Abstract
Anatomical and electrophysiological evidence that gap junctions and electrical coupling occur between neurons was initially confined to invertebrates and nonmammals and was thought to be a primitive form of synaptic transmission. More recent studies revealed that electrical communication is common in the mammalian central nervous system (CNS), often coexisting with chemical synaptic transmission. The subsequent progress indicated that electrical synapses formed by the gap junction protein connexin-36 (Cx36) and its paralogs in nonmammals constitute vital elements in mammalian and fish synaptic circuitry. They govern the collective activity of ensembles of coupled neurons, and Cx36 gap junctions endow them with enormous adaptive plasticity, like that seen at chemical synapses. Moreover, they orchestrate the synchronized neuronal network activity and rhythmic oscillations that underlie the fundamental integrative processes, such as memory and learning. Here, we review the available mechanistic evidence and models that argue for the essential roles of calcium, calmodulin, and the Ca2+/calmodulin-dependent protein kinase II in integrating calcium signals to modulate the strength of electrical synapses through interactions with the gap junction protein Cx36.
1. Introduction
The transmission of signals from one neuron to another is primarily via the release of chemicals from the upstream neuron that activates receptors on the downstream neuron. From electron micrographs of close appositions between pre- and postsynaptic elements of the fish brain, JD Robertson proposed the existence of electrical transmission [1], which was demonstrated through electrophysiology recordings (for review, see [2]). Although it was generally believed that so-called electrical or electrotonic synapses were restricted to invertebrates and lower vertebrate nervous systems, it is now clear that they are present in higher mammals, where they are often found in association with chemical synapses (forming “mixed synapses”) and play essential roles in synchronizing inhibitory interneurons, among other functions [3,4]. The molecular identity of the electrical synaptic protein was initially provided from cDNA libraries of skate retina, which predicted a 35-kDa protein that was named, by convention, Cx35 [5]. Two years later, using the inferior olive as the source material, a new rodent connexin was cloned and subsequently called connexin-36 (Cx36) [6]; this protein had 95% sequence identity to skate Cx35. Condorelli et al. demonstrated a strong mRNA expression in neurons of the inferior olive, the olfactory bulb, the CA3/CA4 hippocampal subfields, brainstem nuclei of a rat brain, and in the retinal ganglion cell and inner nuclear layers. It was the first connexin found as predominantly expressed in mammalian neurons, and the only other tissue in which it is abundant is the endocrine pancreas [7]. The identification of rat Cx36 rapidly led to the cloning of other mammalian isoforms of Cx36 [8], of a group of related connexins in fish [9,10] and in other vertebrates [11]. Following an initial characterization, it became apparent that Cx36/35 constitutes a novel delta subgroup of connexins. As was predicted by Condorelli et al., molecular cloning of the primary neuronal connexin has enabled the study of its role in the physiology and the pathology of the mammalian brain. In this review, we examine the knowledge gains made in the last few decades. Specifically, we will focus on the emerging role of Cx36 and the crucial interaction with calcium-activated calmodulin (CaM) and calmodulin-dependent protein kinase II (CaMKII) partners in the plasticity of electrical synapses.
2. Gap Junction Proteins Forming Electrical Synapses between Neurons and Their Interactions with Other Proteins
The initial characterization of mammalian and teleost Cx36/Cx35 genes showed that the coding sequence is highly conserved. As shown in the amino acid sequence alignment (Supplementary Figure S1), human Cx36 has 98% identity at the protein level with the mouse and rat Cx36 and about 70–80% with the ortholog perch, skate, and the zebrafish Cx35b proteins [5,9,10]. The gene structure of mammalian Cx36 was somewhat unusual for connexins; the protein-coding region is interrupted by a single intron of variable length, separating the coding region 71 bp after the translation initiation site. Where there is a single Cx36 protein in mammals, genome duplications in the teleost lineage give rise to multiple paralogs in the zebrafish [12,13]. Each ortholog is presented by two coding exons in all vertebrates, with separating introns of variable lengths (Figure 1).
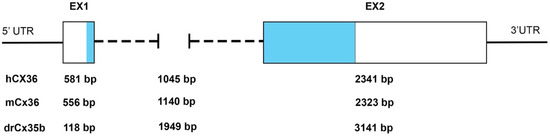
Figure 1.
Organization of the exon–intron structure of Cx35/36 orthologs. Examples of the variable length of Cx35/36 exons (Ex) and introns. The relative position of the protein-coding regions is indicated in light blue. Abbreviations: UTR, untranslated region: hCX36/GJD2, mCx36/Gjd2, and drCx35b/gjd2b. Data source (ENSEMBL releases GRCh38.p13 (human), GRCm39 (mouse), and GRCz11 (zebrafish)).
Mammalian Cx36 and relatives are expressed in neurons and cells of ectodermic origin, such as pancreatic beta-cells [7], microglia cells [14], and chromaffin cells [15]. Although the Cx36 regulatory sequences are not well-characterized, the presence of neuron-restrictive silencer elements (NRSE) in the DNA sequence elements of the mammalian and zebrafish promoter region upstream of the coding region has been taken as a reason to explain why Cx36 is specifically expressed in neurons and in pancreatic beta-cells. NRSEs bind NRSF/REST-silencing transcription factors, thereby repressing the transcriptional activity of neural genes in non-neuronal cells. Cx36 gene expression in insulin-producing beta-cell lines is strictly controlled by the transcriptional repressor NRSF/REST [15].
During development, rodent Cx36 is dynamically expressed over the course of embryonic neurogenesis (for reference, see Supplementary Table S1). Cx36 localizes to the ventricular zone (VZ) during the first wave of neurogenesis [16]. After birth, the Cx36 expression is dramatically reduced during postnatal maturation but remains enriched in subsets of principal neurons and interneurons in the inferior olive, hippocampus, olfactory bulb, cerebellum, and thalamus and is sparsely expressed in the cortex. While reports on Cx36 mRNA expression in rodents’ embryonic brains are detailed, only one study has a reported Cx35 expression in embryonic and larval zebrafish [17]. The authors described Cx35 immunoreactivity in the developing brain using a monoclonal antibody raised against perch Cx35. In whole-mount larval zebrafish, from 1 to 15 dpf Cx35, immunoreactivity was detected in the developing cerebellum, olfactory bulb, habenula, or the hindbrain.
In the adult mammalian nervous system, the distribution of Cx36 mRNA examined by in situ hybridization was most intense in the inferior olivary complex, both in principal and accessory nuclei (for reference, see Supplementary Table S2). Moderate labeling was also observed in several myelencephalic nuclei, in specific cells of the cerebellar cortex, in a relatively large subpopulation of cells in the cerebral cortex, in the hilus of the dentate gyrus, and in the strata radiatum and oriens of the hippocampal subfields. Moreover, labeled cells were revealed in all the lamina of the spinal cord gray matter.
After anti-Cx36 antibodies became available in the early 2000s, rapid advances were made complementing and supplanting the initial RNA-based data. Work from multiple groups demonstrated a widespread localization of the Cx36 protein in major divisions of the nervous system. For example, Cx36 was found in the cerebellar cortex, cerebellum, hippocampus, thalamus, the spinal cord, and trigeminal and dorsal root ganglia of the peripheral nervous system [18,19,20,21,22,23,24]. Cx36 was also discovered in the neurons of the autonomic nervous system [25,26,27]. In sensory systems, Cx36 is expressed in the retina [28,29,30], olfactory [31,32], and auditory systems [33,34,35,36]. Cx36 was found in both the interneurons and excitatory neurons [37]. Advances in high-resolution imaging determined the localization of Cx36 in axo-axonal, axo-dendritic, and dendrodentric contact sites [38]. In confirmed cases, ionotropic and metabotropic NMDA receptors were found in close proximity, suggesting a functional relationship in the post-synapse [23]. Cx36 was also found in presynaptic contacts at mixed synapses of afferent terminals in the spinal cord [39] or hippocampal mossy fibers [40]. The Nagy group recently identified Cx36 at the axon initial segments of neurons in the spinal cord, inferior olive, and cerebral cortex [41].
