AtWAKL10, a Cell Wall Associated Receptor-Like Kinase, Negatively Regulates Leaf Senescence in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
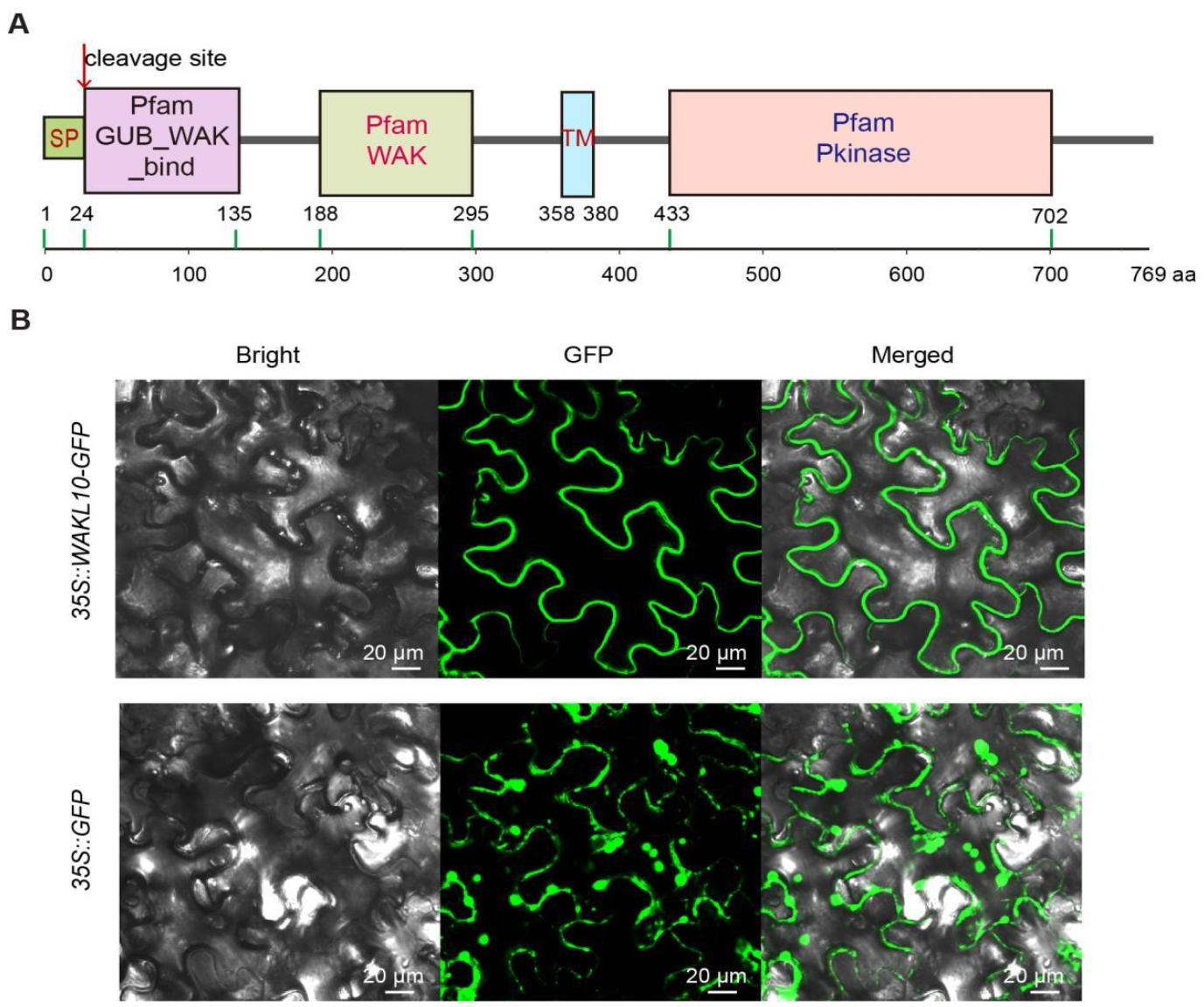
2.1. Functional Domain Analysis and Subcellular Localization of AtWAKL10
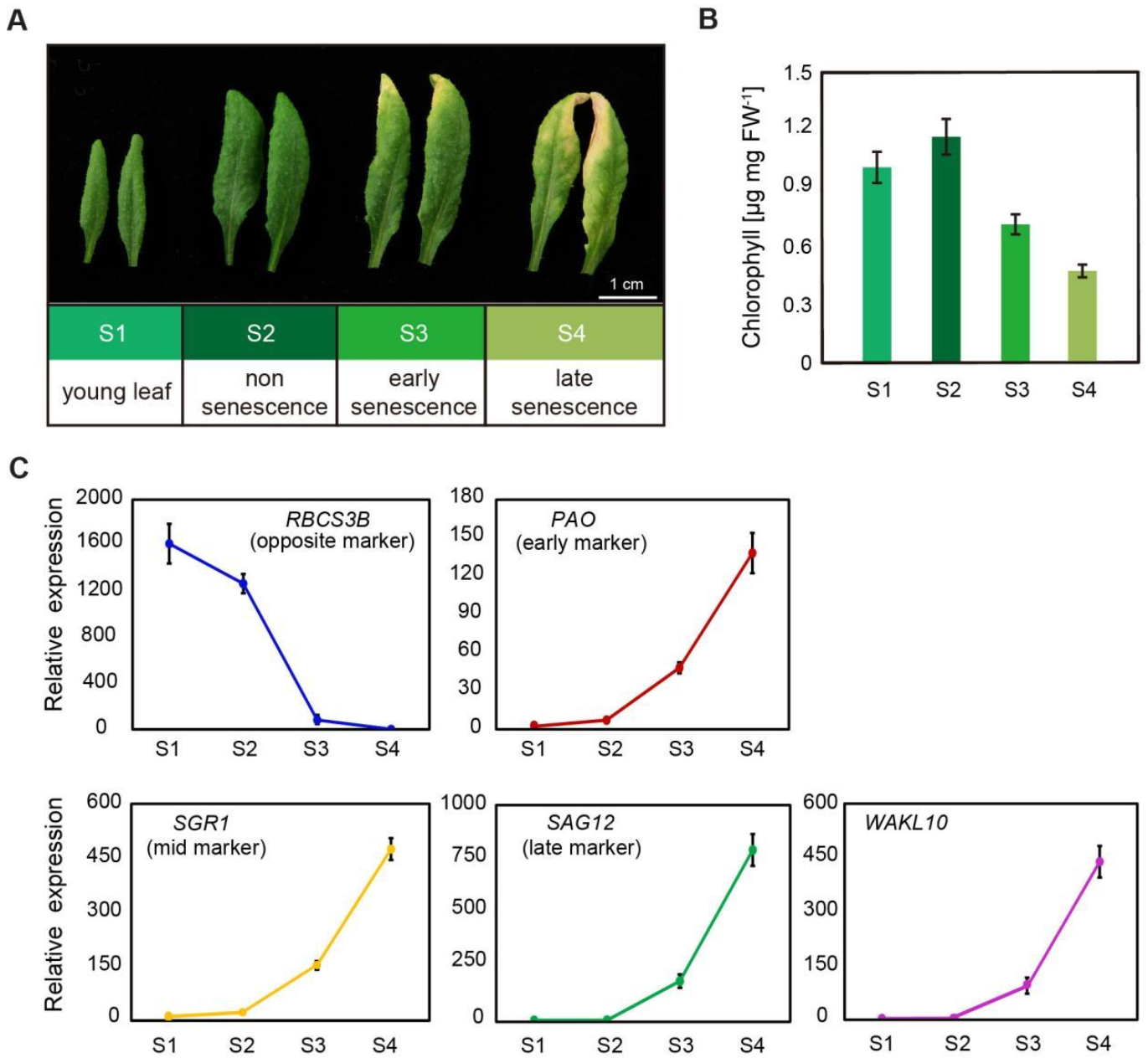
2.2. AtWAKL10 Was Highly Induced during Leaf Senescence
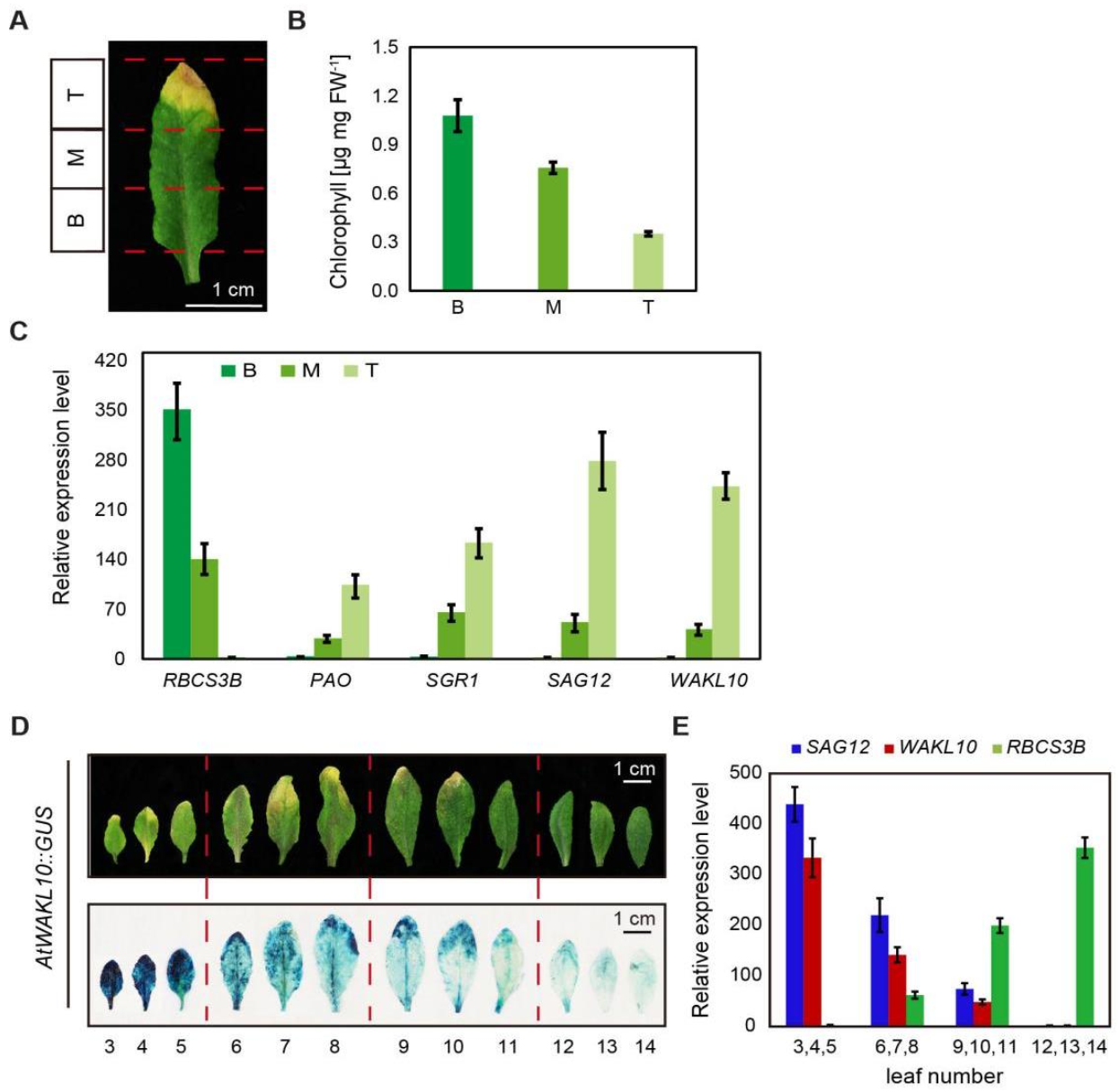
2.3. The Expression Patterns of AtWAKL10 in Various Leaf Tissues
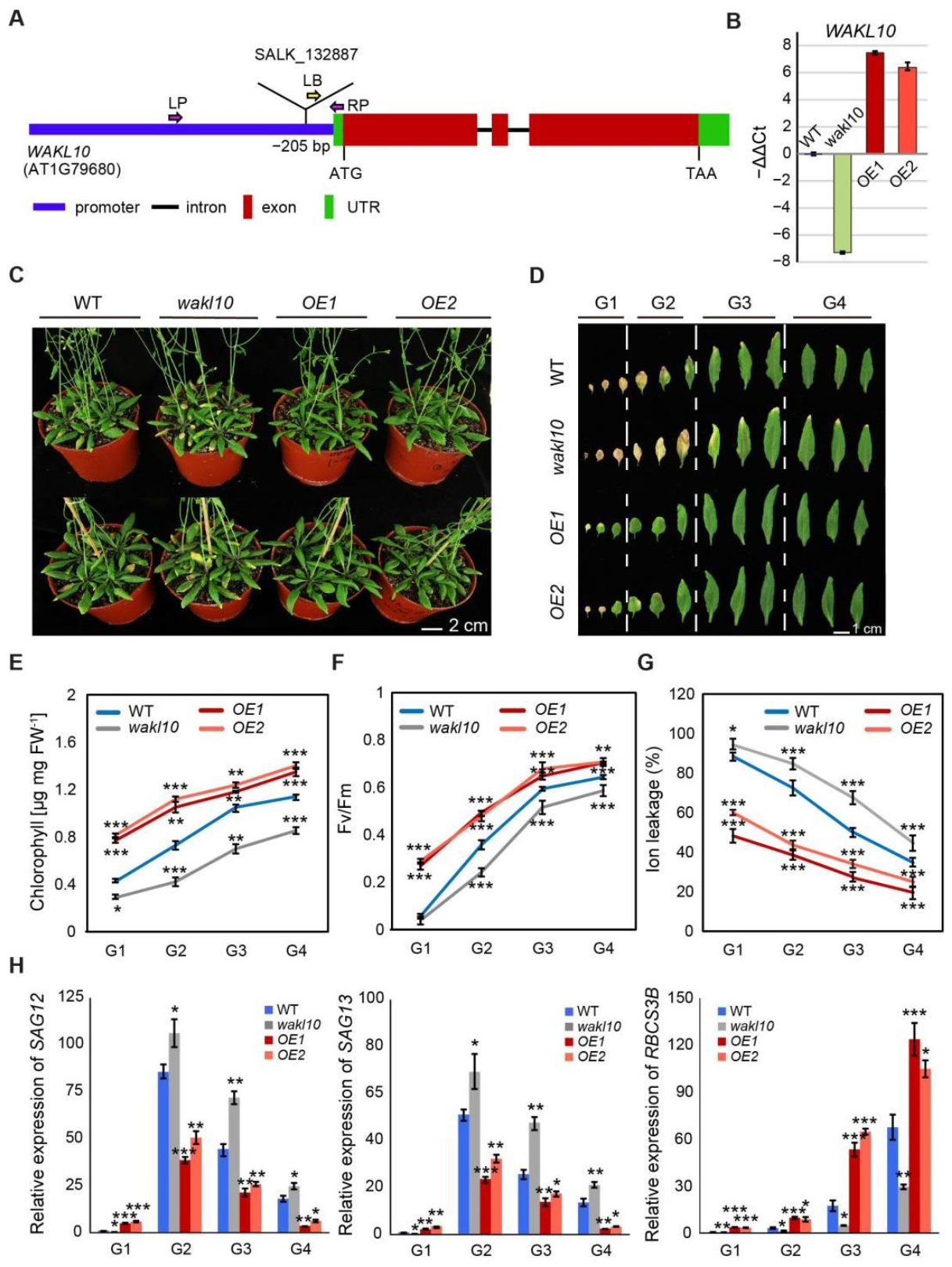
2.4. AtWAKL10 Negatively Regulates Natural Leaf Senescence
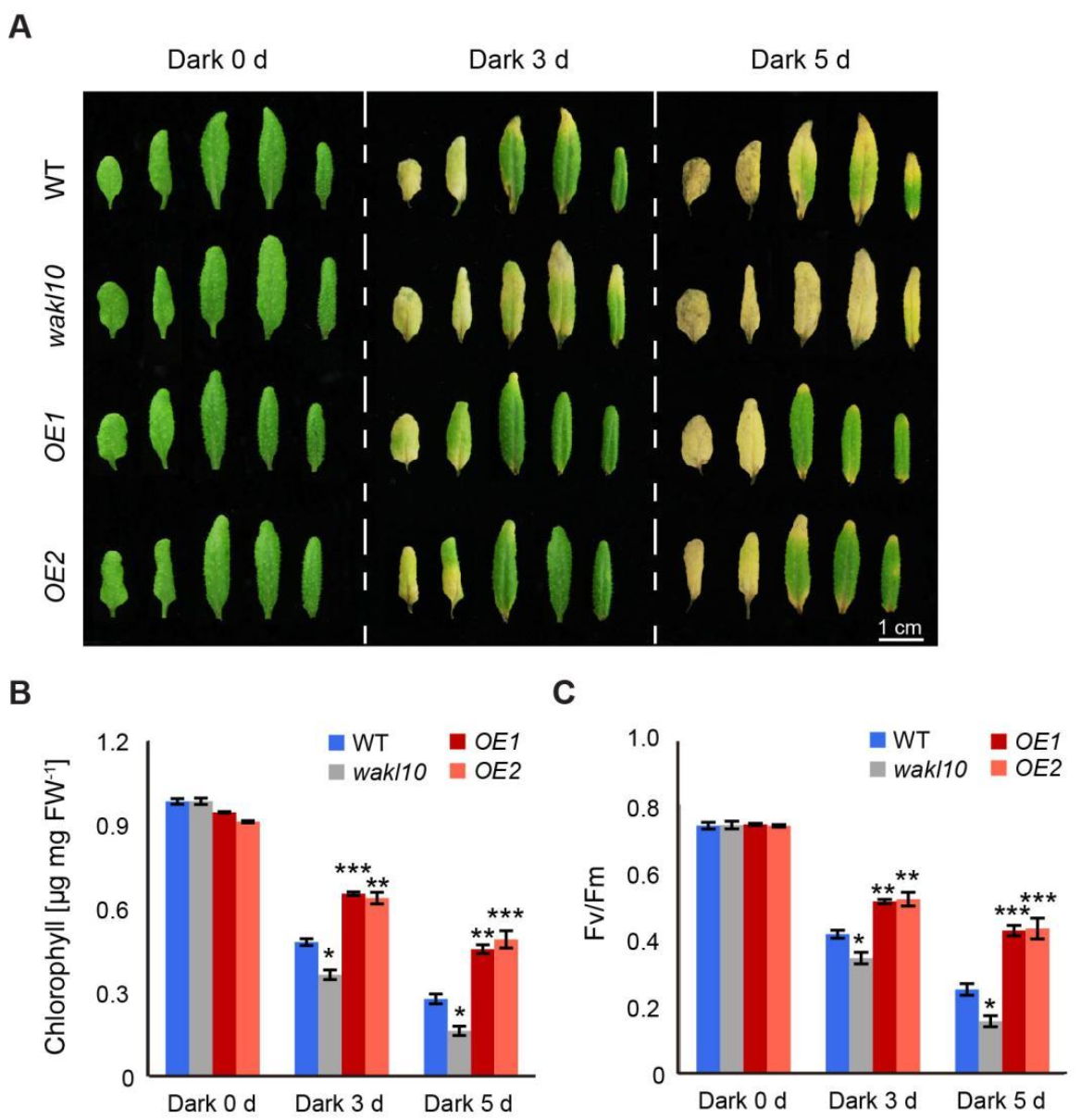
2.5. AtWAKL10 Had a Negative Effect on Dark-Induced Senescence
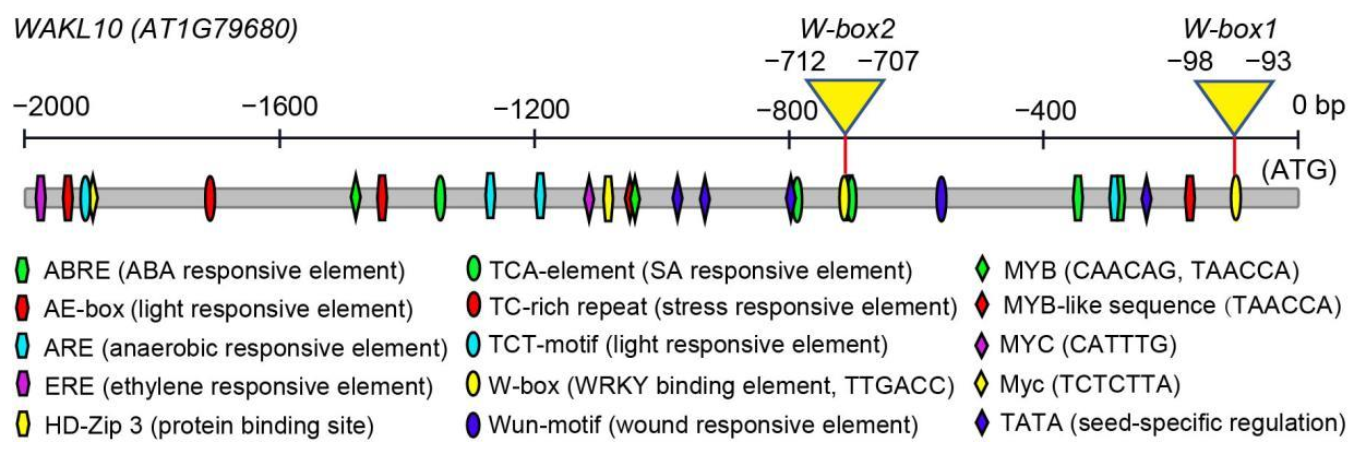
2.6. Promoter Analysis of AtWAKL10
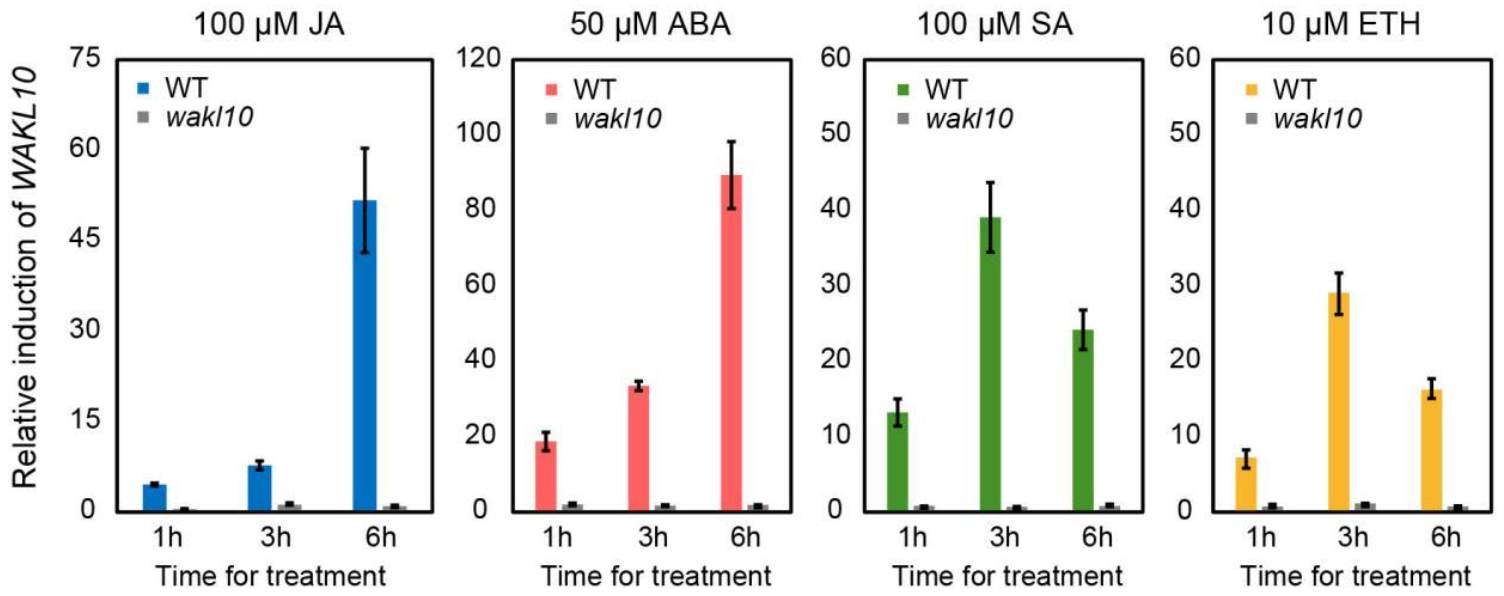
2.7. The Expression Pattern of AtWAKL10 in Response to Various Hormones
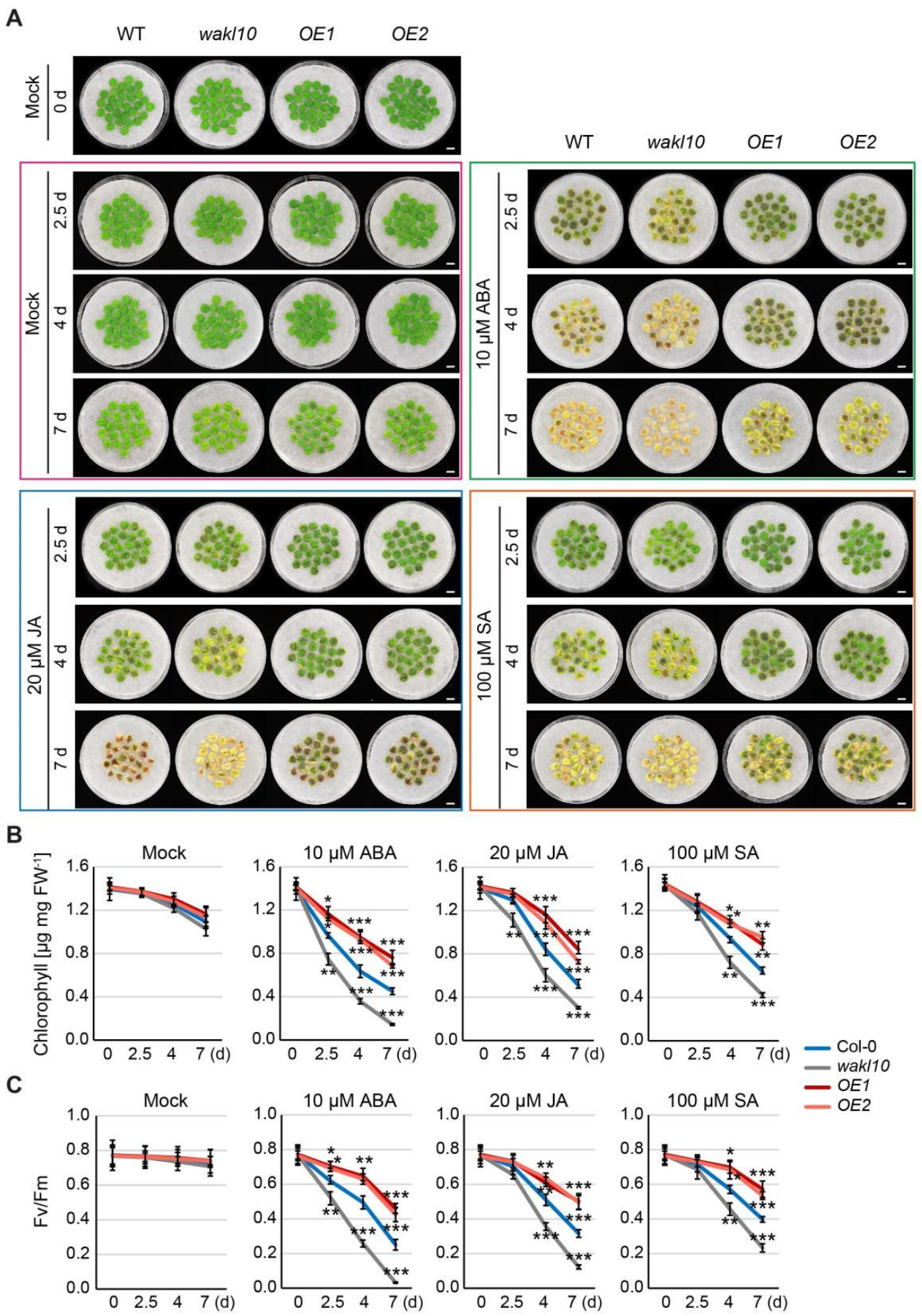
2.8. AtWAKL10 Negatively Regulates Hormone-Induced Senescence
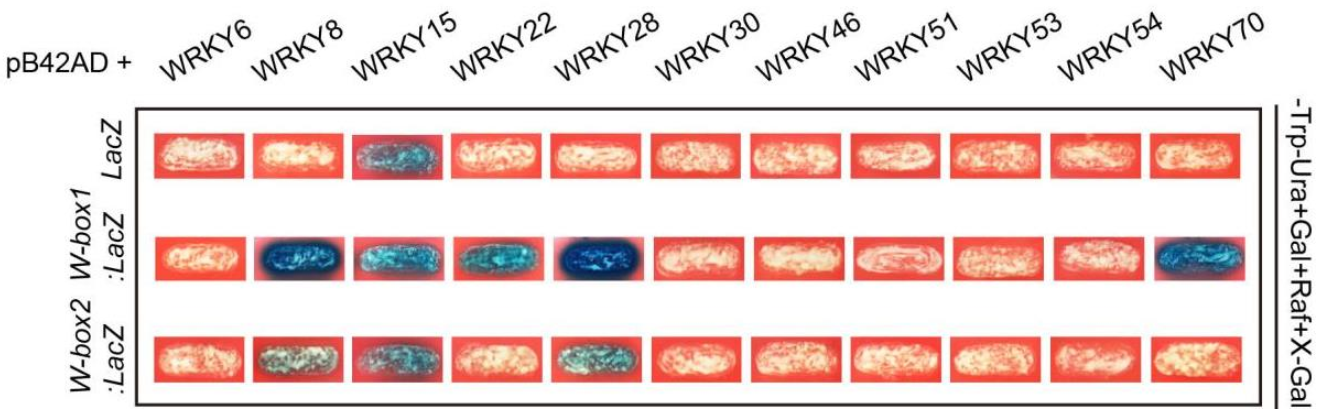
2.9. Identification of Defense and Senescence Related WRKYs Binding to the Promoter of AtWAKL10
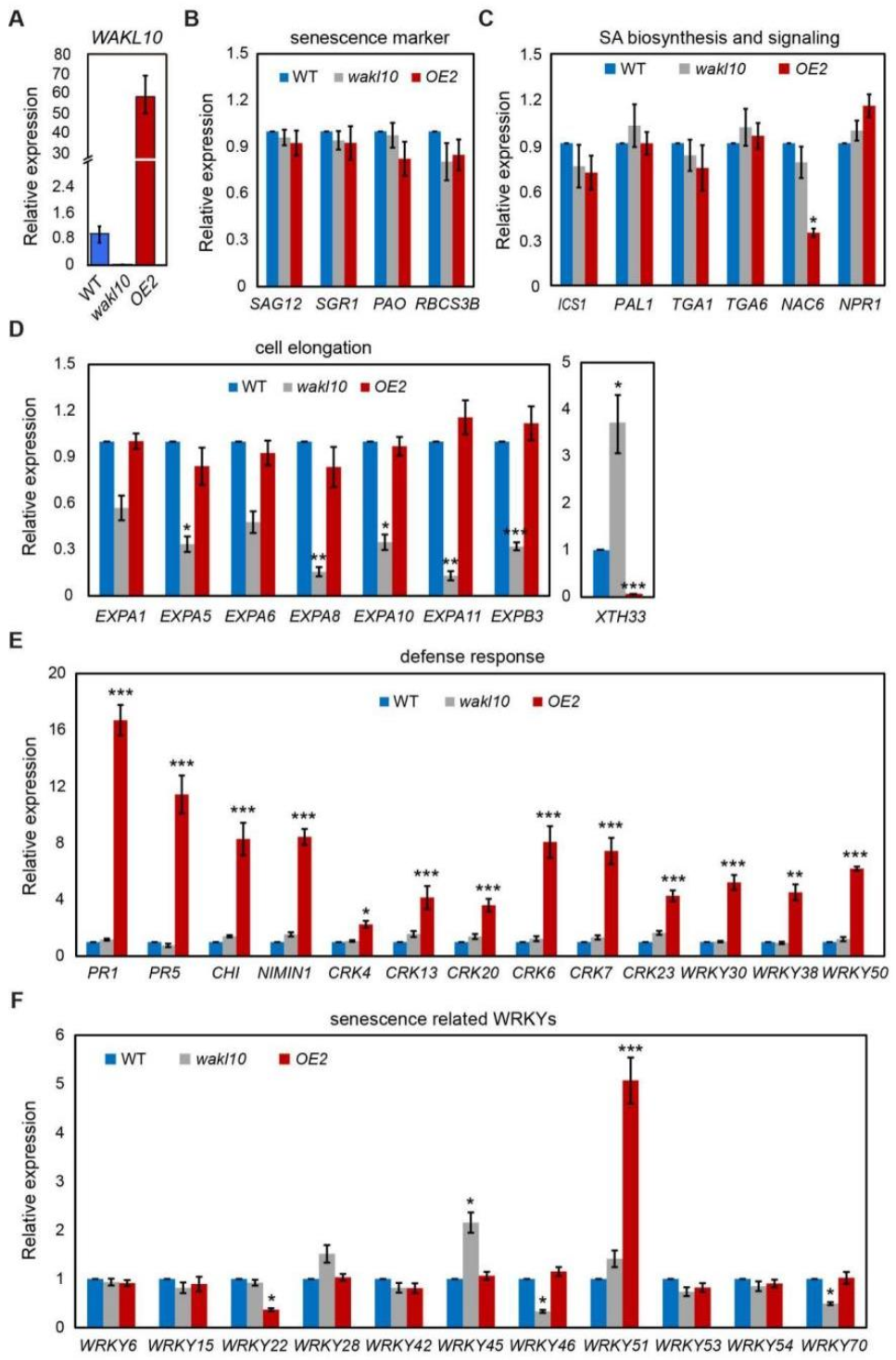
2.10. A Brief Analysis of Potential Transcriptional Changes Specifically Mediated by AtWAKL10 during Natural Leaf Senescence
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Developmental Expression and Promoter Analysis
4.3. RNA Extraction and Quantitative RT-PCR
4.4. Generation of AtWAKL10 Overexpression Lines (OEs) and AtWAKL10::GUS Lines
4.5. Subcellular Localization
4.6. Yeast One-Hybrid Analysis
4.7. Measurement of Chlorophyll Content, Fv/Fm and Ion Leakage
4.8. Dark-Induced Senescence
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gan, S.; Amasino, R.M. Making Sense of Senescence (Molecular Genetic Regulation and Manipulation of Leaf Senescence). Plant Physiol. 1997, 113, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Kim, H.J.; Gil Nam, H. Leaf Senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed]
- Himelblau, E.; Amasino, R.M. Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J. Plant Physiol. 2001, 158, 1317–1323. [Google Scholar] [CrossRef]
- Humbeck, K.; Quast, S.; Krupinska, K. Functional and molecular changes in the photosynthetic apparatus during senescence of flag leaves from field-grown barley plants. Plant Cell Environ. 1996, 19, 337–344. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Earl, S.; Harrison, E.; Mathas, E.; Navabpour, S.; Page, T.; Pink, D. The molecular analysis of leaf senescence—A genomics approach. Plant Biotechnol. J. 2003, 1, 3–22. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, J.H.; Gil Nam, H.; Lim, P.O. The Delayed Leaf Senescence Mutants of Arabidopsis, ore1, ore3, and ore9 are Tolerant to Oxidative Stress. Plant Cell Physiol. 2004, 45, 923–932. [Google Scholar] [CrossRef]
