PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction
Abstract
1. Introduction
2. Results and Discussion
2.1. Drosophila PGRP-LB Isoforms Have a Similar Amidase Activity
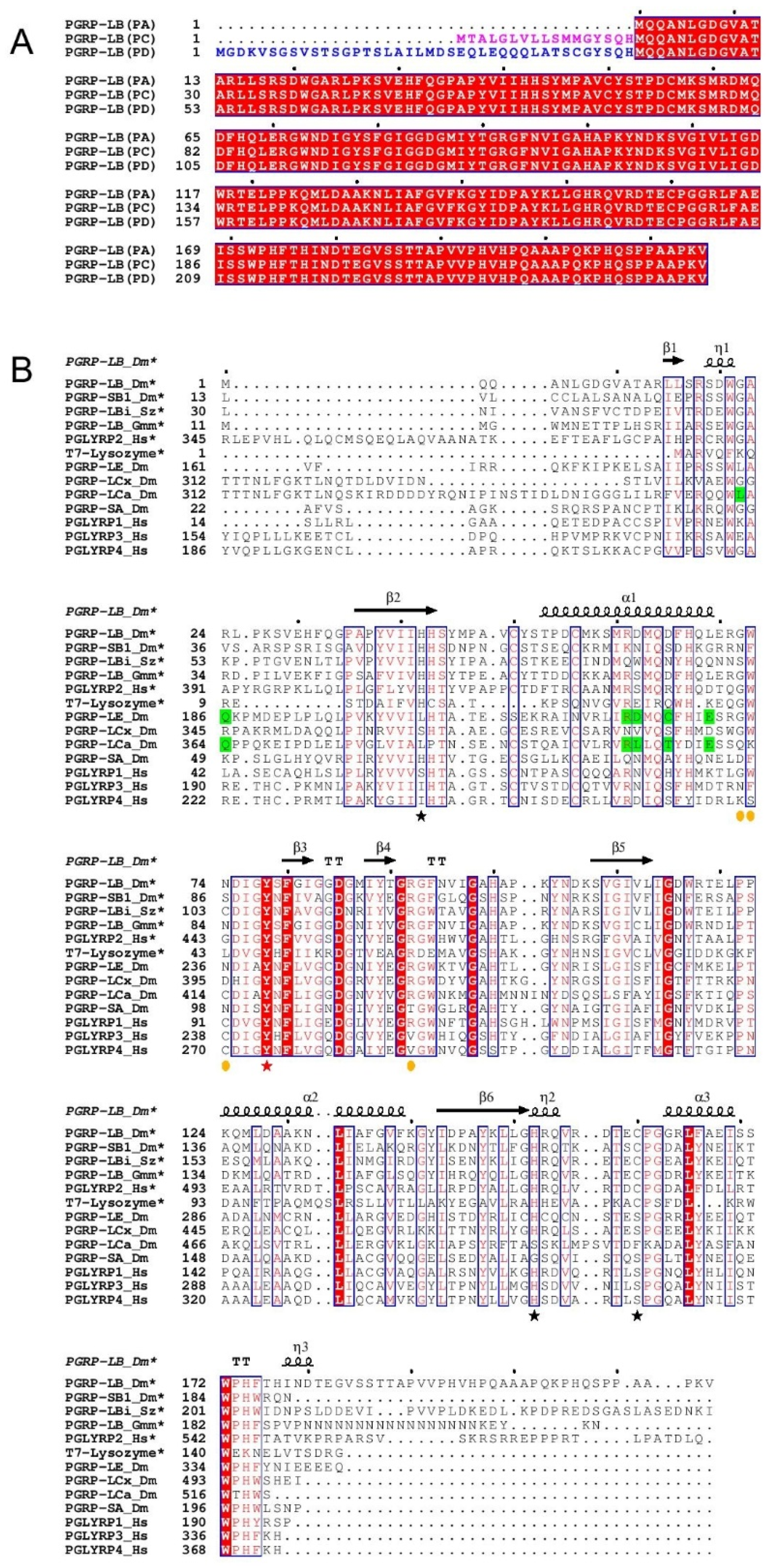
2.2. Identification of Potential Key Residues in PGRP-LB for Amidase Reaction
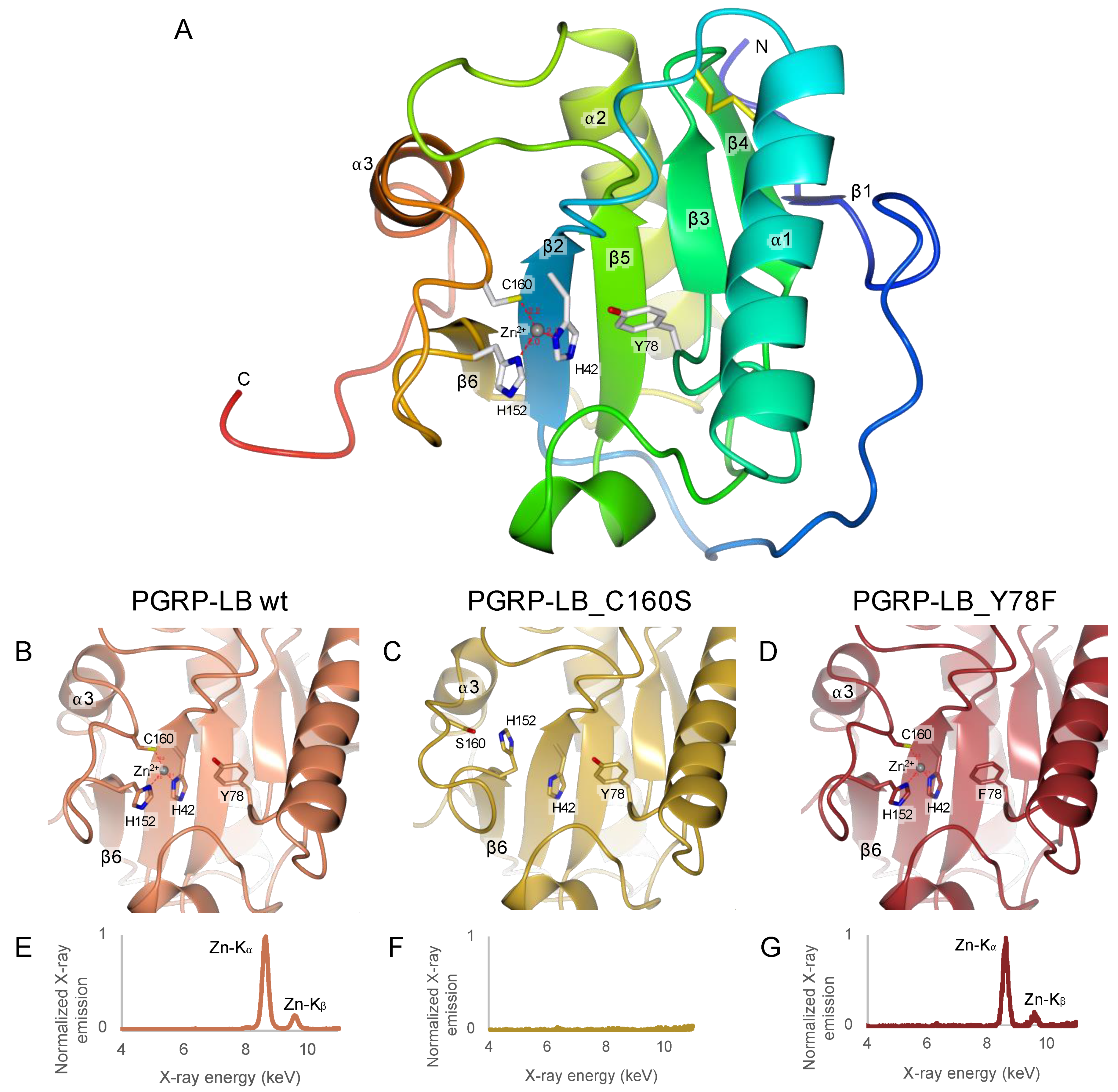
2.3. Only Y78 Residue Is Necessary for the Amidase Reaction
2.4. C160S Is the Most Essential Residue for Zn2+ Chelation
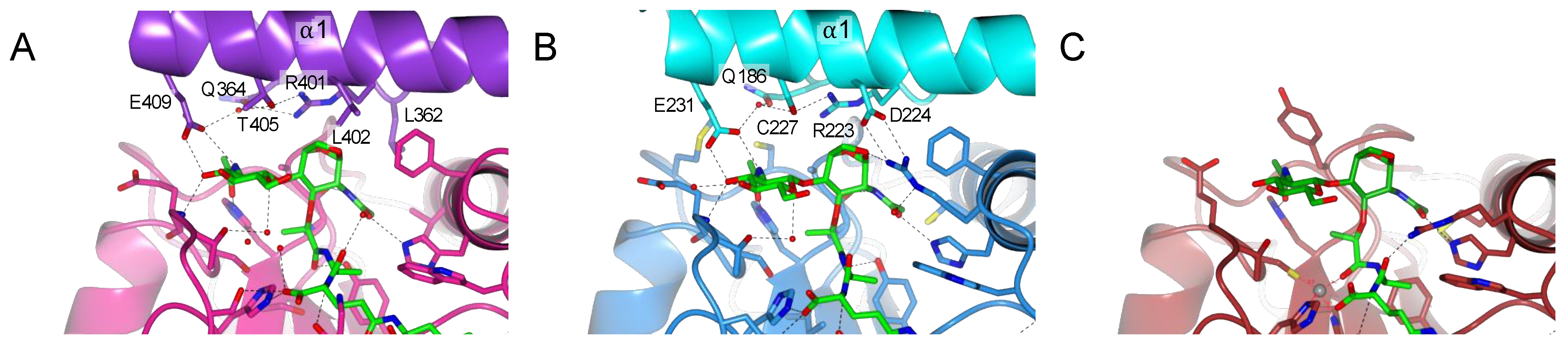
2.5. TCT Makes Two Important Interactions with R92 and Zn2+
2.6. PGRP-LC and PGRP-LE Helix α1 Allows a Stronger Recognition of the Sugar Moiety
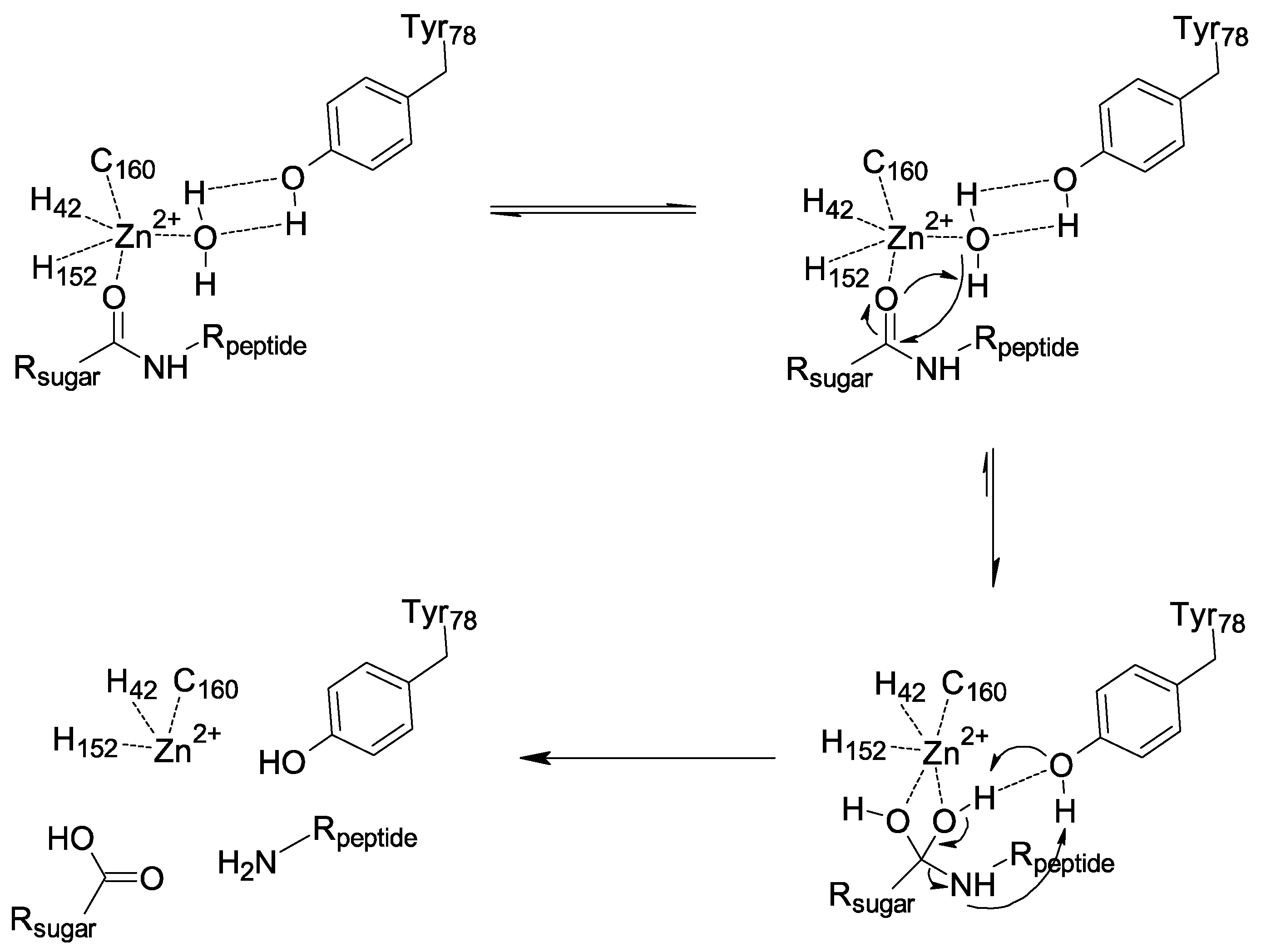
2.7. Amidase PGRP Reaction Mechanism Needs the Dynamic Role of Y78
3. Materials and Methods
3.1. Sequence Alignment
3.2. Protein Cloning, Expression and Purification
3.3. Peptidoglycan and Muropeptides Purification
3.4. Enzymatic Activity
3.5. Interaction with PGN
3.6. Protein Crystallization and Data Collection
3.7. Structure Determination
3.8. 3D Modelling of the Entire TCT
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pazos, M.; Peters, K. Peptidoglycan. In Bacterial Adhesion; Subcellular Biochemistry; Kuhn, A., Ed.; Springer International Publishing: Cham, Switzerland, 2019; Volume 92, pp. 127–168. ISBN 978-3-030-18767-5. [Google Scholar]
- Vollmer, W. Peptidoglycan; Elsevier Ltd.: Amsterdam, The Netherlands, 2015; Volumes 1–3, ISBN 9780123971692. [Google Scholar]
- Stenbak, C.R.; Ryu, J.-H.; Leulier, F.; Pili-Floury, S.; Parquet, C.; Hervé, M.; Chaput, C.; Boneca, I.G.; Lee, W.-J.; Lemaitre, B.; et al. Peptidoglycan Molecular Requirements Allowing Detection by the Drosophila Immune Deficiency Pathway. J. Immunol. 2004, 173, 7339–7348. [Google Scholar] [CrossRef]
- Filipe, S.R.; Tomasz, A.; Ligoxygakis, P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005, 6, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Chaput, C.; Boneca, I.G. Peptidoglycan detection by mammals and flies. Microbes Infect. 2007, 9, 637–647. [Google Scholar] [CrossRef]
