellite instability in colorectal cancer. Acta Biomed. 2018, 89, 97–101. [Google Scholar] [PubMed]
- Yang, G.; Zheng, R.-Y.; Jin, Z.-S. Correlations between microsatellite instability and the biological behaviour of tumours. J. Cancer Res. Clin. Oncol. 2019, 145, 2891–2899. [Google Scholar] [CrossRef]
- Jung, G.; Hernández-Illán, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef]
- De Palma, F.D.E.; D’Argenio, V.; Pol, J.; Kroemer, G.; Maiuri, M.C.; Salvatore, F. The Molecular Hallmarks of the Serrated Pathway in Colorectal Cancer. Cancers 2019, 11, 1017. [Google Scholar] [CrossRef]
- Loke, Y.L.; Chew, M.T.; Ngeow, Y.F.; Lim, W.W.D.; Peh, S.C. Colon Carcinogenesis: The Interplay Between Diet and Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 603086. [Google Scholar] [CrossRef]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Bray, C.; Bell, L.N.; Liang, H.; Collins, D.; Yale, S.H. Colorectal Cancer Screening. WMJ 2017, 116, 27–33. [Google Scholar]
- Schreuders, E.H.; Ruco, A.; Rabeneck, L.; Schoen, R.E.; Sung, J.J.Y.; Young, G.; Kuipers, E.J. Colorectal cancer screening: A global overview of existing programmes. Gut 2015, 64, 1637–1649. [Google Scholar] [CrossRef]
- Calanzani, N.; Chang, A.; Van Melle, M.; Pannebakker, M.M.; Funston, G.; Walter, F. Recognising Colorectal Cancer in Primary Care. Adv. Ther. 2021, 38, 2732–2746. [Google Scholar] [CrossRef]
- Roncucci, L.; Mariani, F. Prevention of colorectal cancer: How many tools do we have in our basket? Eur. J. Intern. Med. 2015, 26, 752–756. [Google Scholar] [CrossRef]
- Nikolouzakis, T.K.; Vassilopoulou, L.; Fragkiadaki, P.; Sapsakos, T.M.; Papadakis, G.Z.; Spandidos, D.; Tsatsakis, A.; Tsiaoussis, J. Improving diagnosis, prognosis and prediction by using biomarkers in CRC patients (Review). Oncol. Rep. 2018, 39, 2455–2472. [Google Scholar] [CrossRef]
- Díaz-Tasende, J. Colorectal cancer screening and survival. Rev. Esp. Enferm. Dig. 2018, 110, 681–683. [Google Scholar] [CrossRef]
- Maida, M.; Macaluso, F.S.; Ianiro, G.; Mangiola, F.; Sinagra, E.; Hold, G.; Maida, C.; Cammarota, G.; Gasbarrini, A.; Scarpulla, G. Screening of colorectal cancer: Present and future. Expert Rev. Anticancer Ther. 2017, 17, 1131–1146. [Google Scholar] [CrossRef]
- The Lancet Gastroenterology Hepatology. Colorectal cancer screening: Is earlier better? Lancet Gastroenterol. Hepatol. 2018, 3, 519. [Google Scholar] [CrossRef]
- Simon, K. Colorectal cancer development and advances in screening. Clin. Interv. Aging 2016, 11, 967–976. [Google Scholar]
- Burt, R.W.; Cannon, J.A.; David, D.S.; Early, D.S.; Ford, J.M.; Giardiello, F.M.; Halverson, A.L.; Hamilton, S.R.; Hampel, H.; Ismail, M.K.; et al. Colorectal cancer screening. J. Natl. Compr. Canc. Netw. 2013, 11, 1538–1575. [Google Scholar] [CrossRef]
- Schönherr, R. Clinical relevance of ion channels for diagnosis and therapy of cancer. J. Membr. Biol. 2005, 205, 175–184. [Google Scholar] [CrossRef]
