Antibody Libraries as Tools to Discover Functional Antibodies and Receptor Pleiotropism
Abstract
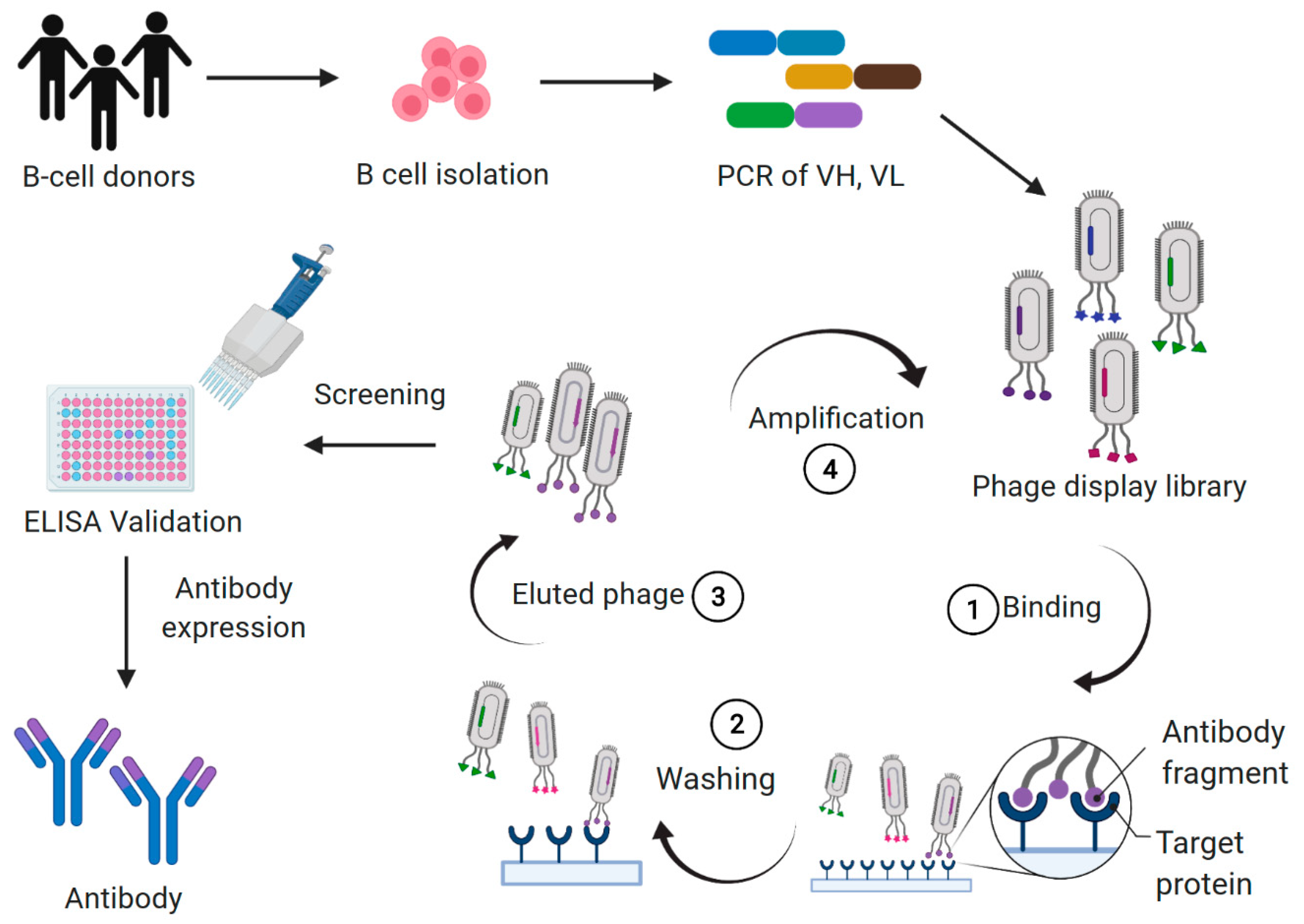
1. Introduction
2. Using Antibody Libraries to Discover Functional Antibodies and Receptor Pleiotropism
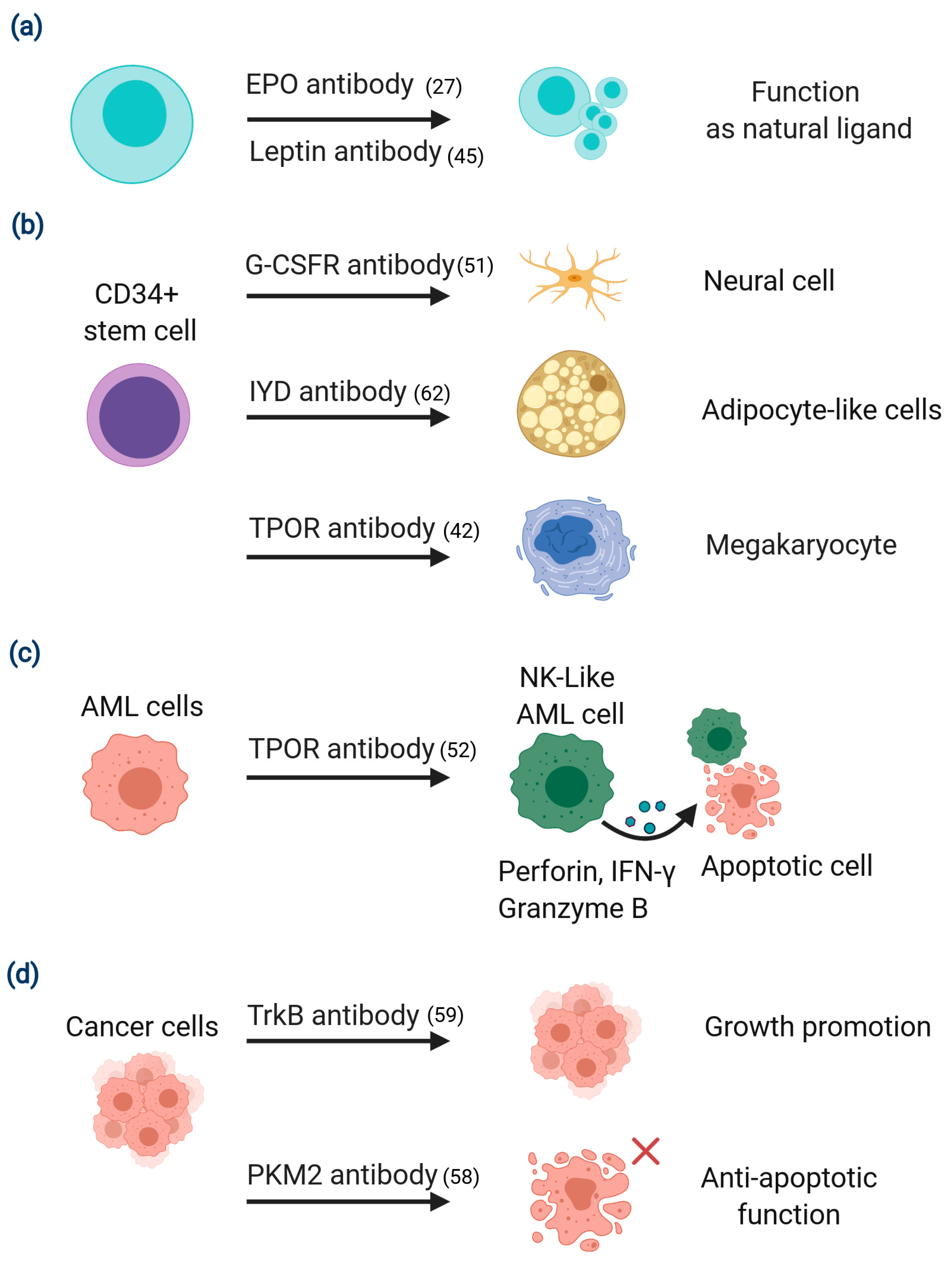
2.1. Target-Based Screening for Agonist Antibodies
2.2. Phenotypic Screening for Isolation of Functional Antibodies That Regulate Cell Fate
3. Antibody Libraries and Emerging Viral Infections
4. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Köhler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Kaplon, H.; Muralidharan, M.; Schneider, Z.; Reichert, J.M. Antibodies to watch in 2020. mAbs 2020, 12, 1703531. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.-J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Huse, W.; Sastry, L.; Iverson, S.; Kang, A.; Alting-Mees, M.; Burton, D.; Benkovic, S.; Lerner, R. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science 1989, 246, 1275–1281. [Google Scholar] [CrossRef]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef]
- Barbas, C.F., III; Kang, A.S.; Lerner, R.A.; Benkovic, S.J. Assembly of combinatorial antibody libraries on phage surfaces: The gene III site. Proc. Natl. Acad. Sci. USA 1991, 88, 7978–7982. [Google Scholar] [CrossRef]
- Clackson, T.P.; Hoogenboom, H.R.; Griffiths, A.D.; Winter, G. Making antibody Fragm. using phage display libraries. Nature 1991, 352, 624–628. [Google Scholar] [CrossRef]
- Breitling, F.; Dübel, S.; Seehaus, T.; Klewinghaus, I.; Little, M. A surface expression vector for antibody screening. Gene 1991, 104, 147–153. [Google Scholar] [CrossRef]
- Kang, A.S.; Barbas, C.F.; Janda, K.D.; Benkovic, S.J.; Lerner, R.A. Linkage of recognition and replication functions by assembling combinatorial antibody Fab libraries along phage surfaces. Proc. Natl. Acad. Sci. USA 1991, 88, 4363–4366. [Google Scholar] [CrossRef]
- Jespers, L.S.; Roberts, A.; Mahler, S.M.; Winter, G.; Hoogenboom, H.R. Guiding the Selection of Human Antibodies from Phage Display Repertoires to a Single Epitope of an Antigen. Nat. Biotechnol. 1994, 12, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Edwards, B.M.; Barash, S.C.; Main, S.H.; Choi, G.H.; Minter, R.; Ullrich, S.; Williams, E.; Du Fou, L.; Wilton, J.; Albert, V.R.; et al. The Remarkable Flexibility of the Human Antibody Repertoire; Isolation of Over One Thousand Different Antibodies to a Single Protein, BLyS. J. Mol. Biol. 2003, 334, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, S. Raxibacumab. mAbs 2009, 1, 531–538. [Google Scholar] [CrossRef]