- Yoshida, H.; Kinoshita, K.; Ashida, M. Purification of a Peptidoglycan Recognition Protein from Hemolymph of the Silkworm, Bombyx mori. J. Biol. Chem. 1996, 271, 13854–13860. [Google Scholar] [CrossRef]
- Royet, J.; Gupta, D.; Dziarski, R. Peptidoglycan recognition proteins: Modulators of the microbiome and inflammation. Nat. Rev. Immunol. 2011, 11, 837–851. [Google Scholar] [CrossRef]
- Lu, Y.; Su, F.; Li, Q.; Zhang, J.; Li, Y.; Tang, T.; Hu, Q.; Yu, X.-Q. Pattern recognition receptors in Drosophila immune responses. Dev. Comp. Immunol. 2020, 102, 103468. [Google Scholar] [CrossRef] [PubMed]
- Kordaczuk, J.; Sułek, M.; Wojda, I. General overview on the role of Peptidoglycan Recognition Proteins in insect immunity. Acta Biochim. Pol. 2020, 67, 319–326. [Google Scholar] [CrossRef]
- Kurata, S. Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 2014, 42, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Kleino, A.; Silverman, N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 2014, 42, 25–35. [Google Scholar] [CrossRef]
- Lindsay, S.A.; Wasserman, S.A. Conventional and non-conventional Drosophila Toll signaling. Dev. Comp. Immunol. 2014, 42, 16–24. [Google Scholar] [CrossRef]
- Zaidman-Rémy, A.; Hervé, M.; Poidevin, M.; Pili-Floury, S.; Kim, M.-S.; Blanot, D.; Oh, B.-H.; Ueda, R.; Mengin-Lecreulx, D.; Lemaitre, B. The Drosophila Amidase PGRP-LB Modulates the Immune Response to Bacterial Infection. Immunity 2006, 24, 463–473. [Google Scholar] [CrossRef]
- Zaidman-Rémy, A.; Poidevin, M.; Hervé, M.; Welchman, D.P.; Paredes, J.C.; Fahlander, C.; Steiner, H.; Mengin-Lecreulx, D.; Lemaitre, B. Drosophila immunity: Analysis of PGRP-SB1 expression, enzymatic activity and function. PLoS ONE 2011, 6, e17231. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-M.; Li, X.; Cocklin, R.R.; Wang, M.; Wang, M.; Fukase, K.; Inamura, S.; Kusumoto, S.; Gupta, D.; Dziarski, R. Human Peptidoglycan Recognition Protein-L Is an N-Acetylmuramoyl-L-alanine Amidase. J. Biol. Chem. 2003, 278, 49044–49052. [Google Scholar] [CrossRef]