- Ougham, H.J.; Hörtensteiner, S.; Armstead, I.P.; Donnison, I.S.; King, I.P.; Thomas, H.; Mur, L.A.J. The control of chlorophyll catabolism and the status of yellowing as a biomarker of leaf senescence. Plant Biol. 2008, 10, 4–14. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. Leaf Senescence: Signals, Execution, and Regulation. Curr. Top. Dev. Biol. 2005, 71, 83–112. [Google Scholar]
- Hopkins, M.; Taylor, C.; Liu, Z.; Ma, F.; McNamara, L.; Wang, T.-W.; Thompson, J.E. Regulation and execution of molecular disassembly and catabolism during senescence. New Phytol. 2007, 175, 201–214. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V. The molecular biology of leaf senescence. J. Exp. Bot. 1997, 48, 181–199. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Gil Nam, H.; Lin, J.-F.; Wu, S.-H.; Swidzinski, J.; Ishizaki, K.; et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef]
- Liebsch, D.; Keech, O. Dark-induced leaf senescence: New insights into a complex light-dependent regulatory pathway. New Phytol. 2016, 212, 563–570. [Google Scholar] [CrossRef]
- Weaver, L.M.; Amasino, R.M. Senescence is induced in individually darkened Arabidopsis leaves but inhibited in whole darkened plants. Plant Physiol. 2001, 127, 876–886. [Google Scholar] [CrossRef]
- Zhang, K.; Gan, S.-S. An Abscisic Acid-AtNAP Transcription Factor-SAG113 Protein Phosphatase 2C Regulatory Chain for Controlling Dehydration in Senescing Arabidopsis Leaves. Plant Physiol. 2012, 158, 961–969. [Google Scholar] [CrossRef]
- Sarwat, M.; Naqvi, A.R.; Ahmad, P.; Ashraf, M.; Akram, N.A. Phytohormones and microRNAs as sensors and regulators of leaf senescence: Assigning macro roles to small molecules. Biotechnol. Adv. 2013, 31, 1153–1171. [Google Scholar] [CrossRef]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.-S.; Penfold, C.A.; Jenkins, D.; et al. High-Resolution Temporal Profiling of Transcripts during Arabidopsis Leaf Senescence Reveals a Distinct Chronology of Processes and Regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence Supporting a Role of Jasmonic Acid in Arabidopsis Leaf Senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef]
- Van Der Graaff, E.; Schwacke, R.; Schneider, A.; DeSimone, M.; Flügge, U.-I.; Kunze, R. Transcription Analysis of Arabidopsis Membrane Transporters and Hormone Pathways during Developmental and Induced Leaf Senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A Novel Nuclear-Localized CCCH-Type Zinc Finger Protein, OsDOS, Is Involved in Delaying Leaf Senescence in Rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef]
- Pruneda-Paz, J.L.; Breton, G.; Nagel, D.H.; Kang, S.E.; Bonaldi, K.; Doherty, C.J.; Ravelo, S.; Galli, M.; Ecker, J.R.; Kay, S.A. A Genome-Scale Resource for the Functional Characterization of Arabidopsis Transcription Factors. Cell Rep. 2014, 8, 622–632. [Google Scholar] [CrossRef]