- Stock, C. How Dysregulated Ion Channels and Transporters Take a Hand in Esophageal, Liver, and Colorectal Cancer. Rev. Physiol. Biochem. Pharmacol. 2021, 181, 129–222. [Google Scholar]
- Zhang, M.; Li, T.; Zhu, J.; Tuo, B.; Liu, X. Physiological and pathophysiological role of ion channels and transporters in the colorectum and colorectal cancer. J. Cell Mol. Med. 2020, 24, 9486–9494. [Google Scholar] [CrossRef]
- Anderson, K.J.; Cormier, R.T.; Scott, P.M. Role of ion channels in gastrointestinal cancer. World J. Gastroenterol. 2019, 25, 5732–5772. [Google Scholar] [CrossRef]
- Bots, M.; Medema, J.P. Granzymes at a glance. J. Cell Sci. 2006, 119 Pt 24, 5011–5014. [Google Scholar] [CrossRef]
- Bovenschen, N.; Kummer, J.A. Orphan granzymes find a home. Immunol. Rev. 2010, 235, 117–127. [Google Scholar] [CrossRef]
- Martínez Cuesta, L.; Pérez, S.E. Perforin and granzymes in neurological infections: From humans to cattle. Comp. Immunol. Microbiol. Infect Dis. 2021, 75, 101610. [Google Scholar] [CrossRef]
- Joeckel, L.T.; Bird, P.I. Are all granzymes cytotoxic in vivo? Biol. Chem. 2014, 395, 181–202. [Google Scholar] [CrossRef]
- Buzza, M.S.; Bird, P.I. Extracellular granzymes: Current perspectives. Biol. Chem. 2006, 387, 827–837. [Google Scholar] [CrossRef]
- Hiroyasu, S.; Hiroyasu, A.; Granville, D.J.; Tsuruta, D. Pathological functions of granzyme B in inflammatory skin diseases. J. Dermatol. Sci. 2021, 104, 76–82. [Google Scholar] [CrossRef]
- Thomas, H.E.; Trapani, J.A.; Kay, T.W.H. The role of perforin and granzymes in diabetes. Cell Death Differ. 2009, 17, 577–585. [Google Scholar] [CrossRef]
- Kamata, Y.; Kimura, U.; Matsuda, H.; Tengara, S.; Kamo, A.; Umehara, Y.; Iizumi, K.; Kawasaki, H.; Suga, Y.; Ogawa, H.; et al. Relationships among plasma granzyme B level, pruritus and dermatitis in patients with atopic dermatitis. J. Dermatol. Sci. 2016, 84, 266–271. [Google Scholar] [CrossRef]
- Garzón-Tituaña, M.; Arias, M.A.; Sierra-Monzón, J.L.; Morte-Romea, E.; Santiago, L.; Ramirez-Labrada, A.G.; Martinez-Lostao, L.; Paño-Pardo, J.R.; Galvez, E.M.; Pardo, J. The Multifaceted Function of Granzymes in Sepsis: Some Facts and a Lot to Discover. Front. Immunol. 2020, 11, 1054. [Google Scholar] [CrossRef]
- Turner, C.; Lim, D.; Granville, D.J. Granzyme B in skin inflammation and disease. Matrix Biol. 2019, 75–76, 126–140. [Google Scholar] [CrossRef]
- Van Daalen, K.R.; Reijneveld, J.F.; Bovenschen, N. Modulation of Inflammation by Extracellular Granzyme A. Front. Immunol. 2020, 11, 931. [Google Scholar] [CrossRef]
- Lavergne, M.; Hernández-Castañeda, M.A.; Mantel, P.-Y.; Martinvalet, D.; Walch, M. Oxidative and Non-Oxidative Antimicrobial Activities of the Granzymes. Front. Immunol. 2021, 12, 750512. [Google Scholar] [CrossRef]
- Romero, V.; Andrade, F. Non-apoptotic functions of granzymes. Tissue Antigens 2008, 71, 409–416. [Google Scholar] [CrossRef]