- Paredes, J.C.; Welchman, D.P.; Poidevin, M.; Lemaitre, B. Negative Regulation by Amidase PGRPs Shapes the Drosophila Antibacterial Response and Protects the Fly from Innocuous Infection. Immunity 2011, 35, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Charroux, B.; Capo, F.; Kurz, C.L.; Peslier, S.; Chaduli, D.; Viallat-lieutaud, A.; Royet, J. Cytosolic and Secreted Peptidoglycan-Degrading Enzymes in Drosophila Respectively Control Local and Systemic Immune Responses to Microbiota. Cell Host Microbe 2018, 23, 215–228.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Aksoy, S. PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse’s offspring. Proc. Natl. Acad. Sci. USA 2012, 109, 10552–10557. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Yang, G.; Aksoy, S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. USA 2009, 106, 12133–12138. [Google Scholar] [CrossRef]
- Maire, J.; Vincent-Monégat, C.; Balmand, S.; Vallier, A.; Hervé, M.; Masson, F.; Parisot, N.; Vigneron, A.; Anselme, C.; Perrin, J.; et al. Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc. Natl. Acad. Sci. USA 2019, 116, 5623–5632. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Byun, M.; Oh, B.-H. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol. 2003, 4, 787–793. [Google Scholar] [CrossRef]
- Guan, R.; Malchiodi, E.L.; Wang, Q.; Schuck, P.; Mariuzza, R.A. Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Iα. J. Biol. Chem. 2004, 279, 31873–31882. [Google Scholar] [CrossRef]