- Jiang, H.; Li, M.; Liang, N.; Yan, H.; Wei, Y.; Xu, X.; Liu, J.; Xu, Z.; Chen, F.; Wu, G. Molecular cloning and function analysis of the stay green gene in rice. Plant J. Cell Mol. Biol. 2007, 52, 197–209. [Google Scholar] [CrossRef]
- Yang, J.; Worley, E.; Udvardi, M. A NAP-AAO3 Regulatory Module Promotes Chlorophyll Degradation via ABA Biosynthesis in Arabidopsis Leaves. Plant Cell 2014, 26, 4862–4874. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Han, S.-H.; Lee, S.-H.; Hörtensteiner, S.; Paek, N.-C. Arabidopsis NAC016 promotes chlorophyll breakdown by directly upregulating STAYGREEN1 transcription. Plant Cell Rep. 2015, 35, 155–166. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4-and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Robatzek, S.; Somssich, I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149. [Google Scholar] [CrossRef]
- Robatzek, S.; Somssich, I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001, 28, 123–133. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Wang, X.; Wang, J.; Fan, K.; Li, Z.; Lin, W. DELLA proteins negatively regulate dark-induced senescence and chlorophyll degradation in Arabidopsis through interaction with the transcription factor WRKY6. Plant Cell Rep. 2018, 37, 981–992. [Google Scholar] [CrossRef]
- Zhang, D.; Zhu, Z.; Gao, J.; Zhou, X.; Zhu, S.; Wang, X.; Wang, X.; Ren, G.; Kuai, B. The NPR1-WRKY46-WRKY6 signaling cascade mediates probenazole/salicylic acid-elicited leaf senescence in Arabidopsis thaliana. J. Integr. Plant Biol. 2020. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.-M. Receptor Kinases in Plant-Pathogen Interactions: More Than Pattern Recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed]
- Cano-Delgado, A.; Penfield, S.; Smith, C.; Catley, M.; Bevan, M. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 2003, 34, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Gish, L.A.; Clark, S.E. The RLK/Pelle family of kinases. Plant J. 2011, 66, 117–127. [Google Scholar] [CrossRef] [PubMed]
- He, Z.H.; Cheeseman, I.; He, D.Z.; Kohorn, B.D. A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol. Biol. 1999, 39, 1189–1196. [Google Scholar] [CrossRef]
- Verica, J.A.; He, Z.-H. The Cell Wall-Associated Kinase (WAK) and WAK-Like Kinase Gene Family. Plant Physiol. 2002, 129, 455–459. [Google Scholar] [CrossRef]
- Wagner, T.A.; Kohorn, B.D. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 2001, 13, 303–318. [Google Scholar] [CrossRef]
- Lally, D.; Ingmire, P.; Tong, H.Y.; He, Z.H. Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 2001, 13, 1317–1331. [Google Scholar]
- He, Z.; He, D.; Kohorn, B.D. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 1998, 14, 55–63. [Google Scholar] [CrossRef]