- Matsubara, J.A.; Tian, Y.; Cui, J.Z.; Zeglinski, M.R.; Hiroyasu, S.; Turner, C.T.; Granville, D.J. Retinal Distribution and Extracellular Activity of Granzyme B: A Serine Protease That Degrades Retinal Pigment Epithelial Tight Junctions and Extracellular Matrix Proteins. Front. Immunol. 2020, 11, 574. [Google Scholar] [CrossRef]
- Andrade, F. Non-cytotoxic antiviral activities of granzymes in the context of the immune antiviral state. Immunol. Rev. 2010, 235, 128–146. [Google Scholar] [CrossRef]
- Kam, C.M.; Hudig, D.; Powers, J.C. Granzymes (lymphocyte serine proteases): Characterization with natural and synthetic substrates and inhibitors. Biochim. Biophys. Acta. 2000, 1477, 307–323. [Google Scholar] [CrossRef]
- Kiselevsky, D.B. Granzymes and Mitochondria. Biochemistry 2020, 85, 131–139. [Google Scholar] [CrossRef]
- Cullen, S.P.; Brunet, M.; Martin, S.J. Granzymes in cancer and immunity. Cell Death Differ. 2010, 17, 616–623. [Google Scholar] [CrossRef]
- Tang, R.; Xu, J.; Zhang, B.; Liu, J.; Liang, C.; Hua, J.; Meng, Q.; Yu, X.; Shi, S. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J. Hematol. Oncol. 2020, 13, 110. [Google Scholar] [CrossRef]
- Martinvalet, D.; Thiery, J.; Chowdhury, D. Chapter Eleven Granzymes and Cell Death. Methods Enzymol. 2008, 442, 213–230. [Google Scholar]
- Voskoboinik, I.; Dunstone, M.A.; Baran, K.; Whisstock, J.C.; Trapani, J.A. Perforin: Structure, function, and role in human immunopathology. Immunol. Rev. 2010, 235, 35–54. [Google Scholar] [CrossRef]
- Obeng, E. Apoptosis (programmed cell death) and its signals—A review. Braz. J. Biol. 2021, 81, 1133–1143. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Tsuchiya, K. Switching from Apoptosis to Pyroptosis: Gasdermin-Elicited Inflammation and Antitumor Immunity. Int. J. Mol. Sci. 2021, 22, 426. [Google Scholar] [CrossRef]
- Xu, X.; Lai, Y.; Hua, Z.-C. Apoptosis and apoptotic body: Disease message and therapeutic target potentials. Biosci. Rep. 2019, 39, BSR20180992. [Google Scholar] [CrossRef]
- Pistritto, G.; Trisciuoglio, D.; Ceci, C.; Garufi, A.; D’Orazi, G. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging 2016, 8, 603–619. [Google Scholar] [CrossRef]
- Majtnerová, P.; Roušar, T. An overview of apoptosis assays detecting DNA fragmentation. Mol. Biol. Rep. 2018, 45, 1469–1478. [Google Scholar] [CrossRef]
- Zhou, Z.; He, H.; Wang, K.; Shi, X.; Wang, Y.; Su, Y.; Wang, Y.; Li, D.; Liu, W.; Zhang, Y.; et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science 2020, 368, eaaz7548. [Google Scholar] [CrossRef]
- Perl, M.; Denk, S.; Kalbitz, M.; Huber-Lang, M. Granzyme B: A New Crossroad of Complement and Apoptosis. Adv. Exp. Med. Biol. 2011, 946, 135–146. [Google Scholar]
- Tibbs, E.; Cao, X. Emerging Canonical and Non-Canonical Roles of Granzyme B in Health and Disease. Cancers 2022, 14, 1436. [Google Scholar] [CrossRef]
- Todoric, J.; Antonucci, L.; Karin, M. Targeting Inflammation in Cancer Prevention and Therapy. Cancer Prev. Res. (Phila.) 2016, 9, 895–905. [Google Scholar]