- Lu, D.; Jimenez, X.; Zhang, H.; Bohlen, P.; Witte, L.; Zhu, Z. Selection of high affinity human neutralizing antibodies to VEGFR2 from a large antibody phage display library for antiangiogenesis therapy. Int. J. Cancer 2001, 97, 393–399. [Google Scholar] [CrossRef]
- De Haard, H.J.; van Neer, N.; Reurs, A.; Hufton, S.E.; Roovers, R.C.; Henderikx, P.; de Bruine, A.P.; Arends, J.W.; Hoogenboom, H.R. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J. Biol. Chem. 1999, 274, 18218–18230. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Kussie, P.; Ferguson, K.M. Structural Basis for EGF Receptor Inhibition by the Therapeutic Antibody IMC-11F8. Structure 2008, 16, 216–227. [Google Scholar] [CrossRef]
- McDermott, D.F.; Sosman, J.A.; Sznol, M.; Massard, C.; Gordon, M.S.; Hamid, O.; Powderly, J.D.; Infante, J.R.; Fassò, M.; Wang, Y.V.; et al. Atezolizumab, an Anti–Programmed Death-Ligand 1 Antibody, in Metastatic Renal Cell Carcinoma: Long-Term Safety, Clinical Activity, and Immune Correlates from a Phase Ia Study. J. Clin. Oncol. 2016, 34, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Soria, J.-C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef]
- Boyerinas, B.; Jochems, C.; Fantini, M.C.; Heery, C.R.; Gulley, J.L.; Tsang, K.Y.; Schlom, J. Antibody-Dependent Cellular Cytotoxicity Activity of a Novel Anti–PD-L1 Antibody Avelumab (MSB0010718C) on Human Tumor Cells. Cancer Immunol. Res. 2015, 3, 1148–1157. [Google Scholar] [CrossRef]
- Markham, A. Guselkumab: First Global Approval. Drugs 2017, 77, 1487–1492. [Google Scholar] [CrossRef]
- Kenniston, J.A.; Faucette, R.R.; Martik, D.; Comeau, S.R.; Lindberg, A.P.; Kopacz, K.J.; Conley, G.P.; Chen, J.; Viswanathan, M.; Kastrapeli, N.; et al. Inhibition of Plasma Kallikrein by a Highly Specific Active Site Blocking Antibody. J. Biol. Chem. 2014, 289, 23596–23608. [Google Scholar] [CrossRef]
- Al-Salama, Z.T. Emapalumab: First Global Approval. Drugs 2019, 79, 99–103. [Google Scholar] [CrossRef]
- Salvatore, G.; Beers, R.; Margulies, I.; Kreitman, R.J.; Pastan, I. Improved cytotoxic activity toward cell lines and fresh leukemia cells of a mutant anti-CD22 immunotoxin obtained by antibody phage display. Clin. Cancer Res. 2002, 8, 995–1002. [Google Scholar] [PubMed]
- Alderson, R.F.; Kreitman, R.J.; Chen, T.; Yeung, P.; Herbst, R.; Fox, J.A.; Pastan, I. CAT-8015: A Second-Generation Pseudomonas Exotoxin A–Based Immunotherapy Targeting CD22-Expressing Hematologic Malignancies. Clin. Cancer Res. 2009, 15, 832–839. [Google Scholar] [CrossRef] [PubMed]