- Maleck, K.; Levine, A.; Eulgem, T.; Morgan, A.; Schmid, J.; Lawton, K.A.; Dangl, J.L.; Dietrich, R.A. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 2000, 26, 403–410. [Google Scholar] [CrossRef]
- Schenk, P.M.; Kazan, K.; Wilson, I.; Anderson, J.P.; Richmond, T.; Somerville, S.C.; Manners, J.M. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 2000, 97, 11655–11660. [Google Scholar] [CrossRef]
- Meier, S.; Ruzvidzo, O.; Morse, M.; Donaldson, L.; Kwezi, L.; Gehring, C.A. The Arabidopsis Wall Associated Kinase-Like 10 Gene Encodes a Functional Guanylyl Cyclase and Is Co-Expressed with Pathogen Defense Related Genes. PLoS ONE 2010, 5, e8904. [Google Scholar] [CrossRef]
- Newton, R.P.; Smith, C.J. Cyclic nucleotides. Phytochemistry 2004, 65, 2423–2437. [Google Scholar] [CrossRef]
- Bot, P.; Mun, B.-G.; Imran, Q.M.; Hussain, A.; Lee, S.-U.; Loake, G.; Yun, B.-W. Differential expression of AtWAKL10 in response to nitric oxide suggests a putative role in biotic and abiotic stress responses. PeerJ 2019, 7, e7383. [Google Scholar] [CrossRef]
- Decreux, A.; Messiaen, J. Wall-associated Kinase WAK1 Interacts with Cell Wall Pectins in a Calcium-induced Conformation. Plant Cell Physiol. 2005, 46, 268–278. [Google Scholar] [CrossRef]
- Decreux, A.; Thomas, A.; Spies, B.; Brasseur, R.; Cutsem, P.; Messiaen, J. In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry 2006, 67, 1068–1079. [Google Scholar] [CrossRef]
- Zhang, K.; Xia, X.; Zhang, Y.; Gan, S.-S. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 2011, 69, 667–678. [Google Scholar] [CrossRef]
- Feller, U.; Fischer, A. Nitrogen metabolism in senescing leaves. Crit. Rev. Plant Sci. 1994, 13, 241–273. [Google Scholar] [CrossRef]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.-M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Ndamukong, I.; Chetram, A.; Saleh, A.; Avramova, Z. Wall-modifying genes regulated by the Arabidopsis homolog of trithorax, ATX1: Repression of the XTH33 gene as a test case. Plant J. 2009, 58, 541–553. [Google Scholar] [CrossRef]
- Divol, F.; Vilaine, F.; Thibivilliers, S.; Kusiak, C.; Sauge, M.H.; Dinant, S. Involvement of the xyloglucan endotransglycosylase/hydrolases encoded by celery XTH1 and Arabidopsis XTH33 in the phloem response to aphids. Plant Cell Environ. 2007, 30, 187–201. [Google Scholar] [CrossRef]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 Interacts with the DELLA Protein RGL1 to Positively Regulate Age-Triggered Leaf Senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef]
- Zhou, X.; Jiang, Y.; Yu, D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 2011, 31, 303–313. [Google Scholar] [CrossRef]