Ultrastructural imaging studies in the 1960s and the 1970s described that gap junctions are found near electron-dense structures. Sotelo et al. (1974) described in their studies on the inferior olive of cats a specialized junctional zone that was named an attachment plate [42]. This structure, sometimes called Nexus or electrical synapse density (ESD), resembles the postsynaptic density found at chemical synapses. Although no comprehensive OMICS study of the molecular components of ESDs has been reported yet, proteins interacting with Cx36 have been identified. Broadly, these proteins fall into the following classes: cytoskeletal/transport (tubulin [43]); anchoring (ZO1 family, AF6, and MUPP1 [44,45,46,47,48]); or kinases (PKA and CaMKII [49,50,51,52,53]); binding proteins (CaM [54,55]); and proteins that retrieve Cx36 from the gap junction plaque [56,57]. Although the rich diversity of the molecules that assemble at electrical synapses parallel the diversity of the proteins involved in forming pre- and postsynaptic compartments at chemical synapses, the molecular and functional relationships of these proteins in mixed synaptic communication have yet to be resolved. In general, the activity of the Cx36 channels appears closely regulated, notably by posttranslational modifications such as phosphorylation. Protein kinases such as PKA and CaMKII appear to regulate the gap junction at several levels, including the assembly of channels in the plasma membrane, connexin turnover, and directly affecting the opening and closure (“gating”) of the channels. Further, Cx36 subunits bind to auxiliary proteins that play essential roles in channel localization and activity or form networks in which these proteins frequently anchor to the cytoskeleton.
The protein complexes at the chemical and electrical synapses share the molecular machinery involving CaMKII and CaM that achieves functional plasticity [53]. Cx36, CaM, and CaMKII can all be found in both pre- and postsynaptic compartments, from which it might be assumed that electrical synapses are functionally symmetrical. However, the presence of other proteins can lead to an exciting asymmetry across the synapse. For example, the localization of NMDA receptors, a major source of an activity-dependent influx of calcium, in postsynaptic compartments suggests that the Cx36 signaling complex in the CNS neurons is not a simple mirror image across the gap junction plaque. Additionally, axonal or dendritic transport utilizes distinct mechanisms. Together, there is growing evidence for functional asymmetry potentially arising from differences in posttranslational modifications of individual Cx36 proteins [49] by asymmetrically organized signaling complexes of Cx36-associated proteins found at the ESD [44,47,58], as well as by asymmetric properties of pre- and postsynaptic membranes (for review, see [59,60]).
3. Cx36 Interaction with Calmodulin Connects Calcium Signals with Gap Junction Communication
The importance of calmodulin-regulated gap junction communication has been reviewed recently [61,62,63]. In the nervous system, calcium (Ca2+) is an essential ligand that binds its primary intracellular receptor calmodulin (CaM) to trigger multiple downstream processes and pathways in both pre- and postsynaptic compartments. CaM binding to GST–Cx36 fusion proteins was first demonstrated for mammalian Cx36 and fish orthologs and involved the C-terminal domains [54,55] (Figure 2a). Recently, a second CaM-binding site was found in the cytoplasmic loop of the perch Cx35. This site was not analyzed in detail after showing weak binding characteristics and rapid kinetics in surface plasmon resonance experiments [64]. The in vitro binding and dissociation kinetics were in the µM concentration range and Ca2+-dependent. The rapid on and off binding rates implies that the interaction of Cx36 with CaM may change dynamically in neurons when transient increases of Ca2+ occur in or near the activated synapses. Siu et al. [54] demonstrated in Neuro2a cells that Ca2+-loaded CaM binds a site in the carboxy-terminus of Cx36 overlapping with a previously identified CaMKII-binding site [52,53]. This extended the findings by Burr et al. [55], who argued that the CaM-binding site’s characteristics might confer sensitivity to the competition with other proteins with higher CaM affinities.
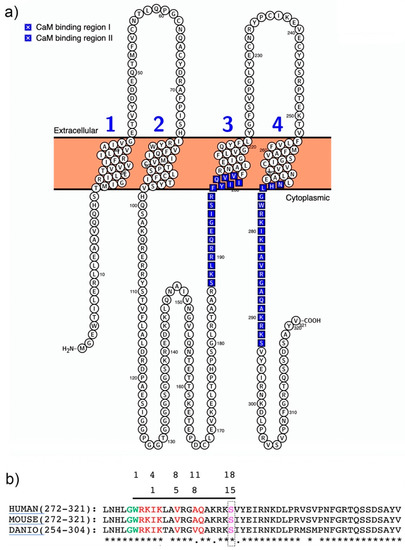
Figure 2.
Structure of the Cx36–CaM-binding motifs. (a) Visualization of the predicted murine Cx36 structure using Protter software [73]. The CaM-binding regions I and II are indicated in blue. (b) Protein sequence alignment of the carboxyterminal domains of mammalian and zebrafish (Cx35b). The sequence homology and the two overlapping CaM-binding motifs described by [54,64] are highlighted in light green and red. The PKA phosphorylation site in position S293 (pink) is located at the boundary of the core CaM-binding motif [50].
It can be reasoned that local differences in the intracellular calcium concentrations open up ample possibilities for the spatial and temporal sequestering of interactions between Cx36 and CaM. This was demonstrated in Neuro2a cells, where Cx36 was able to bind CaM outside of the gap junction plaque in the ER/Golgi complex [54]. Similarly, another connexin, Cx32, interacts with CaM before GJs are formed [65]. The binding of Cx proteins to CaM has diverse functional outcomes. In Neuro2a cells, Siu et al. (2016) showed that Ca2+-loaded CaM increased the coupling across GJs formed by Cx36 channels. This outcome is different from other connexins closed after binding [66,67]. The NMR solution structure demonstrated that CaM binds Cx36 in a compact state with significant hydrophobic contributions from amino acid W277 at anchor position 1 and V284 at position 8 of Cx36 [54] (Figure 2b). Different functional outcomes of Cx36 binding to CaM were reported by Aseervatham et al. [64] in Hela cells expressing Cx35, where CaM binding induced Ca2+-mediated uncoupling. The opposite effect on the coupling is surprising, since both studies used site-directed mutagenesis and the pharmacological blocking of CaM. In general, Hela cells and Neuro2a cells are not thought to express significant amounts of endogenous connexins or form gap junctions. However, coupling in HeLa cells is documented [68,69], which could confound the interpretation of the results. It stands to be demonstrated whether the different intracellular environments and degrees of endogenous gap junction coupling of human cervical carcinoma (Hela) and rodent neuroblastoma (Neuro2a) cell lines account for the reported opposite outcomes.