- Lieberman, J. Granzyme A activates another way to die. Immunol. Rev. 2010, 235, 93–104. [Google Scholar] [CrossRef]
- Van Eck, J.A.; Shan, L.; Meeldijk, J.; Hack, C.E.; Bovenschen, N. A novel proinflammatory role for granzyme A. Cell Death Dis. 2017, 8, e2630. [Google Scholar] [CrossRef]
- Yeung, Y.T.; Aziz, F.; Guerrero-Castilla, A.; Argüelles, S. Signaling Pathways in Inflammation and Anti-inflammatory Therapies. Curr. Pharm. Des. 2018, 24, 1449–1484. [Google Scholar] [CrossRef]
- Van Opdenbosch, N.; Lamkanfi, M. Caspases in Cell Death, Inflammation, and Disease. Immunity 2019, 50, 1352–1364. [Google Scholar] [CrossRef]
- Germolec, D.R.; Shipkowski, K.A.; Frawley, R.P.; Evans, E. Markers of Inflammation. Methods Mol. Biol. 2018, 1803, 57–79. [Google Scholar]
- Kunnumakkara, A.B.; Sailo, B.L.; Banik, K.; Harsha, C.; Prasad, S.; Gupta, S.C.; Bharti, A.C.; Aggarwal, B.B. Chronic diseases, inflammation, and spices: How are they linked? J. Transl. Med. 2018, 16, 14. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Mancek-Keber, M. Inflammation-mediating proteases: Structure, function in (patho) physiology and inhibition. Protein Pept. Lett. 2014, 21, 1209–1229. [Google Scholar] [CrossRef]
- Greene, C.M.; McElvaney, N.G. Proteases and antiproteases in chronic neutrophilic lung disease—Relevance to drug discovery. Br. J. Pharmacol. 2009, 158, 1048–1058. [Google Scholar] [CrossRef]
- Velotti, F.; Barchetta, I.; Cimini, F.A.; Cavallo, M.G. Granzyme B in Inflammatory Diseases: Apoptosis, Inflammation, Extracellular Matrix Remodeling, Epithelial-to-Mesenchymal Transition and Fibrosis. Front. Immunol. 2020, 11, 587581. [Google Scholar] [CrossRef]
- Haneklaus, M.; Gerlic, M.; O’Neill, L.A.; Masters, S.L. miR-223: Infection, inflammation and cancer. J. Intern. Med. 2013, 274, 215–226. [Google Scholar] [CrossRef]
- Cimini, F.A.; Barchetta, I.; Ceccarelli, V.; Chiappetta, C.; Di Biasio, A.; Bertoccini, L.; Sentinelli, F.; Leonetti, F.; Silecchia, G.; Di Cristofano, C.; et al. Granzyme B Expression in Visceral Adipose Tissue Associates With Local Inflammation and Glyco-Metabolic Alterations in Obesity. Front. Immunol. 2020, 11, 589188. [Google Scholar] [CrossRef]
- Hiebert, P.R.; Granville, D.J. Granzyme B in injury, inflammation, and repair. Trends Mol. Med. 2012, 18, 732–741. [Google Scholar] [CrossRef]
- Cooper, D.M.; Pechkovsky, D.; Hackett, T.L.; Knight, D.A.; Granville, D.J. Granzyme K Activates Protease-Activated Receptor-1. PLoS ONE 2011, 6, e21484. [Google Scholar] [CrossRef]
- Wensink, A.C.; Hack, C.E.; Bovenschen, N. Granzymes regulate proinflammatory cytokine responses. J. Immunol. 2015, 194, 491–497. [Google Scholar] [CrossRef]
- Suarez-Carmona, M.; Lesage, J.; Cataldo, D.; Gilles, C. EMT and inflammation: Inseparable actors of cancer progression. Mol. Oncol. 2017, 11, 805–823. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Diakos, C.I.; Charles, K.A.; McMillan, D.C.; Clarke, S.J. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014, 15, e493–e503. [Google Scholar] [CrossRef]