- Diévart, A.; Clark, S.E. Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 2003, 6, 507–516. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, D.; Xu, Y.; Jin, S.; Zhang, L.; Zhang, S.; Yang, G.; Huang, J.; Yan, K.; Wu, C.; et al. CEPR2 phosphorylates and accelerates the degradation of PYR/PYLs in Arabidopsis. J. Exp. Bot. 2019, 70, 5457–5469. [Google Scholar] [CrossRef]
- Chen, L.-J.; Wuriyanghan, H.; Zhang, Y.-Q.; Duan, K.-X.; Chen, H.-W.; Li, Q.-T.; Lu, X.; He, S.-J.; Ma, B.; Zhang, W.-K.; et al. An S-Domain Receptor-Like Kinase, OsSIK2, Confers Abiotic Stress Tolerance and Delays Dark-Induced Leaf Senescence in Rice. Plant Physiol. 2013, 163, 1752–1765. [Google Scholar] [CrossRef]
- Xiao, D.; Cui, Y.; Xu, F.; Xu, X.; Gao, G.; Wang, Y.; Guo, Z.; Wang, D.; Wang, N.N. Senescence-Suppressed Protein Phosphatase Directly Interacts with the Cytoplasmic Domain of Senescence-Associated Receptor-Like Kinase and Negatively Regulates Leaf Senescence in Arabidopsis. Plant Physiol. 2015, 169, 1275–1291. [Google Scholar] [CrossRef]
- Lee, I.C.; Hong, S.W.; Whang, S.S.; Lim, P.O.; Gil Nam, H.; Koo, J.C. Age-Dependent Action of an ABA-Inducible Receptor Kinase, RPK1, as a Positive Regulator of Senescence in Arabidopsis Leaves. Plant Cell Physiol. 2011, 52, 651–662. [Google Scholar] [CrossRef]
- Li, X.; Ahmad, S.; Ali, A.; Guo, C.; Li, H.; Yu, J.; Zhang, Y.; Gao, X.; Guo, Y. Characterization of Somatic Embryogenesis Receptor-Like Kinase 4 as a Negative Regulator of Leaf Senescence in Arabidopsis. Cells 2019, 8, 50. [Google Scholar] [CrossRef]
- Li, X.-P.; Gan, R.; Li, P.-L.; Ma, Y.-Y.; Zhang, L.-W.; Zhang, R.; Wang, Y.; Wang, N.N. Identification and functional characterization of a leucine-rich repeat receptor-like kinase gene that is involved in regulation of soybean leaf senescence. Plant Mol. Biol. 2006, 61, 829–844. [Google Scholar] [CrossRef]
- Hou, X.; Tong, H.; Selby, J.; DeWitt, J.; Peng, X.; He, Z.-H. Involvement of a Cell Wall-Associated Kinase, WAKL4, in Arabidopsis Mineral Responses. Plant Physiol. 2005, 139, 1704–1716. [Google Scholar] [CrossRef] [PubMed]
- Verica, J.A.; Chae, L.; Tong, H.; Ingmire, P.; He, Z.-H. Tissue-Specific and Developmentally Regulated Expression of a Cluster of Tandemly Arrayed Cell Wall-Associated Kinase-Like Kinase Genes in Arabidopsis. Plant Physiol. 2003, 133, 1732–1746. [Google Scholar] [CrossRef] [PubMed]
- Larkan, N.J.; Ma, L.; Haddadi, P.; Buchwaldt, M.; Parkin, I.A.P.; Djavaheri, M.; Borhan, M.H. The Brassica napus wall-associated kinase-like (WAKL) gene Rlm9 provides race-specific blackleg resistance. Plant J. Cell Mol. Biol. 2020, 104, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.E.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S.-S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2011, 35, 644–655. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Holm, P.B. Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol. J. 2007, 5, 192–206. [Google Scholar] [CrossRef]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef]