The study by Aseervatham et al. [64] further indicated that the CaM-binding motif in the carboxyterminal domain of Cx35/Cx36 allows for structural flexibility. Aseervatham et al. reported a 1-5-10-binding motif, which is distinct from the previously reported 1-4-8-11 motif [54]. The differences between the two predicted CaM-binding motifs may reflect the distinct computational and analytic approaches used in separate studies. Equally well, the CaM-binding motifs of Cx35/6 could also undergo the rapid structural adaptation of protein interfaces that adjust the binding properties when local calcium concentrations are dynamic. In favor of this argument are the rapid structural adaptations of CaM, which were exploited to develop fast-acting and sensitive biosensors for intracellular calcium dynamics. Furthermore, the Cx36 carboxyterminal domain structure is predicted to be intrinsically disordered, like other connexins [67,70,71].
The use of alternative CaM-binding motifs might also prepare Cx36 for different protein interactions. An example is the interaction of Cx36 with tubulin. The Cx36-binding motif overlaps with the CaM-binding site but is distinct from those found in Cx43 [43,72]. The mechanistic details of the trafficking to the gap-junction plaque (GJP) were demonstrated using a pharmacological and mutational analysis, demonstrating that amino acids K279, I280, and K281 were critical. The potentiation of the communication strength between coupled cells was demonstrated, reinforcing the role of and protein transport to fine-tuned gap junction communication.
4. Building a Cx36-CaMKII Interaction Complex
The research summarized in the previous section established Cx36 as a hub binding Ca2+-loaded CaM, and they identified this interaction as a critical step with implications for functions preceding the initiation of calcium/calmodulin-dependent kinase II (CaMKII)-mediated plasticity at electrical synapses. Here, we will demonstrate that the molecular interaction of Cx36 with CaMKII at electrical synapses has a resemblance to interactions of CaMKII with NMDA receptors, enabling chemical synaptic potentiation, which is at the core of plasticity and learning and memory [74,75]. CaMKII is a dodecameric holoenzyme [76]. CaMKII comprises a family of >25 isoforms that are derived from four genes (α, β, γ, and δ). The α- and β-subunits are the predominant isoforms in the brain, where they form holoenzymes that are composed of either one or both subunit types [77]. Each CaMKII isoform consists of a catalytic domain, an autoinhibitory domain, a variable segment, and a self-association domain (Figure 3a). The catalytic domain is responsible for the kinase activity of CaMKII [78,79]. The regulatory domain is required for autophosphorylation and CaM binding [80]. The association domain forms a hub-like, tetrameric assembly, composed of two rings of seven protomers each, which are stacked head-to-head and held together by extensive interfaces [81,82].
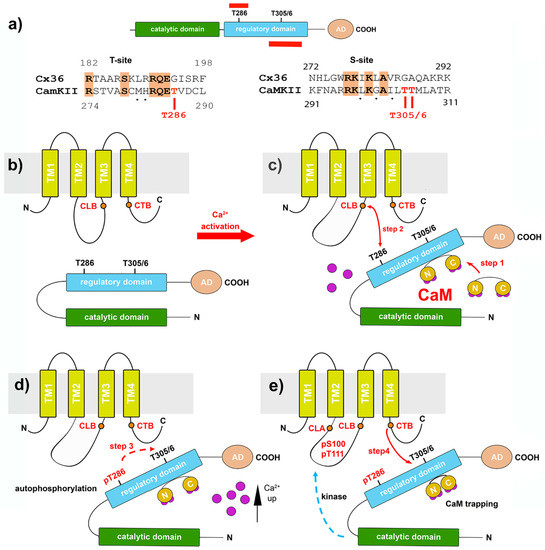
Figure 3.
The Cx36–CaMKII-binding complex. (a) The catalytic, regulatory, and accessory domain of CaMKII. The autophosphorylation and T-site (top) and the CaM-binding site at the S-site are shown (bottom) in red. T286 and T305/6 indicated phosphorylation sites in the regulatory domain. The sequence alignments below show conserved amino acids in the T- and S-sites of Cx36 and CaMKII. (b–e) Model of the activation sequence in which CaMKII is opened by calcium-activated CaM, followed by the sequential binding and phosphorylation of Cx36 (modified from reference [53]). (c) In step 1, elevated intracellular Ca2+ activates CaM, and the binding of Ca2+/CaM to the autoinhibitory domains displaces the gate and, in step 2, allows the CaMKII-binding domain in the cytoplasmic loop of Cx36 (CLB) to interact with the kinase’s target site, CaMKII. (d) Autophosphorylation at T286 (step 3) (e) enables the kinase to bind to a domain in the carboxyl terminus of Cx36 (labeled CTB) (step 4). Phosphorylation of the cytoplasmic loop sites follow (dashed blue arrow in (e)), as well as in the carboxyl terminal domain.
Central to the ability of CaMKII to generate sustained changes in the postsynaptic efficacy after stimulation is the interaction of the subunits with the Ca2+/CaM complex. In the “off state”, CaMKII is inactive, because the enzyme’s autoinhibitory regulatory domain buries the catalytic region, preventing autoactivation and substrate binding (Figure 3b) [83,84]. Interestingly, both the autoactivation (T-site) and substrate binding (S-site) domains of CaMKII share sequence homology with sequences in the cytoplasmic loop (CLB) and carboxyterminal (CTB) domain of Cx36 [53]. The binding of Ca2+/CaM releases CaMKII from autoinhibition by exposing the T-site and the S-site of the enzyme. This opening of the CaMKII generates autonomous kinase activity by an intraholoenzyme, causing intersubunit phosphorylation of the threonine residue at position T286 [79,80,85]. Once phosphorylated at T286, the regulatory domain with its T- and S-sites can no longer interact with the catalytic site’s corresponding domains, allowing the catalytic domain to access and phosphorylate substrates [86]. At this step, sustained autonomous CaMKII activity is independent of Ca2+/CaM and keeps the kinase active after the initial Ca2+ stimulus has subsided.
One of the most prominent substrates of CaMKII at postsynaptic sites is the NR2B subunit of the NMDA receptor. The autophosphorylation-dependent binding of CaMKII to NR2B locks CaMKII in the active conformation [87,88]. CaMKII binding to the NR2B protein requires two binding sites [75,89]. Based on similarities of the interaction of the Cx36 and CaMKII-binding sites with properties of the NR2B subunit, a model was proposed where the CLB and CTB sites of Cx36 function in a bipartite fashion, like the interaction of the NR2B subunit with CaMKII [53]. The proposed sequence of events put forward in Figure 3b–e is based on the autoinhibitory “gate” hypothesis of the regulatory and catalytic domains of CaMKII. The following sequence of events was predicted. In step 1, the binding of Ca2+/CaM to the autoinhibitory domains displaces the gate and allows the CLB domain (step 2) to interact with the kinase’s target site. The phosphorylation of CaMKII is not required for this initial step. The subsequent autophosphorylation at T286 (step 3) enables the kinase to interact with the target or substrate proteins and bind Cx36–CTB to its corresponding substrate recognition motif (step 4). An argument for the subsequential processing of both Cx36 segments during interactions with the kinase can be made from the discovery of phosphorylation found at amino acid residues S110 and T111 within the cytoplasmic loop (CLA) and at S315 of the carboxyl-terminal domain [53]. Based on the model shown, it is reasonable to speculate that the phosphorylation of S110/T111 is a direct consequence of fully activating CaMKII, even though the exact timing has not been determined (steps 3 and 4). Although CaMKII does not phosphorylate Cx36 at position S315 in vitro, this modification is notable. At S315, Cx36 is phosphorylated by PKA in a position where structural changes after phosphorylation could affect the dynamic coupling–uncoupling of Cx36 from the interaction complexes. This effect would explain the antagonistic nature of CaMKII and PKA phosphorylation on Cx36-mediated gap junction coupling in the cerebellum [49] or the rodent [50] or zebrafish retina [51,90].