- AlFaleh, M.A.; Alsaab, H.O.; Mahmoud, A.B.; Alkayyal, A.A.; Jones, M.L.; Mahler, S.M.; Hashem, A.M. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Urquhart, L. Market watch: Top drugs and companies by sales in 2017. Nat. Rev. Drug Discov. 2018, 17, 232. [Google Scholar] [CrossRef]
- Zhang, H.; Wilson, I.A.; Lerner, R.A. Selection of antibodies that regulate phenotype from intracellular combinatorial antibody libraries. Proc. Natl. Acad. Sci. USA 2012, 109, 15728–15733. [Google Scholar] [CrossRef]
- Visintin, M.; Tse, E.; Axelson, H.; Rabbitts, T.H.; Cattaneo, A. Selection of antibodies for intracellular function using a two-hybrid in vivo system. Proc. Natl. Acad. Sci. USA 1999, 96, 11723–11728. [Google Scholar] [CrossRef]
- Harmansa, S.; Alborelli, I.; Bieli, D.; Caussinus, E.; Affolter, M. A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. eLife 2017, 6, e22549. [Google Scholar] [CrossRef]
- Moutel, S.; Bery, N.; Bernard, V.; Keller, L.; Lemesre, E.; de Marco, A.; Ligat, L.; Rain, J.C.; Favre, G.; Olichon, A.; et al. NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. eLife 2016, 5, e16228. [Google Scholar] [CrossRef] [PubMed]
- Scholler, P.; Nevoltris, D.; de Bundel, D.; Bossi, S.; Moreno-Delgado, D.; Rovira, X.; Møller, T.C.; El Moustaine, D.; Mathieu, M.; Blanc, E.; et al. Allosteric nanobodies uncover a role of hippocampal mGlu2 receptor homodimers in contextual fear con-solidation. Nat. Commun. 2017, 8, 1967. [Google Scholar] [CrossRef]
- Manglik, A.; Kobilka, B.K.; Steyaert, J. Nanobodies to Study G Protein–Coupled Receptor Structure and Function. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Cheloha, R.W.; Fischer, F.A.; Woodham, A.W.; Daley, E.; Suminski, N.; Gardella, T.J.; Ploegh, H.L. Improved GPCR ligands from nanobody tethering. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.; Biocca, S. The selection of intracellular antibodies. Trends Biotechnol. 1999, 17, 115–121. [Google Scholar] [CrossRef]
- Schmidt, F.I.; Hanke, L.; Morin, B.; Brewer, R.; Brusic, V.; Whelan, S.P.; Ploegh, H.L. Phenotypic lentivirus screens to identify functional single domain antibodies. Nat. Microbiol. 2016, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Marasco, A.W. Intrabodies: Turning the humoral immune system outside in for intracellular immunization. Gene Ther. 1997, 4, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Lerner, R.A. Combinatorial antibody libraries: New advances, new immunological insights. Nat. Rev. Immunol. 2016, 16, 498–508. [Google Scholar] [CrossRef]