- Chang, C.-I. Structure of Tracheal Cytotoxin in Complex with a Heterodimeric Pattern-Recognition Receptor. Science 2006, 311, 1761–1764. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-H.; Kim, M.-S.; Kim, H.-E.; Yano, T.; Oshima, Y.; Aggarwal, K.; Goldman, W.E.; Silverman, N.; Kurata, S.; Oh, B.-H. Structural Basis for Preferential Recognition of Diaminopimelic Acid-type Peptidoglycan by a Subset of Peptidoglycan Recognition Proteins. J. Biol. Chem. 2006, 281, 8286–8295. [Google Scholar] [CrossRef]
- Chang, C.-I.; Pili-Floury, S.; Hervé, M.; Parquet, C.; Chelliah, Y.; Lemaitre, B.; Mengin-Lecreulx, D.; Deisenhofer, J. A Drosophila Pattern Recognition Receptor Contains a Peptidoglycan Docking Groove and Unusual L,D-Carboxypeptidase Activity. PLoS Biol. 2004, 2, e277. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Roychowdhury, A.; Ember, B.; Kumar, S.; Boons, G.-J.; Mariuzza, R.A. Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17168–17173. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Brown, P.H.; Swaminathan, C.P.; Roychowdhury, A.; Boons, G.-J.; Mariuzza, R.A. Crystal structure of human peptidoglycan recognition protein Iα bound to a muramyl pentapeptide from Gram-positive bacteria. Protein Sci. 2006, 15, 1199–1206. [Google Scholar] [CrossRef]
- Cho, S.; Wang, Q.; Swaminathan, C.P.; Hesek, D.; Lee, M.; Boons, G.-J.; Mobashery, S.; Mariuzza, R.A. Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 8761–8766. [Google Scholar] [CrossRef] [PubMed]
- Reiser, J.-B.; Teyton, L.; Wilson, I.A. Crystal Structure of the Drosophila Peptidoglycan Recognition Protein (PGRP)-SA at 1.56 Å Resolution. J. Mol. Biol. 2004, 340, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhang, X.; Pflugrath, J.W.; Studier, F.W. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 1994, 91, 4034–4038. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef]
- Wang, W.-J.; Cheng, W.; Luo, M.; Yan, Q.; Yu, H.-M.; Li, Q.; Cao, D.-D.; Huang, S.; Xu, A.; Mariuzza, R.A.; et al. Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain. PLoS ONE 2015, 10, e0140953. [Google Scholar] [CrossRef]