Having established that phosphorylation by PKA and CaMKII keeps Cx36 coupling and uncoupling in a physiological range, the next question arises as to what the role is of the two CaMKII-binding regions. It has been demonstrated that the binding and extent of phosphorylation of the Cx36 cytoplasmic domains varied according to the autophosphorylated state of CaMKII and, thus, resembled the behavior of fragments of the NR2B subunits of the NMDA receptor. The autophosphorylation of CaMKII is less effective at the Cx36–CLB site in terms of binding and phosphorylation. The Cx36–CTB site exhibited a higher efficacy toward both effects when CaMKII was autophosphorylated. Since CaM bound to the CT of Cx36 would inhibit the interaction with the substrate-binding site of the kinase, the several orders of magnitude increase in affinity of CaMKII to Ca2+/CaM after autophosphorylation of T286 (so-called Ca2+/CaM trapping) (Figure 3e), could explain how CaM competes with Cx36 for the overlapping CTB-binding motif. It suggests that when CaM is displaced from the CTB site, Cx36 is released from its inhibitions and able to bind to CaMKII.
The initial electrophysiological studies of Cx36 revealed that the gap junction channels have a remarkably low conductance and are highly insensitive to transjunctional voltage compared to other connexins [91,92]. These properties optimize transmissions during significant voltage changes and endow the electrical synapses with the requirement that substantial changes in the conductance are accompanied by a significant increase or decrease of the number of open or closed channels. A substantial increase in junctional conductance was recorded between pairs of voltage-clamped cells after the whole-cell mode was achieved, generally rising about tenfold for ten minutes [52,93] (see Figure 4a–c, black traces). This phenomenon of adaptive plasticity was coined a “run-up”. It is unique for cell pairs expressing Cx36 tested in Neuro2a cell cultures [52]. It illustrated the time-dependent changes in Cx36 junctional conductance (Gj) and was explained as the increased open probability following altered phosphorylation or transport of Cx36 to the gap junction plaque (GJP). Experimental ablation of the CLB and CTB-binding regions in Cx36 (Figure 4a,b) [52] or inhibition of CaMKII was used to test for a weakening of the “run-up” phenomenon previously described [93]. In these studies, the run-up was virtually absent in mutants lacking the CaMKII-binding regions. It was proposed that the run-up process involved CaMKII increasing the channel open time rather than affecting the channel insertion internalization [52]. The pharmacological blockade of CaMKII using KN-93 and with cognate peptides applied intracellularly, together with site-directed mutagenesis of the previously described CaMKII-binding and phosphorylation sites of Cx36, connected the previously reported direct interaction of CaMKII with its binding sites CLB and CTB, as well as the CaMKII phosphorylation sites at S110/T111 of Cx36 [53].
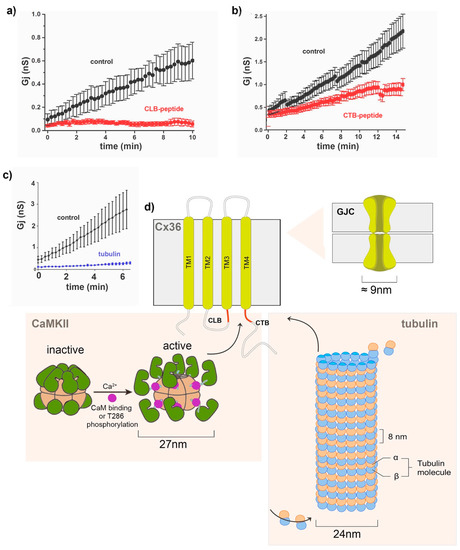
Figure 4.
Cx36 interactions with macromolecular protein complexes. (a–c) Dual patch–clamp recordings of Neuro2a cell pairs expressing Cx36 typically reveal an initial 5–10-fold increase in the junctional conductance (Gj), termed run-up (black symbols), modified from reference [52]. The extent of the run-up was reduced by the intracellular application of peptides (100 µM) blocking the CLB (aa175–195) and CTB (aa272–292) sites of Cx36 (curves with red symbols). (c) The intracellular application of monomeric tubulin (50 µM) reduced the run-up plasticity (blue trace). Modified from reference [43]. (d) Summary of macromolecular interactions between Cx36 and the CaMKII holoenzyme or microtubules. As illustrated in Figure 3, CaMKII is activated by binding to the Ca2+/CaM complex or following T286 phosphorylation (modified from reference [85]. Active CaMKII binds to the intracellular CTB domain, a binding target shared with the CaM and microtubules. Details of the spatiotemporal regulation of how the macromolecular complexes bind are presently unknown; however, the linkage of Cx36 to microtubules likely facilitates its delivery to [87] the GJP, a function that is lost when microtubules depolymerize.
The control of Cx36 Gj was not restricted to the interaction with CaMKII. Brown et al. [43] determined that Cx36 binds to tubulin using a motif overlapping with the afore-described CaM and CaMKII-binding motifs. Dual patch–clamp recordings demonstrated that pharmacological interference of the cytoskeleton abolished Cx36 plasticity (Figure 4c). Mechanistic details of Cx36 trafficking demonstrated that tubulin-dependent transport potentiates the synaptic strength by delivering channels to GJPs. The extent to which the microtubule-mediated delivery in Cx36 accounts for the run-up compared to other stimuli remains to be determined.
The binding of three calcium-dependent proteins to overlapping domains of Cx36 emphasizes that the plasticity of the electrical synapses is controlled at multiple levels and that local calcium fluctuations are a critical factor in the processes. It also points out that these interactions involve binding hexametric gap junction channels to the macromolecular complexes formed by the CaMKII holoenzyme or the microtubule superstructure (Figure 4d). While structural and mechanistic constraints imposed by interactions of these large protein assemblies remain to be fully resolved, the machinery, protein–protein interactions and post-translational modifications involved in long-term electrical synapse changes may be as complex as those at chemical synapses.
As pointed out by reference [64], gap junctions formed by Cx36 that function as electrical synapses within networks of neurons will routinely encounter large fluctuations in the local cytoplasmic calcium concentration. Presently, we have only started to uncover the multi-faceted details of how dynamic calcium signaling modulates the functions of mixed synapses. The complexity of the problem of studying calcium signals separated in time and space is immense. Postsynaptic dendritic spines are home to >1000 different proteins, many of which are calcium-dependent and require demand transport and activation, such as AMPA/NMDA receptors or Cx36. The molecular composition of presynaptic contacts is equally challenging. Sourcing calcium and building often short-term (ms) to second signals requires mechanisms not fully detailed yet. The elevation of intracellular calcium can arise from multiple sources, such as voltage-gated calcium channels, AMPA/NMDARs, or the axonal or spine ER. Identifying the relevant spatiotemporal calcium signals that drive Cx36-mediated plasticity will require a new generation of tools that combine the sensitivity of genetically encoded biosensors with advanced and dynamic high-resolution microscopy. The O’Brien group’s recent work is a significant step in the direction needed [94]. In this work, the investigators demonstrated how changes in the local Ca2+ signaling regulates plasticity using a GCaMP Ca2+ biosensor fused to Cx36. The Cx36–GCaMP biosensor robustly reported local Ca2+ increases in response to the addition of a Ca2+ ionophore with increases in the fluorescence that recovered during washout. Recovery was strongly dependent on Na+-Ca2+ exchange activity. Cx36–GCaMP revealed transient and concentration-dependent increases in local Ca2+ after the application of glutamate in cells transfected with NMDA receptor subunits. The mechanism was dependent, in part, on the CaMKII activity. In summary, Cx36–GCaMP is an effective tool to measure the changes in the Ca2+ microenvironment around the Cx36 gap junctions. Based on this framework, it seems reasonable to build a toolbox of customized genetically encoded fluorescent biosensors or FRET sensors to dissect the calcium signaling pathways in the context of Cx36 microdomains to shed light on the mechanistic details of how electrical synapses operate. Finally, the recent advances in cryoEM suggest that resolving the structure of Cx36–CaM or Cx36–CaMKII in its different stages, in similar detail as the recently reported structures of pannexins and innexins, is a timely objective.