- Murata, M. Inflammation and cancer. Environ. Health Prev. Med. 2018, 23, 50. [Google Scholar] [CrossRef]
- Khandia, R.; Munjal, A. Interplay between inflammation and cancer. Adv. Protein Chem. Struct Biol. 2020, 119, 199–245. [Google Scholar]
- Qu, X.; Tang, Y.; Hua, S. Immunological Approaches Towards Cancer and Inflammation: A Cross Talk. Front. Immunol. 2018, 9, 563. [Google Scholar] [CrossRef]
- Qiao, L.; Li, X. Role of chronic inflammation in cancers of the gastrointestinal system and the liver: Where we are now. Cancer Lett. 2014, 345, 150–152. [Google Scholar] [CrossRef]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef]
- Nathan, C. Points of control in inflammation. Nature 2002, 420, 846–852. [Google Scholar] [CrossRef]
- Watson, N.; Ding, B.; Zhu, X.; Frisina, R.D. Chronic inflammation—Inflammaging—In the ageing cochlea: A novel target for future presbycusis therapy. Ageing Res. Rev. 2017, 40, 142–148. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Vendramini-Costa, D.B.; Carvalho, J.E. Molecular link mechanisms between inflammation and cancer. Curr. Pharm. Des. 2012, 18, 3831–3852. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Yashiro, M. Ulcerative colitis-associated colorectal cancer. World J. Gastroenterol. 2014, 20, 16389–16397. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ling, Z.; Li, L. The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 2020, 11, 615056. [Google Scholar] [CrossRef] [PubMed]
- Shawki, S.; Ashburn, J.; Signs, S.A.; Huang, E. Colon Cancer: Inflammation-Associated Cancer. Surg. Oncol. Clin. N. Am. 2018, 27, 269–287. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zhang, J.; Zeng, D.; Sun, H.; Rong, X.; Shi, M.; Bin, J.; Liao, Y.; Liao, W. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I–III colon cancer. Cancer Immunol. Immunother. 2018, 68, 433–442. [Google Scholar] [CrossRef]
- Iyengar, N.M.; Gucalp, A.; Dannenberg, A.J.; Hudis, C.A. Obesity and Cancer Mechanisms: Tumor Microenvironment and Inflammation. J. Clin. Oncol. 2016, 34, 4270–4276. [Google Scholar] [CrossRef]
- Gupta, R.; Bhatt, L.K.; Johnston, T.P.; Prabhavalkar, K.S. Colon cancer stem cells: Potential target for the treatment of colorectal cancer. Cancer Biol. Ther. 2019, 20, 1068–1082. [Google Scholar] [CrossRef]
- Ruan, H.; Leibowitz, B.J.; Zhang, L.; Yu, J. Immunogenic cell death in colon cancer prevention and therapy. Mol. Carcinog. 2020, 59, 783–793. [Google Scholar] [CrossRef]
- Ye, L.; Zhang, T.; Kang, Z.; Guo, G.; Sun, Y.; Lin, K.; Huang, Q.; Shi, X.; Ni, Z.; Ding, N.; et al. Tumor-Infiltrating Immune Cells Act as a Marker for Prognosis in Colorectal Cancer. Front. Immunol. 2019, 10, 2368. [Google Scholar] [CrossRef]
- Park, S.; Anderson, N.L.; Canaria, D.A.; Olson, M.R. Granzyme-Producing CD4 T Cells in Cancer and Autoimmune Disease. ImmunoHorizons 2021, 5, 909–917. [Google Scholar] [CrossRef]