- Ülker, B.; Mukhtar, M.S.; Somssich, I.E. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 2007, 226, 125–137. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nat. Cell Biol. 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Van Verk, M.C.; Bol, J.F.; Linthorst, H.J. WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 2011, 11, 89. [Google Scholar] [CrossRef]
- Tian, T.; Ma, L.; Liu, Y.; Xu, D.; Chen, Q.; Li, G. Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 Integrates Age and Light Signals to Negatively Regulate Leaf Senescence. Plant Cell 2020, 32, 1574–1588. [Google Scholar] [CrossRef]
- Hinckley, W.E.; Brusslan, J.A. Gene expression changes occurring at bolting time are associated with leaf senescence in Arabidopsis. Plant Direct 2020, 4, e00279. [Google Scholar] [CrossRef]
- Morris, K.; Mackerness, S.A.-H.; Page, T.; John, C.F.; Murphy, A.M.; Carr, J.P.; Buchanan-Wollaston, V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 2000, 23, 677–685. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54 and WRKY70 Transcription Factors are Involved in Brassinosteroid-Regulated Plant Growth and Drought Response. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef]
- Li, J.; Besseau, S.; Toronen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, E.T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013, 200, 455–472. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 Transcription Factor: A Node of Convergence for Jasmonate-Mediated and Salicylate-Mediated Signals in Plant Defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef]
- Li, J.; Zhong, R.; Palva, E.T. WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLoS ONE 2017, 12, e0183731. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar] [CrossRef]
- Ishihama, N.; Yamada, R.; Yoshioka, M.; Katou, S.; Yoshioka, H. Phosphorylation of the Nicotiana benthamiana WRKY8 Transcription Factor by MAPK Functions in the Defense Response. Plant Cell 2011, 23, 1153–1170. [Google Scholar] [CrossRef]
- Kim, S.-H.; Lee, J.-H.; Seo, K.-I.; Ryu, B.; Sung, Y.; Chung, T.; Deng, X.W.; Lee, J.-H. Characterization of a Novel DWD protein that participates in heat stress response in Arabidopsis. Mol. Cells 2014, 37, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.-C.; Chou, M.-Y.; Chou, S.-J.; Li, Y.-R.; Peng, H.-P.; Shih, M.-C. Submergence Confers Immunity Mediated by the WRKY22 Transcription Factor in Arabidopsis. Plant Cell 2013, 25, 2699–2713. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.-J.; Lai, Z.; Lee, S.; Yun, D.J.; Mengiste, T. Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell 2016, 28, 1662–1681. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.; Cui, X.; Zhao, P.; Sun, M.; Yang, B.; Deyholos, M.K.; Li, Y.; Zhao, X.; Jiang, Y.-Q. WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesis in Arabidopsis thaliana. Plant J. 2020, 104, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Li, Z.; Huang, P.; Li, B.; Fang, S.; Chu, J.; Guo, H. A Tripartite Amplification Loop Involving the Transcription Factor WRKY75, Salicylic Acid, and Reactive Oxygen Species Accelerates Leaf Senescence. Plant Cell 2017, 29, 2854–2870. [Google Scholar] [CrossRef]
- Pasternak, T.; Groot, E.P.; Kazantsev, F.V.; Teale, W.; Omelyanchuk, N.; Kovrizhnykh, V.; Palme, K.; Mironova, V.V. Salicylic Acid Affects Root Meristem Patterning via Auxin Distribution in a Concentration-Dependent Manner. Plant Physiol. 2019, 180, 1725–1739. [Google Scholar] [CrossRef]
- Chai, J.; Liu, J.; Zhou, J.; Xing, D. Mitogen-activated protein kinase 6 regulates NPR1 gene expression and activation during leaf senescence induced by salicylic acid. J. Exp. Bot. 2014, 65, 6513–6528. [Google Scholar] [CrossRef]
- Goh, H.-H.; Sloan, J.; Dorca-Fornell, C.; Fleming, A. Inducible Repression of Multiple Expansin Genes Leads to Growth Suppression during Leaf Development. Plant Physiol. 2012, 159, 1759–1770. [Google Scholar] [CrossRef]
- Cho, H.T.; Kende, H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 1997, 9, 1661–1671. [Google Scholar]
- Cho, H.-T.; Cosgrove, D.J. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 9783–9788. [Google Scholar] [CrossRef]
- Zhao, M.-R.; Li, F.; Fang, Y.; Gao, Q.; Wang, W. Expansin-regulated cell elongation is involved in the drought tolerance in wheat. Protoplasma 2011, 248, 313–323. [Google Scholar] [CrossRef]
- Qin, L.; Kudla, U.; Roze, E.H.A.; Goverse, A.; Popeijus, H.; Nieuwland, J.; Overmars, H.; Jones, J.T.; Schots, A.; Smant, G.; et al. Plant degradation: A nematode expansin acting on plants. Nature 2004, 427, 30. [Google Scholar] [CrossRef]
- Tan, J.; Wang, M.; Shi, Z.; Miao, X. OsEXPA10 mediates the balance between growth and resistance to biotic stress in rice. Plant Cell Rep. 2018, 37, 993–1002. [Google Scholar] [CrossRef]
- Ju, Y.-L.; Min, Z.; Yue, X.-F.; Zhang, Y.-L.; Zhang, J.-X.; Zhang, Z.-Q.; Fang, Y.-L. Overexpression of grapevine VvNAC08 enhances drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 151, 214–222. [Google Scholar] [CrossRef]
- Collin, A.; Daszkowska-Golec, A.; Kurowska, M.; Szarejko, I. Barley ABI5 (Abscisic Acid INSENSITIVE 5) is Involved in Abscisic Acid-Dependent Drought Response. Front. Plant Sci. 2020, 11, 1138. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, X.; Wang, C.; Jing, R. Overexpression of a Common Wheat Gene TaSnRK2.8 Enhances Tolerance to Drought, Salt and Low Temperature in Arabidopsis. PLoS ONE 2010, 5, e16041. [Google Scholar] [CrossRef]