5. Conclusions
Regardless of the lack of a precise understanding of the conformational changes underlying the events described above, Cx36 might quickly sample CaM, CaMKII, and other protein-mediating functions such as transport or maintaining the local scaffold. Primed by local calcium dynamics, CaM may translate the numerous signaling contexts Cx36 encounters throughout the passage from the ER/Golgi complex to the synaptic contacts. Beyond doubt, the decoding of calcium signals is essential as proteins stimulated by the calmodulin–calcium complex are protein kinases, phosphatases, or accessory proteins with multiple functions. How these complex interactions are governed is presently unknown.
Supplementary Materials
The following materials are available online and can be found at https://www.mdpi.com/article/10.3390/ijms22094473/s1: Supplementary Figure S1; Supplementary Table S2: Cx36 mRNA expression during early development of the mouse brain; Supplementary Table S2: Cx36 mRNA and protein expression in the adult rodent nervous system.
Author Contributions
Conceptualization, writing—review and editing, and visualization, D.C.S. and G.R.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This review was funded by the Canada Research Chairs program (CRCP; G.R.Z.); Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Program (G.R.Z.); and National Institutes of Health NS092466, AR070547, and NS116892 (D.C.S.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank all trainees and collaborators with whom we studied interactions of the gap junctions with calmodulin and CAMKII, including Rolf Dermietzel and Eliana Scemes, Cantas Alev, Cherie Brown, Cristiane del Corsso, Ryan C. Siu, Midituru Srinivas, and Randy F. Stout Jr.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Robertson, J.D. The occurrence of a subunit pattern in the unit membranes of club endings in mauthner cell synapses in goldfish brains. J. Cell Biol. 1963, 19, 201–221. [Google Scholar] [CrossRef] [PubMed]
- Faber, D.S.; Pereda, A.E. Two Forms of Electrical Transmission between Neurons. Front. Mol. Neurosci. 2018, 11, 427. [Google Scholar] [CrossRef]
- Nagy, J.I.; Pereda, A.E.; Rash, J.E. Electrical synapses in mammalian CNS: Past eras, present focus and future directions. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 102–123. [Google Scholar] [CrossRef] [PubMed]
- Nagy, J.I.; Pereda, A.E.; Rash, J.E. On the occurrence and enigmatic functions of mixed (chemical plus electrical) synapses in the mammalian CNS. Neurosci. Lett. 2019, 695, 53–64. [Google Scholar] [CrossRef]
- O’Brien, J.; Al-Ubaidi, M.R.; Ripps, H. Connexin 35: A gap-junctional protein expressed preferentially in the skate retina. Mol. Biol. Cell 1996, 7, 233–243. [Google Scholar] [CrossRef][Green Version]
- Condorelli, D.F.; Parenti, R.; Spinella, F.; Salinaro, A.T.; Belluardo, N.; Cardile, V.; Cicirata, F. Cloning of a new gap junction gene (Cx36) highly expressed in mammalian brain neurons. Eur. J. Neurosci. 1998, 10, 1202–1208. [Google Scholar] [CrossRef] [PubMed]
- Serre-Beinier, V.; Le Gurun, S.; Belluardo, N.; Trovato-Salinaro, A.; Charollais, A.; Haefliger, J.A.; Condorelli, D.F.; Meda, P. Cx36 preferentially connects beta-cells within pancreatic islets. Diabetes 2000, 49, 727–734. [Google Scholar] [CrossRef]
- Belluardo, N.; Trovato-Salinaro, A.; Mudo, G.; Hurd, Y.L.; Condorelli, D.F. Structure, chromosomal localization, and brain expression of human Cx36 gene. J. Neurosci. Res. 1999, 57, 740–752. [Google Scholar] [CrossRef]
- O’Brien, J.; Bruzzone, R.; White, T.W.; Al-Ubaidi, M.R.; Ripps, H. Cloning and Expression of Two Related Connexins from the Perch Retina Define a Distinct Subgroup of the Connexin Family. J. Neurosci. 1998, 18, 7625–7637. [Google Scholar] [CrossRef]
- McLachlan, E.; White, T.W.; Ugonabo, C.; Olson, C.; Nagy, J.I.; Valdimarsson, G. Zebrafish Cx35: Cloning and characterization of a gap junction gene highly expressed in the retina. J. Neurosci. Res. 2003, 73, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, V.M.; Singh, R.; Minogue, P.J.; Ragsdale, C.W.; Beyer, E.C. Highly restricted pattern of connexin36 expression in chick somite development. Anat. Embryol. 2004, 209, 11–18. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eastman, S.D.; Chen, T.H.-P.; Falk, M.M.; Mendelson, T.C.; Iovine, M.K. Phylogenetic analysis of three complete gap junction gene families reveals lineage-specific duplications and highly supported gene classes. Genomics 2006, 87, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.C.; Whitebirch, A.C.; Shah, A.N.; Marsden, K.C.; Granato, M.; O’Brien, J.; Moens, C.B. A genetic basis for molecular asymmetry at vertebrate electrical synapses. eLife 2017, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Dobrenis, K.; Chang, H.-Y.; Pina-Benabou, M.; Woodroffe, A.; Lee, S.; Rozental, R.; Spray, D.; Scemes, E. Human and mouse microglia express connexin36, and functional gap junctions are formed between rodent microglia and neurons. J. Neurosci. Res. 2005, 82, 306–315. [Google Scholar] [CrossRef]
- Martin, D.; Tawadros, T.; Meylan, L.; Abderrahmani, A.; Condorelli, D.F.; Waeber, G.; Haefliger, J.-A. Critical Role of the Transcriptional Repressor Neuron-restrictive Silencer Factor in the Specific Control of Connexin36 in Insulin-producing Cell Lines. J. Biol. Chem. 2003, 278, 53082–53089. [Google Scholar] [CrossRef]
- Swayne, L.A.; Bennett, S.A.L. Connexins and pannexins in neuronal development and adult neurogenesis. BMC Cell Biol. 2016, 17, 39–49. [Google Scholar] [CrossRef]
- Jabeen, S.; Thirumalai, V. Distribution of the gap junction protein connexin 35 in the central nervous system of developing zebrafish larvae. Front. Neural Circuits 2013, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Zappalà, A.; Cicero, D.; Serapide, M.; Paz, C.; Catania, M.; Falchi, M.; Parenti, R.; Pantò, M.; La Delia, F.; Cicirata, F. Expression of pannexin1 in the CNS of adult mouse: Cellular localization and effect of 4-aminopyridine-induced seizures. Neuroscience 2006, 141, 167–178. [Google Scholar] [CrossRef]