- McNicholas, S.; Potterton, E.; Wilson, K.S.; Noble, M.E.M. Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Solé, V.A.; Papillon, E.; Cotte, M.; Walter, P.; Susini, J. A multiplatform code for the analysis of energy-dispersive X-ray fluorescence spectra. Spectrochim. Acta Part B At. Spectrosc. 2007, 62, 63–68. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Kleino, A.; Ramia, N.F.; Bozkurt, G.; Shen, Y.; Nailwal, H.; Huang, J.; Napetschnig, J.; Gangloff, M.; Chan, F.K.M.; Wu, H.; et al. Peptidoglycan-Sensing Receptors Trigger the Formation of Functional Amyloids of the Adaptor Protein Imd to Initiate Drosophila NF-κB Signaling. Immunity 2017, 47, 635–647.e6. [Google Scholar] [CrossRef]
- Kleino, A.; Silverman, N. Regulation of the Drosophila Imd pathway by signaling amyloids. Insect Biochem. Mol. Biol. 2019, 108, 16–23. [Google Scholar] [CrossRef]
- Laitaoja, M.; Valjakka, J.; Jänis, J. Zinc coordination spheres in protein structures. Inorg. Chem. 2013, 52, 10983–10991. [Google Scholar] [CrossRef]
- Kerff, F.; Petrella, S.; Mercier, F.; Sauvage, E.; Herman, R.; Pennartz, A.; Zervosen, A.; Luxen, A.; Frère, J.M.; Joris, B.; et al. Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family. J. Mol. Biol. 2010, 397, 249–259. [Google Scholar] [CrossRef]
- Büttner, F.M.; Zoll, S.; Nega, M.; Götz, F.; Stehle, T. Structure-function analysis of Staphylococcus aureus amidase reveals the determinants of peptidoglycan recognition and cleavage. J. Biol. Chem. 2014, 289, 11083–11094. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Berrow, N.S.; Alderton, D.; Sainsbury, S.; Nettleship, J.; Assenberg, R.; Rahman, N.; Stuart, D.I.; Owens, R.J. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.; Waterman, D.G.; Parkhurst, J.M.; Brewster, A.S.; Gildea, R.J.; Gerstel, M.; Fuentes-Montero, L.; Vollmar, M.; Michels-Clark, T.; Young, I.D.; et al. DIALS: Implementation and evaluation of a new integration package. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 85–97. [Google Scholar] [CrossRef]