- Slastnikova, T.A.; Ulasov, A.V.; Rosenkranz, A.A.; Sobolev, A.S. Targeted Intracellular Delivery of Antibodies: The State of the Art. Front. Pharmacol. 2018, 9, 1208. [Google Scholar] [CrossRef]
- Schultz, P.G.; Lerner, A.R. From molecular diversity to catalysis: Lessons from the immune system. Science 1995, 269, 1835–1842. [Google Scholar] [CrossRef]
- Mayes, P.A.; Hance, K.W.; Hoos, A. The promise and challenges of immune agonist antibody development in cancer. Nat. Rev. Drug Discov. 2018, 17, 509–527. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, Y.; Liu, Y.; Wang, Y.; Jia, H.; Kang, M.; Luo, X.; Caballero, D.; González, J.; Sherwood, L.; et al. Functional human antibody CDR fusions as long-acting therapeutic endocrine agonists. Proc. Natl. Acad. Sci. USA 2015, 112, 1356–1361. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yea, K.; Xie, J.; Ruiz, D.; Wilson, I.A.; Lerner, R.A. Selecting Agonists from Single Cells Infected with Combinatorial Antibody Libraries. Chem. Biol. 2013, 20, 734–741. [Google Scholar] [CrossRef]
- Zhang, H.; Xie, J.; Lerner, R.A. A proximity based general method for identification of ligand and receptor interactions in living cells. Biochem. Biophys. Res. Commun. 2014, 454, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Merkouris, S.; Barde, Y.-A.; Binley, K.E.; Allen, N.D.; Stepanov, A.V.; Wu, N.C.; Grande, G.; Lin, C.-W.; Li, M.; Nan, X.; et al. Fully human agonist antibodies to TrkB using autocrine cell-based selection from a combinatorial antibody library. Proc. Natl. Acad. Sci. USA 2018, 115, E7023–E7032. [Google Scholar] [CrossRef] [PubMed]
- Tao, P.; Kuang, Y.; Li, Y.; Li, W.; Gao, Z.; Liu, L.; Qiang, M.; Zha, Z.; Fan, K.; Ma, P.; et al. Selection of a Full Agonist Combinatorial Antibody that Rescues Leptin Deficiency In Vivo. Adv. Sci. 2020, 7, 2000818. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Li, J.; Zhang, N.; Hu, L.A.; Ma, Y.; Tagari, P.; Xu, J.; Zhang, M.-Y. Function-based high-throughput screening for antibody antagonists and agonists against G protein-coupled receptors. Commun. Biol. 2020, 3, 1–10. [Google Scholar] [CrossRef]
- Sasso, E.; D’Avino, C.; Passariello, M.; D’Alise, A.M.; Siciliano, D.; Esposito, M.L.; Froechlich, G.; Cortese, R.; Scarselli, E.; Zambrano, N.; et al. Massive parallel screening of phage libraries for the generation of repertoires of human immunomodulatory monoclonal antibodies. mAbs 2018, 10, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.G.; Vincent, F.; Lee, J.A.; Eder, J.; Prunotto, M. Opportunities and challenges in phenotypic drug discovery: An industry perspective. Nat. Rev. Drug Discov. 2017, 16, 531–543. [Google Scholar] [CrossRef]