- Nagy, J.I.; Rash, J.E. Cx36, Cx43 and Cx45 in mouse and rat cerebellar cortex: Species-specific expression, compensation in Cx36 null mice and co-localization in neurons vs. glia. Eur. J. Neurosci. 2017, 46, 1790–1804. [Google Scholar] [CrossRef]
- Meier, C.; Petrasch-Parwez, E.; Habbes, H.-W.; Teubner, B.; Güldenagel, M.; Degen, J.; Söhl, G.; Willecke, K.; Dermietzel, R. Immunohistochemical detection of the neuronal connexin36 in the mouse central nervous system in comparison to connexin36-deficient tissues. Histochem. Cell Biol. 2002, 117, 461–471. [Google Scholar] [CrossRef]
- Armendariz, E.M.P.; Norcini, M.; Hernández-Tellez, B.; Castell-Rodríguez, A.; Coronel-Cruz, C.; Alquicira, R.G.; Sideris, A.; Recio-Pinto, E. Neurons and satellite glial cells in adult rat lumbar dorsal root ganglia express connexin 36. Acta Histochem. 2018, 120, 168–178. [Google Scholar] [CrossRef]
- Nagy, J.; Lynn, B.; Senecal, J.; Stecina, K. Connexin36 Expression in Primary Afferent Neurons in Relation to the Axon Reflex and Modality Coding of Somatic Sensation. Neuroscience 2018, 383, 216–234. [Google Scholar] [CrossRef]
- Rash, J.E.; Staines, W.A.; Yasumura, T.; Patel, D.; Furman, C.S.; Stelmack, G.L.; Nagy, J.I. Immunogold evidence that neuronal gap junctions in adult rat brain and spinal cord contain connexin-36 but not connexin-32 or connexin-43. Proc. Natl. Acad. Sci. USA 2000, 97, 7573–7578. [Google Scholar] [CrossRef] [PubMed]
- Rozental, R.; Srinivas, M.; Gökhan, S.; Urban, M.; Dermietzel, R.; Kessler, J.; Spray, D.; Mehler, M. Temporal expression of neuronal connexins during hippocampal ontogeny. Brain Res. Rev. 2000, 32, 57–71. [Google Scholar] [CrossRef]
- Frinchi, M.; Di Liberto, V.; Turimella, S.; D’Antoni, F.; Theis, M.; Belluardo, N.; Mudò, G. Connexin36 (Cx36) expression and protein detection in the mouse carotid body and myenteric plexus. Acta Histochem. 2013, 115, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Lall, V.K.; Bruce, G.; Voytenko, L.; Drinkhill, M.; Wellershaus, K.; Willecke, K.; Deuchars, J.; Deuchars, S.A. Physiologic regulation of heart rate and blood pressure involves connexin 36–containing gap junctions. FASEB J. 2017, 31, 3966–3977. [Google Scholar] [CrossRef]
- Marina, N.; Becker, D.L.; Gilbey, M.P. Immunohistochemical detection of connexin36 in sympathetic preganglionic and somatic motoneurons in the adult rat. Auton. Neurosci. 2008, 139, 15–23. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Güldenagel, M.; Söhl, G.; Plum, A.; Traub, O.; Teubner, B.; Weiler, R.; Willecke, K. Expression patterns of connexin genes in mouse retina. J. Comp. Neurol. 2000, 425, 193–201. [Google Scholar] [CrossRef]
- Feigenspan, A.; Teubner, B.; Willecke, K.; Weiler, R. Expression of Neuronal Connexin36 in AII Amacrine Cells of the Mammalian Retina. J. Neurosci. 2001, 21, 230–239. [Google Scholar] [CrossRef]
- Massey, S.C.; O’Brien, J.J.; Trexler, E.B.; Li, W.; Keung, J.W.; Mills, S.L.; O’Brien, J. Multiple neuronal connexins in the mammalian retina. Cell Commun. Adhes. 2003, 10, 425–430. [Google Scholar] [CrossRef]
- Zhang, C.; Restrepo, D. Heterogeneous expression of connexin 36 in the olfactory epithelium and glomerular layer of the olfactory bulb. J. Comp. Neurol. 2003, 459, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Degen, J.; Meier, C.; Van Der Giessen, R.S.; Söhl, G.; Petrasch-Parwez, E.; Urschel, S.; Dermietzel, R.; Schilling, K.; De Zeeuw, C.I.; Willecke, K. Expression pattern of lacZ reporter gene representing connexin36 in transgenic mice. J. Comp. Neurol. 2004, 473, 511–525. [Google Scholar] [CrossRef]
- Horowitz, S.S.; Stamper, S.A.; Simmons, J.A. Neuronal connexin expression in the cochlear nucleus of big brown bats. Brain Res. 2008, 1197, 76–84. [Google Scholar] [CrossRef]
- Liu, W.; Boström, M.; Kinnefors, A.; Rask-Andersen, H. Unique expression of connexins in the human cochlea. Hear. Res. 2009, 250, 55–62. [Google Scholar] [CrossRef]
- Blakley, B.W.; Garcia, C.E.A.; Da Sliva, S.R.; Florêncio, V.M.B.; I Nagy, J. Elevated auditory brainstem response thresholds in mice with Connexin36 gene ablation. Acta Oto-Laryngol. 2015, 135, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.; Nagy, J. Connexin36 expression in major centers of the auditory system in the CNS of mouse and rat: Evidence for neurons forming purely electrical synapses and morphologically mixed synapses. Neuroscience 2015, 303, 604–629. [Google Scholar] [CrossRef] [PubMed]
- Hamzei-Sichani, F.; Davidson, K.G.V.; Yasumura, T.; Janssen, W.G.M.; Wearne, S.L.; Hof, P.R.; Traub, R.D.; Gutierrez, R.; Ottersen, O.P.; Rash, J.E. Mixed Electrical–Chemical Synapses in Adult Rat Hippocampus are Primarily Glutamatergic and Coupled by Connexin-36. Front. Neuroanat. 2012, 6, 13. [Google Scholar] [CrossRef]
- Hidaka, S.; Kato, T.; Miyachi, E.-I. Expression of gap junction connexin36 in adult rat retinal ganglion cells. J. Integr. Neurosci. 2002, 1, 3–22. [Google Scholar] [CrossRef]
- Bautista, W.; McCrea, D.A.; Nagy, J.I. Connexin36 identified at morphologically mixed chemical/electrical synapses on trigeminal motoneurons and at primary afferent terminals on spinal cord neurons in adult mouse and rat. Neuroscience 2014, 263, 159–180. [Google Scholar] [CrossRef]
- Nagy, J.I. Evidence for connexin36 localization at hippocampal mossy fiber terminals suggesting mixed chemical/electrical transmission by granule cells. Brain Res. 2012, 1487, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.; Senecal, J.M.; Lynn, B.D.; Traub, R.D.; I Nagy, J. Connexin36 localization along axon initial segments in the mammalian CNS. Int. J. Physiol. Pathophysiol. Pharmacol. 2020, 12, 153–165. [Google Scholar]
- Sotelo, C.; Llinás, R.; Baker, R. Structural study of inferior olivary nucleus of the cat: Morphological correlates of electrotonic coupling. J. Neurophysiol. 1974, 37, 541–559. [Google Scholar] [CrossRef]