- Schmitt, M.; Greten, F.R. The inflammatory pathogenesis of colorectal cancer. Nat. Rev. Immunol. 2021, 21, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Hibino, S.; Kawazoe, T.; Kasahara, H.; Itoh, S.; Ishimoto, T.; Sakata-Yanagimoto, M.; Taniguchi, K. Inflammation-Induced Tumorigenesis and Metastasis. Int. J. Mol. Sci. 2021, 22, 5421. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kawada, K.; Obama, K. Inflammation-Related Biomarkers for the Prediction of Prognosis in Colorectal Cancer Patients. Int. J. Mol. Sci. 2021, 22, 8002. [Google Scholar] [CrossRef]
- Santiago, L.; Castro, M.; Sanz-Pamplona, R.; Garzón, M.; Ramirez-Labrada, A.; Tapia, E.; Moreno, V.; Layunta, E.; Gil-Gómez, G.; Garrido, M.; et al. Extracellular Granzyme A Promotes Colorectal Cancer Development by Enhancing Gut Inflammation. Cell Rep. 2020, 32, 107847. [Google Scholar] [CrossRef]
- Turner, C.T.; Zeglinski, M.R.; Richardson, K.C.; Santacruz, S.; Hiroyasu, S.; Wang, C.; Zhao, H.; Shen, Y.; Sehmi, R.; Lima, H.; et al. Granzyme B Contributes to Barrier Dysfunction in Oxazolone-Induced Skin Inflammation through E-Cadherin and FLG Cleavage. J. Investig. Dermatol. 2020, 141, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Prakash, M.D.; Bird, C.H.; Bird, P.I. Active and zymogen forms of granzyme B are constitutively released from cytotoxic lymphocytes in the absence of target cell engagement. Immunol. Cell Biol. 2009, 87, 249–254. [Google Scholar] [CrossRef]
- Božič, J.; Dolenc, I. Feedback Regulation of Cathepsin C by the Propeptide Dipeptides of Granzymes A and B. Acta Chim. Slov. 2019, 66, 501–509. [Google Scholar] [CrossRef]
- Kountouras, J.; Diamantidis, M.; Tsapournas, G.; Zavos, C. New aspects of regulatory signaling pathways and novel therapies in pancreatic cancer. Curr. Mol. Med. 2008, 8, 12–37. [Google Scholar] [CrossRef]
- Van Ham, S.M.; Heutinck, K.M.; Jorritsma, T.; Bemelman, F.J.; Strik, M.C.; Vos, W.; Muris, J.J.; Florquin, S.; Ten Berge, I.J.; Rowshani, A.T. Urinary granzyme A mRNA is a biomarker to diagnose subclinical and acute cellular rejection in kidney transplant recipients. Kidney Int. 2010, 78, 1033–1040. [Google Scholar] [CrossRef]
- Szekely, B.; Bossuyt, V.; Li, X.; Wali, V.; Patwardhan, G.; Frederick, C.; Silber, A.; Park, T.; Harigopal, M.; Pelekanou, V.; et al. Immunological differences between primary and metastatic breast cancer. Ann. Oncol. 2018, 29, 2232–2239. [Google Scholar] [CrossRef] [PubMed]
- Pagès, F.; Berger, A.; Camus, M.; Sanchez-Cabo, F.; Costes, A.; Molidor, R.; Mlecnik, B.; Kirilovsky, A.; Nilsson, M.; Damotte, D.; et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N. Engl. J. Med. 2005, 353, 2654–2666. [Google Scholar] [CrossRef] [PubMed]
- Salama, P.; Phillips, M.; Platell, C.; Iacopetta, B. Low expression of Granzyme B in colorectal cancer is associated with signs of early metastastic invasion. Histopathology 2011, 59, 207–215. [Google Scholar] [CrossRef]