- Sidney, L.E.; Branch, M.J.; Dunphy, S.E.; Dua, H.S.; Hopkinson, A. Concise Review: Evidence for CD34 as a Common Marker for Diverse Progenitors. Stem Cells 2014, 32, 1380–1389. [Google Scholar] [CrossRef]
- Watt, F.M.; Hogan, B.L. Out of Eden: Stem cells and their niches. Science 2000, 287, 1427–1430. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, H.; Yea, K.; Lerner, R.A. Autocrine signaling based selection of combinatorial antibodies that transdifferentiate human stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 8099–8104. [Google Scholar] [CrossRef]
- Yea, K.; Zhang, H.; Xie, J.; Jones, T.M.; Lin, C.-W.; Francesconi, W.; Berton, F.; Fallahi, M.; Sauer, K.; Lerner, R.A. Agonist antibody that induces human malignant cells to kill one another. Proc. Natl. Acad. Sci. USA 2015, 112, E6158–E6165. [Google Scholar] [CrossRef]
- Lerner, R.A.; Grover, R.K.; Zhang, H.; Xie, J.; Han, K.H.; Peng, Y.; Yea, K. Antibodies from combinatorial libraries use functional receptor pleiotropism to regulate cell fates. Q. Rev. Biophys. 2015, 48, 389–394. [Google Scholar] [CrossRef]
- Melidoni, A.N.; Dyson, M.R.; Wormald, S.; McCafferty, J. Selecting antagonistic antibodies that control differentiation through inducible expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 17802–17807. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, J.W.; Xie, J.; El-Mecharrafie, N.; Gross, S.; Lee, S.; Lerner, A.R.; Baldwin, K.K. Replacing reprogramming factors with antibodies selected from combinatorial antibody libraries. Nat. Biotechnol. 2017, 35, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Yea, K.; Zhang, H.; Moldt, B.; He, L.; Zhu, J.; Lerner, R.A. Prevention of Cell Death by Antibodies Selected from Intracellular Combinatorial Libraries. Chem. Biol. 2014, 21, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Liu, T.; Kuwana, T.; Zhang, H.; Heiden, M.G.V.; Lerner, R.A.; Newmeyer, D.D. Phenotypic selection with an intrabody library reveals an anti-apoptotic function of PKM2 requiring Mitofusin-1. PLoS Biol. 2019, 17, e2004413. [Google Scholar] [CrossRef]