- Brown, C.A.; Del Corsso, C.; Zoidl, C.; Donaldson, L.W.; Spray, D.C. Tubulin-Dependent Transport of Connexin-36 Potentiates the Size and Strength of Electrical Synapses. Cells 2019, 8, 1146. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Olson, C.; Lu, S.; Kamasawa, N.; Yasumura, T.; Rash, J.E.; Nagy, J.I. Neuronal connexin36 association with zonula occludens-1 protein (ZO-1) in mouse brain and interaction with the first PDZ domain of ZO-1. Eur. J. Neurosci. 2004, 19, 2132–2146. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Olson, C.; Lu, S.; Nagy, J.I. Association of connexin36 with zonula occludens-1 in HeLa cells? TC-3 cells, pancreas, and adrenal gland. Histochem. Cell Biol. 2000, 122, 485–498. [Google Scholar] [CrossRef]
- Li, X.; Lu, S.; Nagy, J.I. Direct association of connexin36 with zonula occludens-2 and zonula occludens-3. Neurochem. Int. 2009, 54, 393–402. [Google Scholar] [CrossRef]
- Li, X.; Lynn, B.D.; Nagy, J.I. The effector and scaffolding proteins AF6 and MUPP1 interact with connexin36 and localize at gap junctions that form electrical synapses in rodent brain. Eur. J. Neurosci. 2012, 35, 166–181. [Google Scholar] [CrossRef]
- Tetenborg, S.; Wang, H.Y.; Nemitz, L.; Depping, A.; Espejo, A.B.; Aseervatham, J.; Bedford, M.T.; Janssen-Bienhold, U.; O’Brien, J.; Dedek, K. Phosphorylation of Connexin36 near the C-terminus switches binding affinities for PDZ-domain and 14–3–3 proteins in vitro. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Bazzigaluppi, P.; Isenia, S.C.; Haasdijk, E.D.; Elgersma, Y.; De Zeeuw, C.I.; Van Der Giessen, R.S.; De Jeu, M.T.G. Modulation of Murine Olivary Connexin 36 Gap Junctions by PKA and CaMKII. Front. Cell. Neurosci. 2017, 11, 397. [Google Scholar] [CrossRef]
- Urschel, S.; Höher, T.; Schubert, T.; Alev, C.; Söhl, G.; Wörsdörfer, P.; Asahara, T.; Dermietzel, R.; Weiler, R.; Willecke, K. Protein Kinase A-mediated Phosphorylation of Connexin36 in Mouse Retina Results in Decreased Gap Junctional Communication between AII Amacrine Cells. J. Biol. Chem. 2006, 281, 33163–33171. [Google Scholar] [CrossRef]
- Kothmann, W.W.; Li, X.; Burr, G.S.; O’Brien, J. Connexin 35/36 is phosphorylated at regulatory sites in the retina. Vis. Neurosci. 2007, 24, 363–375. [Google Scholar] [CrossRef]
- Del Corsso, C.; Iglesias, R.; Zoidl, G.; Dermietzel, R.; Spray, D.C. Calmodulin dependent protein kinase increases conductance at gap junctions formed by the neuronal gap junction protein connexin36. Brain Res. 2012, 1487, 69–77. [Google Scholar] [CrossRef]
- Alev, C.; Urschel, S.; Sonntag, S.; Zoidl, G.; Fort, A.G.; Höher, T.; Matsubara, M.; Willecke, K.; Spray, D.C.; Dermietzel, R. The neuronal connexin36 interacts with and is phosphorylated by CaMKII in a way similar to CaMKII interaction with glutamate receptors. Proc. Natl. Acad. Sci. USA 2008, 105, 20964–20969. [Google Scholar] [CrossRef] [PubMed]
- Siu, R.C.F.; Smirnova, E.; Brown, C.A.; Zoidl, C.; Spray, D.C.; Donaldson, L.W.; Zoidl, G. Structural and Functional Consequences of Connexin 36 (Cx36) Interaction with Calmodulin. Front. Mol. Neurosci. 2016, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Burr, G.S.; Mitchell, C.K.; Keflemariam, Y.J.; Heidelberger, R.; O’Brien, J. Calcium-dependent binding of calmodulin to neuronal gap junction proteins. Biochem. Biophys. Res. Commun. 2005, 335, 1191–1198. [Google Scholar] [CrossRef][Green Version]
- Kotova, A.; Timonina, K.; Zoidl, G.R. Endocytosis of Connexin 36 is Mediated by Interaction with Caveolin-1. Int. J. Mol. Sci. 2020, 21, 5401. [Google Scholar] [CrossRef] [PubMed]
- Schubert, A.-L.; Schubert, W.; Spray, A.D.C.; Lisanti, M.P. Connexin Family Members Target to Lipid Raft Domains and Interact with Caveolin-1. Biochemistry 2002, 41, 5754–5764. [Google Scholar] [CrossRef]
- Nagy, J.; Lynn, B. Structural and Intermolecular Associations between Connexin36 and Protein Components of the Adherens Junction-Neuronal Gap Junction Complex. Neuroscience 2018, 384, 241–261. [Google Scholar] [CrossRef]
- O’Brien, J. Design principles of electrical synaptic plasticity. Neurosci. Lett. 2019, 695, 4–11. [Google Scholar] [CrossRef]
- Miller, A.C.; Pereda, A.E. The electrical synapse: Molecular complexities at the gap and beyond. Dev. Neurobiol. 2017, 77, 562–574. [Google Scholar] [CrossRef]
- Peracchia, C. Calmodulin-Mediated Regulation of Gap Junction Channels. Int. J. Mol. Sci. 2020, 21, 485. [Google Scholar] [CrossRef] [PubMed]
- Peracchia, C. Calmodulin-Cork Model of Gap Junction Channel Gating—One Molecule, Two Mechanisms. Int. J. Mol. Sci. 2020, 21, 4938. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Riquelme, M.; Gu, S.; Jiang, J. Regulation of Connexin Gap Junctions and Hemichannels by Calcium and Calcium Binding Protein Calmodulin. Int. J. Mol. Sci. 2020, 21, 8194. [Google Scholar] [CrossRef] [PubMed]
- Aseervatham, J.; Li, X.; Mitchell, C.K.; Lin, Y.-P.; Heidelberger, R.; O’Brien, J. Calmodulin Binding to Connexin 35: Specializations to Function as an Electrical Synapse. Int. J. Mol. Sci. 2020, 21, 6346. [Google Scholar] [CrossRef] [PubMed]
- Peracchia, C.; Sotkis, A.; Wang, X.G.; Peracchia, L.L.; Persechini, A. Calmodulin Directly Gates Gap Junction Channels. J. Biol. Chem. 2000, 275, 26220–26224. [Google Scholar] [CrossRef]
- Török, K.; Stauffer, K.; Evans, W.H. Connexin 32 of gap junctions contains two cytoplasmic calmodulin-binding domains. Biochem. J. 1997, 326, 479–483. [Google Scholar] [CrossRef]
- Stauch, K.; Kieken, F.; Sorgen, P. Characterization of the Structure and Intermolecular Interactions between the Connexin 32 Carboxyl-terminal Domain and the Protein Partners Synapse-associated Protein 97 and Calmodulin. J. Biol. Chem. 2012, 287, 27771–27788. [Google Scholar] [CrossRef]
- Choi, E.J.; Palacios-Prado, N.; Sáez, J.C.; Lee, J. Identification of Cx45 as a Major Component of GJs in HeLa Cells. Biomolecules 2020, 10, 1389. [Google Scholar] [CrossRef]