- Mulder, W.M.; Bloemena, E.; Stukart, M.J.; Kummer, J.A.; Wagstaff, J.; Scheper, R.J. T cell receptor-zeta and granzyme B expression in mononuclear cell infiltrates in normal colon mucosa and colon carcinoma. Gut 1997, 40, 113–119. [Google Scholar] [CrossRef][Green Version]
- Wang, H.; Sun, Q.; Wu, Y.; Wang, L.; Zhou, C.; Ma, W.; Zhang, Y.; Wang, S.; Zhang, S. Granzyme M expressed by tumor cells promotes chemoresistance and EMT in vitro and metastasis in vivo associated with STAT3 activation. Oncotarget 2015, 6, 5818–5831. [Google Scholar] [CrossRef]
- Souza-Fonseca-Guimaraes, F.; Krasnova, Y.; Putoczki, T.; Miles, K.; MacDonald, K.; Town, L.; Shi, W.; Gobe, G.C.; McDade, L.; Mielke, L.; et al. Granzyme M has a critical role in providing innate immune protection in ulcerative colitis. Cell Death Dis. 2016, 7, e2302. [Google Scholar] [CrossRef]
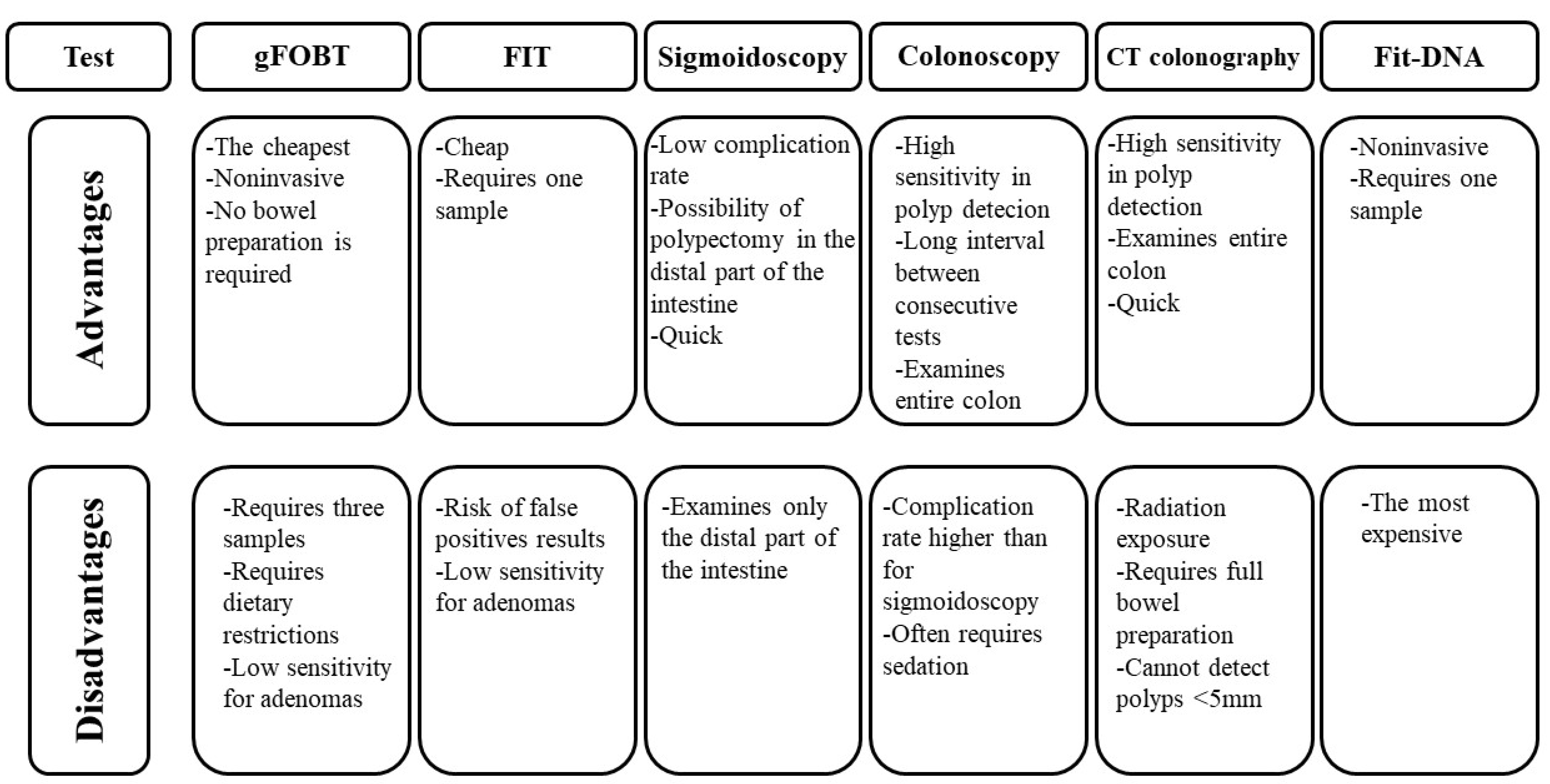
| Non-Modifiable Risk Factors | Modifiable Risk Factors |
|---|---|
| age sex ethnicity family history personal history of adenomas polyposis syndromes inflammatory bowel disease BRCA gene mutations | red meat consumption processed meat consumption smoking tobacco alcohol abuse low-fibre diet overweight and obesity lack of physical activity |
| Protein | Type of Expression | Method of Detection | Effect | Correlation with Inflammatory Genes | Suggested Role | Ref. |
|---|---|---|---|---|---|---|
| Granzyme A | mRNA expression | RT-PCR | - significantly elevated expression was observed in CRC and inflammatory samples in comparison to control group | yes | - promotion of tumour development - progression of CRC | [105] |
| Granzyme B | mRNA expression | RT-PCR | - increased levels were associated with absence of pathological signs of early metastasis invasion | nd | - antitumour activity | [114] |
| protein expression | IHC | - low expression was associated with early signs of metastasis - high expression was associated with better survival | nd | - prognostic factor | [113] | |
| mRNA expression | RT-PCR | nd | no | nd | [114] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pączek, S.; Łukaszewicz-Zając, M.; Mroczko, B. Granzymes—Their Role in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 5277. https://doi.org/10.3390/ijms23095277
Pączek S, Łukaszewicz-Zając M, Mroczko B. Granzymes—Their Role in Colorectal Cancer. International Journal of Molecular Sciences. 2022; 23(9):5277. https://doi.org/10.3390/ijms23095277
Chicago/Turabian StylePączek, Sara, Marta Łukaszewicz-Zając, and Barbara Mroczko. 2022. "Granzymes—Their Role in Colorectal Cancer" International Journal of Molecular Sciences 23, no. 9: 5277. https://doi.org/10.3390/ijms23095277
APA StylePączek, S., Łukaszewicz-Zając, M., & Mroczko, B. (2022). Granzymes—Their Role in Colorectal Cancer. International Journal of Molecular Sciences, 23(9), 5277. https://doi.org/10.3390/ijms23095277