- Xu, X.; Wan, W.; Jiang, G.; Xi, Y.; Huang, H.; Cai, J.; Chang, Y.; Duan, C.-G.; Mangrauthia, S.K.; Peng, X.; et al. Nucleocytoplasmic Trafficking of the Arabidopsis WD40 Repeat Protein XIW1 Regulates ABI5 Stability and Abscisic Acid Responses. Mol. Plant 2019, 12, 1598–1611. [Google Scholar] [CrossRef]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Kinkema, M.; Fan, W.; Dong, X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 2000, 12, 2339–2350. [Google Scholar] [CrossRef]
- Despres, C.; DeLong, C.; Glaze, S.; Liu, E.; Fobert, P.R. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 2000, 12, 279–290. [Google Scholar] [CrossRef]
- Wang, C.; Dai, S.; Zhang, Z.; Lao, W.; Wang, R.; Meng, X.; Zhou, X. Ethylene and salicylic acid synergistically accelerate leaf senescence in Arabidopsis. J. Integr. Plant Biol. 2021, 2021. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Sheludko, Y.; Sindarovska, Y.; Gerasymenko, I.; Bannikova, M.; Kuchuk, N. Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression. Biotechnol. Bioeng. 2006, 96, 608–614. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and Carotenoids: Pigments of Photosynthetic Biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Rossel, J.B.; Walter, P.B.; Hendrickson, L.; Chow, W.S.; Poole, A.; Mullineaux, P.M.; Pogson, B.J. A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 2006, 29, 269–281. [Google Scholar] [CrossRef]
- Li, L.; Kubiszewski-Jakubiak, S.; Radomiljac, J.; Wang, Y.; Law, S.R.; Keech, O.; Narsai, R.; Berkowitz, O.; Duncan, O.; Murcha, M.W.; et al. Characterization of a novel beta-barrel protein (AtOM47) from the mitochondrial outer membrane of Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6061–6075. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Li, K.; Ali, A.; Guo, Y. AtWAKL10, a Cell Wall Associated Receptor-Like Kinase, Negatively Regulates Leaf Senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 4885. https://doi.org/10.3390/ijms22094885
Li L, Li K, Ali A, Guo Y. AtWAKL10, a Cell Wall Associated Receptor-Like Kinase, Negatively Regulates Leaf Senescence in Arabidopsis thaliana. International Journal of Molecular Sciences. 2021; 22(9):4885. https://doi.org/10.3390/ijms22094885
Chicago/Turabian StyleLi, Lu, Kui Li, Akhtar Ali, and Yongfeng Guo. 2021. "AtWAKL10, a Cell Wall Associated Receptor-Like Kinase, Negatively Regulates Leaf Senescence in Arabidopsis thaliana" International Journal of Molecular Sciences 22, no. 9: 4885. https://doi.org/10.3390/ijms22094885
APA StyleLi, L., Li, K., Ali, A., & Guo, Y. (2021). AtWAKL10, a Cell Wall Associated Receptor-Like Kinase, Negatively Regulates Leaf Senescence in Arabidopsis thaliana. International Journal of Molecular Sciences, 22(9), 4885. https://doi.org/10.3390/ijms22094885