- Lin, C.-W.; Xie, J.; Zhang, D.; Han, K.H.; Grande, G.; Wu, N.C.; Yang, Z.; Yea, K.; Lerner, R.A. Immunity against cancer cells may promote their proliferation and metastasis. Proc. Natl. Acad. Sci. USA 2019, 117, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Ljungars, A.; Mårtensson, L.; Mattsson, J.; Kovacek, M.; Sundberg, A.; Tornberg, U.-C.; Jansson, B.; Persson, N.; Emruli, V.K.; Ek, S.; et al. A platform for phenotypic discovery of therapeutic antibodies and targets applied on Chronic Lymphocytic Leukemia. npj Precis. Oncol. 2018, 2, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Arlian, B.M.; Macauley, M.S.; Paulson, J.C.; Lerner, R.A. Migration-based selections of antibodies that convert bone marrow into trafficking microglia-like cells that reduce brain amyloid beta. Proc. Natl. Acad. Sci. USA 2018, 115, E372–E381. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Arlian, B.M.; Lin, C.-W.; Jin, H.Y.; Kang, G.-H.; Lee, S.; Lee, P.C.-W.; Lerner, R.A. Agonist Antibody Converts Stem Cells into Migrating Brown Adipocyte-Like Cells in Heart. Cells 2020, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Gonzalez-Quintial, R.; Peng, Y.; Baccala, R.; Theofilopoulos, A.N.; Lerner, R.A. An agonist antibody that blocks autoimmunity by inducing anti-inflammatory macrophages. FASEB J. 2016, 30, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Ying, T.; Du, L.; Ju, T.W.; Prabakaran, P.; Lau, C.C.Y.; Lu, L.; Liu, Q.; Wang, L.; Feng, Y.; Wang, Y.; et al. Exceptionally Potent Neutralization of Middle East Respiratory Syndrome Coronavirus by Human Monoclonal Antibodies. J. Virol. 2014, 88, 7796–7805. [Google Scholar] [CrossRef]
- Sui, J.; Hwang, W.C.; Perez, S.; Wei, G.; Aird, D.; Chen, L.-M.; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009, 16, 265–273. [Google Scholar] [CrossRef]
- Kashyap, A.K.; Steel, J.; Oner, A.F.; Dillon, M.A.; Swale, R.E.; Wall, K.M.; Perry, K.J.; Faynboym, A.; Ilhan, M.; Horowitz, M.; et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. USA 2008, 105, 5986–5991. [Google Scholar] [CrossRef] [PubMed]
- Qiang, M.; Ma, P.; Li, Y.; Liu, H.; Harding, A.; Min, C.; Liu, L.; Yuan, M.; Ji, Q.; Tao, P.; et al. Potent SARS-CoV-2 neutralizing antibodies selected from a human antibody library constructed decades ago. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ng, K.W.; Faulkner, N.; Cornish, G.H.; Rosa, A.; Harvey, R.; Hussain, S.; Ulferts, R.; Earl, C.; Wrobel, A.G.; Benton, D.J.; et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 2020, 370, 1339–1343. [Google Scholar] [CrossRef]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.C.D.; So, R.T.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef]
- Noy-Porat, T.; Makdasi, E.; Alcalay, R.; Mechaly, A.; Levy, Y.; Bercovich-Kinori, A.; Zauberman, A.; Tamir, H.; Yahalom-Ronen, Y.; Israeli, M.; et al. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020, 11, 4303. [Google Scholar] [CrossRef]