- Evans, P.R. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]

| Specific Activity (nmol.min−1.mg of Protein−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein | Substrate | ||||||||
| E. coli PGN (DAP Type) | GM(anh)- tetraDAP (TCT) | GM(anh)- tetraDAP dimer * | GM(anh)- tetraDAP dimer ** | GM(anh)-pentaLys | GM-tetraDAP | M(anh)- tetraDAP | M-tetraDAP | GM(anh)- tetraDAP (No Zn2+) | |
| PGRP-LBPA/PC | 1570 ± 40 | 4525 ± 682 | 1807 ± 341 | 992 ± 671 | 167 ± 34 | 1132 ± 230 | 1456 ± 542 | 669 ± 215 | 3468 ± 624 |
| PGRP-LBPD | 660 ± 27 | 3730 ± 597 | 1055 ± 220 | 330 ± 312 | 126 ± 28 | 832 ± 279 | 1441 ± 235 | 266 ± 84 | 4908 ± 1351 |
| PGRP-LBPA/PC_H42A | NA | 13 ± 4 | 11 ± 2 | NA | NA | 15 ± 6 | NA | NA | 10 ± 4 |
| PGRP-LBPA/PC_Y78F | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| PGRP-LBPA/PC_H152A | NA | 7 ± 2 | 4 ± 1 | NA | NA | NA | NA | NA | 7 ± 3 |
| PGRP-LBPA/PC_C160S | NA | 26 ± 5 | 28 ± 8 | 7 ± 9 | NA | NA | 10 ± 4 | NA | NA |
| PGRP-LBPA/PC | PGRP-LBPA/PC_C160S | PGRP-LBPA/PC_Y78F | PGRP-LBPA/PC_Y78F + TCT | |
|---|---|---|---|---|
| PDB code | 7NSX | 7NSY | 7NSZ | 7NT0 |
| Data collection | ||||
| Space Group | C2221 | P1 | P6122 | P1 |
| Cell parameters | ||||
| a, b, c (Å) | 39.87, 70.55, 112.83 | 41.02, 49.19, 52.14 | 40.50, 40.50, 338.94 | 38.59, 52.17, 55.58 |
| α, β, γ (°) | 90, 90, 90 | 71.82, 79, 66.84 | 90, 90, 120 | 92.11, 105.12, 111.51 |
| Resolution range | 56.48–1.90 (1.94–1.90) | 49.39–1.40 (1.40–1.42) | 56.55–1.30 (1.32–1.30) | 53.07–1.80 (1.84–1.80) |
| Unique reflection | 12,942 (808) | 67,209 (3218) | 42,802 (1995) | 34,852 (2038) |
| Rmeas | 0.154 (1.406) | 0.043 (0.420) | 0.099 (0.476) | 0.099 (0.564) |
| Completeness (%) | 99.8 (99.2) | 96.1 (92.2) | 99.9 (98.9) | 97.7 (97.2) |
| Multiplicity | 7.9 (7.2) | 3.4 (2.6) | 31.5 (13.0) | 6.9 (6.8) |
| I/σ(I) | 8.5 (1.4) | 14.1 (2.5) | 20.8 (2.8) | 11.9 (3.3) |