- Eckert, R.; Dunina-Barkovskaya, A. Biophysical characterization of gap-junction channels in HeLa cells. Pflügers Arch. Eur. J. Physiol. 1993, 424, 335–342. [Google Scholar] [CrossRef]
- Li, H.; Spagnol, G.; Pontifex, T.K.; Burt, J.M.; Sorgen, P.L. Chemical shift assignments of the connexin37 carboxyl terminal domain. Biomol. NMR Assign. 2017, 11, 137–141. [Google Scholar] [CrossRef]
- Spagnol, G.; Al-Mugotir, M.; Kopanic, J.L.; Zach, S.; Li, H.; Trease, A.J.; Stauch, K.L.; Grosely, R.; Cervantes, M.; Sorgen, P.L. Secondary structural analysis of the carboxyl-terminal domain from different connexin isoforms. Biopolymers 2016, 105, 143–162. [Google Scholar] [CrossRef]
- Giepmans, B.N.; Verlaan, I.; Hengeveld, T.; Janssen, H.; Calafat, J.; Falk, M.M.; Moolenaar, W.H. Gap junction protein connexin-43 interacts directly with microtubules. Curr. Biol. 2001, 11, 1364–1368. [Google Scholar] [CrossRef]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Gardoni, F.; Polli, F.; Cattabeni, F.; Di Luca, M. Calcium-calmodulin-dependent protein kinase II phosphorylation modulates PSD-95 binding to NMDA receptors. Eur. J. Neurosci. 2006, 24, 2694–2704. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.S.; Bayer, K.-U.; Merrill, M.A.; Lim, I.A.; Shea, M.A.; Schulman, H.; Hell, J.W. Regulation of Calcium/Calmodulin-dependent Protein Kinase II Docking toN-Methyl-d-aspartate Receptors by Calcium/Calmodulin and α-Actinin. J. Biol. Chem. 2002, 277, 48441–48448. [Google Scholar] [CrossRef] [PubMed]
- Hudmon, A.; Schulman, H. Structure–function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem. J. 2002, 364, 593–611. [Google Scholar] [CrossRef] [PubMed]
- Brocke, L.; Chiang, L.W.; Wagner, P.D.; Schulman, H. Functional Implications of the Subunit Composition of Neuronal CaM Kinase II. J. Biol. Chem. 1999, 274, 22713–22722. [Google Scholar] [CrossRef] [PubMed]
- Thaler, C.; Koushik, S.V.; Puhl, H.L.; Blank, P.S.; Vogel, S.S. Structural rearrangement of CaMKII catalytic domains encodes activation. Proc. Natl. Acad. Sci. USA 2009, 106, 6369–6374. [Google Scholar] [CrossRef]
- Rosenberg, O.S.; Deindl, S.; Sung, R.-J.; Nairn, A.C.; Kuriyan, J. Structure of the Autoinhibited Kinase Domain of CaMKII and SAXS Analysis of the Holoenzyme. Cell 2005, 123, 849–860. [Google Scholar] [CrossRef]
- Chao, L.H.; Pellicena, P.; Deindl, S.; Barclay, L.A.; Schulman, H.; Kuriyan, J. Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation. Nat. Struct. Mol. Biol. 2010, 17, 264–272. [Google Scholar] [CrossRef]
- Rosenberg, O.S.; Deindl, S.; Comolli, L.R.; Hoelz, A.; Downing, K.H.; Nairn, A.C.; Kuriyan, J. Oligomerization states of the association domain and the holoenyzme of Ca2+/CaM kinase II. FEBS J. 2006, 273, 682–694. [Google Scholar] [CrossRef]
- Hoelz, A.; Nairn, A.C.; Kuriyan, J. Crystal Structure of a Tetradecameric Assembly of the Association Domain of Ca2+/Calmodulin-Dependent Kinase II. Mol. Cell 2003, 11, 1241–1251. [Google Scholar] [CrossRef]
- Mukherji, S.; Brickey, D.A.; Soderling, T.R. Mutational analysis of secondary structure in the autoinhibitory and autophosphorylation domains of calmodulin kinase II. J. Biol. Chem. 1994, 269, 20733–20738. [Google Scholar] [CrossRef]
- Stratton, M.M.; Chao, L.H.; Schulman, H.; Kuriyan, J. Structural studies on the regulation of Ca2+/Calmodulin dependent protein kinase II. Curr. Opin. Struct. Biol. 2013, 23, 292–301. [Google Scholar] [CrossRef]
- Stratton, M.; Lee, I.H.; Bhattacharyya, M.; Christensen, S.M.; Chao, L.H.; Schulman, H.; Groves, J.T.; Kuriyan, J. Activation-triggered subunit exchange between CaMKII holoenzymes facilitates the spread of kinase activity. eLife 2014, 3, e01610. [Google Scholar] [CrossRef]
- Chao, L.H.; Stratton, M.M.; Lee, I.-H.; Rosenberg, O.S.; Levitz, J.; Mandell, D.J.; Kortemme, T.; Groves, J.T.; Schulman, H.; Kuriyan, J. A Mechanism for Tunable Autoinhibition in the Structure of a Human Ca2+/Calmodulin- Dependent Kinase II Holoenzyme. Cell 2011, 146, 732–745. [Google Scholar] [CrossRef]
- Bayer, K.-U.; De Koninck, P.; Leonard, A.S.; Hell, J.W.; Schulman, H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nat. Cell Biol. 2001, 411, 801–805. [Google Scholar] [CrossRef]
- Strack, S.; Colbran, R.J. Autophosphorylation-dependent Targeting of Calcium/ Calmodulin-dependent Protein Kinase II by the NR2B Subunit of theN-Methyl-d-aspartate Receptor. J. Biol. Chem. 1998, 273, 20689–20692. [Google Scholar] [CrossRef] [PubMed]
- Mayadevi, M.; Praseeda, M.; Kumar, K.S.; Omkumar, R.V. Sequence determinants on the NR2A and NR2B subunits of NMDA receptor responsible for specificity of phosphorylation by CaMKII. Biochim. Biophys. Acta (BBA) Bioenergy 2002, 1598, 40–45. [Google Scholar] [CrossRef]
- Li, H.; Chuang, A.Z.; O’Brien, J. Photoreceptor Coupling Is Controlled by Connexin 35 Phosphorylation in Zebrafish Retina. J. Neurosci. 2009, 29, 15178–15186. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, M.; Rozental, R.; Kojima, T.; Dermietzel, R.; Mehler, M.; Condorelli, D.F.; Kessler, J.A.; Spray, D.C. Functional Properties of Channels Formed by the Neuronal Gap Junction Protein Connexin36. J. Neurosci. 1999, 19, 9848–9855. [Google Scholar] [CrossRef] [PubMed]
- Teubner, B.; Degen, J.; Sohl, G.; Guldenagel, M.; Bukauskas, F.F.; Trexler, E.B.; Verselis, V.K.; De Zeeuw, C.I.; Lee, C.G.; Kozak, C.A.; et al. Functional expression of the murine connexin 36 gene coding for a neuron-specific gap junctional protein. J. Membr. Biol. 2000, 176, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Zoidl, G.; Meier, C.; Petrasch-Parwez, E.; Zoidl, C.; Habbes, H.-W.; Kremer, M.; Srinivas, M.; Spray, D.; Dermietzel, R. Evidence for a role of the N-terminal domain in subcellular localization of the neuronal connexin36 (Cx36). J. Neurosci. Res. 2002, 69, 448–465. [Google Scholar] [CrossRef]
- Moore, K.B.; Mitchell, C.K.; Lin, Y.-P.; Lee, Y.-H.; Shihabeddin, E.; O’Brien, J. Localized Calcium Signaling and the Control of Coupling at Cx36 Gap Junctions. eNeuro 2020, 7. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).