- Li, W.; Chen, C.; Drelich, A.; Martinez, D.R.; Gralinski, L.E.; Sun, Z.; Schäfer, A.; Kulkarni, S.S.; Liu, X.; Leist, S.R.; et al. Rapid identification of a human antibody with high prophylactic and therapeutic efficacy in three animal models of SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2020, 117, 29832–29838. [Google Scholar] [CrossRef] [PubMed]
| Antibody | Brand Name | Target | Indications | Approved | Company | Ref. |
|---|---|---|---|---|---|---|
| Adalimumab | Humira | TNFα | Rheumatoid arthritis | 2002 | AbbVie | [11] |
| Belimumab | Benlysta | BLyS | Systemic lupus erythematosus | 2011 | GlaxoSmithKline/Human Genome Sciences | [12] |
| Raxibacumab | Abthrax | B. anthrasis PA | Anthrax infection | 2012 | GlaxoSmithKline/Human Genome Sciences | [13] |
| Ramucirumab | Cyramza | VEGFR2 | Gastric cancer | 2014 | Eli Lilly/ImClone Systems | [14] |
| Necitumumab | Portrazza | EGFR | Non-small cell lung cancer | 2015 | Eli Lilly/ImClone Systems Inc. | [15,16] |
| Atezolizumab | Tecentriq | PD-L1 | Metastatic lung cancer | 2016 | Genentech | [17,18] |
| Avelumab | Bavencio | PD-L1 | Merkel cell carcinoma | 2017 | Merck Serono International S.A./Pfizer | [19] |
| Guselkumab | Tremfya | IL-23 | Plaque psoriasis | 2017 | MorphoSys/Janssen Biotech Inc. | [20] |
| Lanadelumab | Takhzyro | PKaI | Hereditary angioedema attacks | 2018 | Dyax Corp/Shire | [21] |
| Emapalumab | Gamifant | IFNγ | Primary hemophagocytic lymphohistiocytosis | 2018 | NovImmmune | [22] |
| Moxetumomab pasudodox | Lumoxiti | CD22 | Hairy cell leukemia | 2018 | MedImmune/AstraZeneca | [23,24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-W.; Lerner, R.A. Antibody Libraries as Tools to Discover Functional Antibodies and Receptor Pleiotropism. Int. J. Mol. Sci. 2021, 22, 4123. https://doi.org/10.3390/ijms22084123
Lin C-W, Lerner RA. Antibody Libraries as Tools to Discover Functional Antibodies and Receptor Pleiotropism. International Journal of Molecular Sciences. 2021; 22(8):4123. https://doi.org/10.3390/ijms22084123
Chicago/Turabian StyleLin, Chih-Wei, and Richard A. Lerner. 2021. "Antibody Libraries as Tools to Discover Functional Antibodies and Receptor Pleiotropism" International Journal of Molecular Sciences 22, no. 8: 4123. https://doi.org/10.3390/ijms22084123
APA StyleLin, C.-W., & Lerner, R. A. (2021). Antibody Libraries as Tools to Discover Functional Antibodies and Receptor Pleiotropism. International Journal of Molecular Sciences, 22(8), 4123. https://doi.org/10.3390/ijms22084123