| CC1/2 | 0.993 (0.562) | 0.999 (0.875) | 0.998 (0 0.955) | 0.998 (0.891) |
| Refinement | ||||
| Rwork/Rfree | 0.180/0.213 | 0.167/0.188 | 0.186/0.221 | 0.145/0.180 |
| RMSD | ||||
| Bond length (Å) | 0.0098 | 0.0145 | 0.0143 | 0.0131 |
| Bond angle (°) | 1.597 | 1.906 | 1.904 | 1.684 |
| B factors (Å2) | ||||
| Protein | 29.09 | 18.05 | 14.32 | 20.44 |
| Ion | 19.64 | - | 11.35 | 14.68 |
| Ligand | - | - | - | 25.35 |
| Water | 33.55 | 30.97 | 26.97 | 30.78 |
| Ramachandran (%) | ||||
| Favored | 94.64 | 94.22 | 94.71 | 93.96 |
| Allowed | 4.17 | 4.56 | 4.71 | 5.44 |
| Outliers | 1.19 | 1.22 | 0.59 | 0.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orlans, J.; Vincent-Monegat, C.; Rahioui, I.; Sivignon, C.; Butryn, A.; Soulère, L.; Zaidman-Remy, A.; Orville, A.M.; Heddi, A.; Aller, P.; et al. PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction. Int. J. Mol. Sci. 2021, 22, 4957. https://doi.org/10.3390/ijms22094957
Orlans J, Vincent-Monegat C, Rahioui I, Sivignon C, Butryn A, Soulère L, Zaidman-Remy A, Orville AM, Heddi A, Aller P, et al. PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction. International Journal of Molecular Sciences. 2021; 22(9):4957. https://doi.org/10.3390/ijms22094957
Chicago/Turabian StyleOrlans, Julien, Carole Vincent-Monegat, Isabelle Rahioui, Catherine Sivignon, Agata Butryn, Laurent Soulère, Anna Zaidman-Remy, Allen M. Orville, Abdelaziz Heddi, Pierre Aller, and et al. 2021. "PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction" International Journal of Molecular Sciences 22, no. 9: 4957. https://doi.org/10.3390/ijms22094957
APA StyleOrlans, J., Vincent-Monegat, C., Rahioui, I., Sivignon, C., Butryn, A., Soulère, L., Zaidman-Remy, A., Orville, A. M., Heddi, A., Aller, P., & Da Silva, P. (2021). PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction. International Journal of Molecular Sciences, 22(9), 4957. https://doi.org/10.3390/ijms22094957






