Bioassay Development for Bispecific Antibodies—Challenges and Opportunities
Abstract
1. Introduction
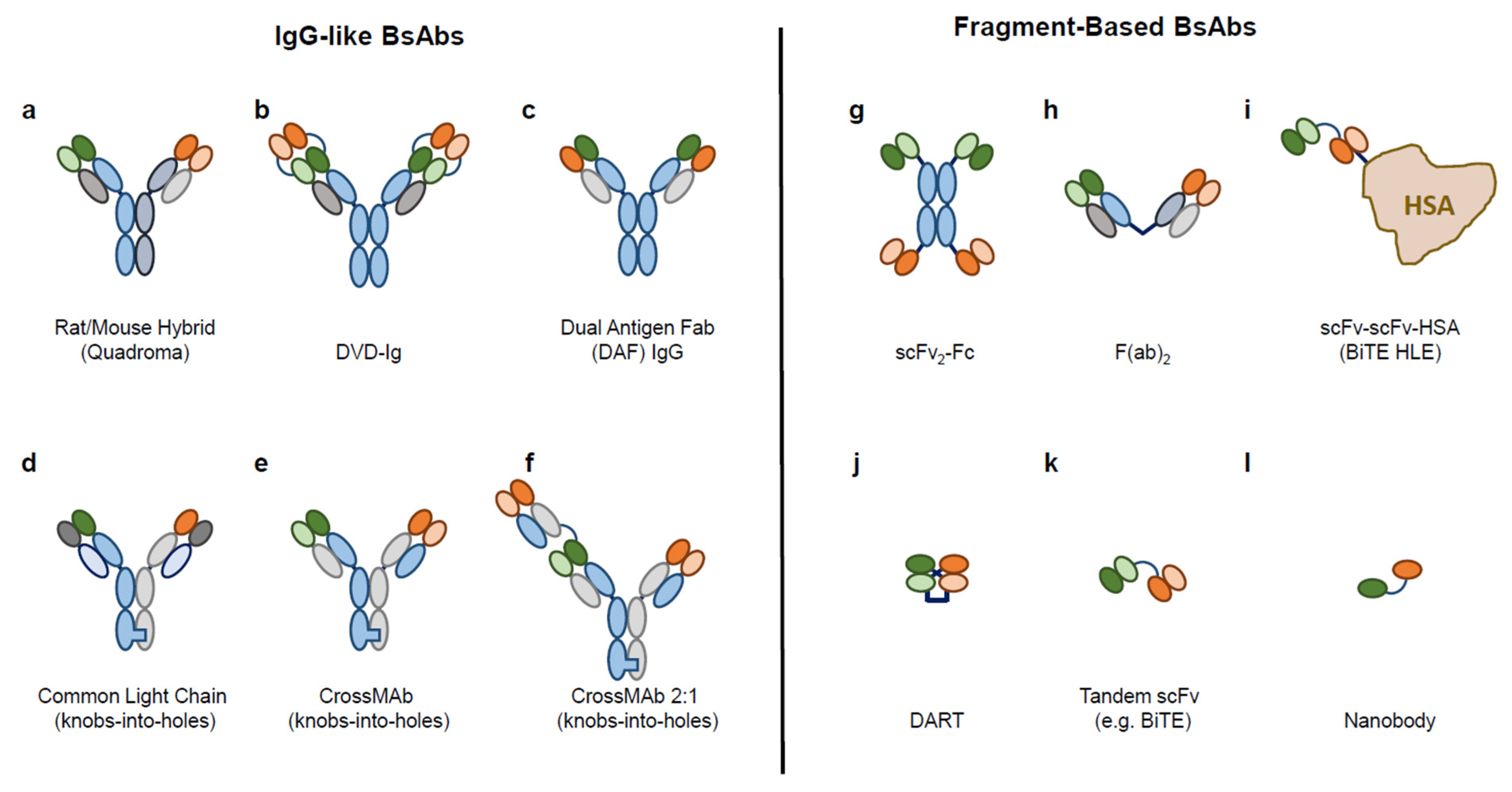
1.1. Diverse Formats of BsAb
1.2. Mechanisms of Action of BsAb
1.2.1. MoA Type 1—Cell-bridging BsAbs
1.2.2. MoA Type 2—Receptor/Ligand-Blocking or -Activating BsAbs
1.2.3. MoA Type 3—Cofactor Mimicking BsAbs
1.2.4. MoA Type 4—“Homing” BsAbs
1.3. Challenges and Opportunities of BsAb Bioassay Development
2. Strategies and Considerations for BsAb Bioassay Development
2.1. Phase-Appropriate Approach
2.2. Mechanism of Action
2.3. Overall BsAb Characterization Strategy
3. Bioassays for Bispecific Antibodies and Case Studies
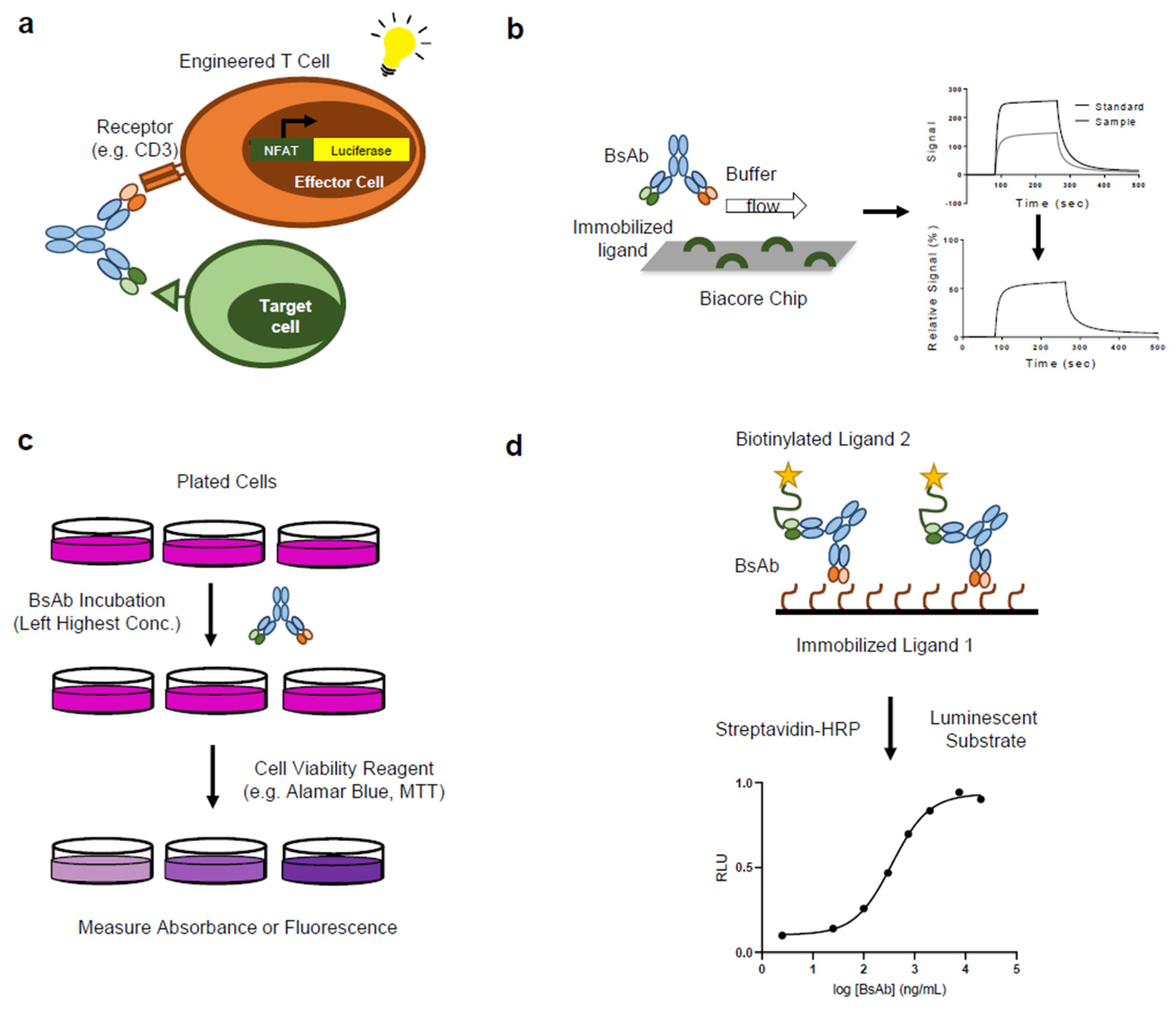
3.1. Bioassays for Biotherapeutics
3.2. BsAb Bioassay: Binding Assays
- Case Study: ELISA to detect simultaneous target binding by a tetravalent BsAb [86]: The authors developed a tetravalent BsAb composed of two different variable heavy (VH) domains on an IgG framework (tetra-VH IgG). The structure was meant to be an alternative to dual-action Fab (DAF) molecules, which are often difficult to generate via mutagenesis and additionally may not be able to bind both targets simultaneously, due to overlapping binding surfaces. Three bispecific tetra-VH IgGs were created (CD40/OX40, 4-1BB/CD40, OX40/4-1BB). In order to confirm that the BsAbs were able to bind both targets simultaneously, a competition ELISA assay was developed. The BsAb was immobilized on a plate and incubated with non-biotinylated ligand (for example OX40), followed by biotinylated ligand (for example OX40 or CD40). For each BsAb, binding of the biotinylated ligand was only inhibited by binding of the biotinylated ligand of the same specificity, while the ligand of the other specificity retained binding, suggesting that the BsAb is able to bind both targets simultaneously. To confirm that each arm of the BsAb (VHOX40-VH4-1BB Fab) can simultaneously bind each target, a bridging ELISA assay was used. An assay plate was coated with OX40 and allowed to bind to the VHOX40-VH41BB Fab before being incubated with varying concentrations of biotinylated 4-1BB or biotinylated OX40. Dose-dependent binding of 4-1BB was observed, while no binding of OX40 was observed, supporting the conclusion that each arm of the BsAb is able to simultaneously bind both targets.
- Case Study: SPR to measure kinetics and binding affinity of Ang-2/VEGF BsAb [88,89]: The authors evaluated a humanized bivalent-BsAb generated for the neutralization of angiopoietin-2 (Ang-2) and vascular endothelial growth factor A (VEGF-A) [88]. Using SPR technology to characterize the kinetics and affinity of binding, the authors developed an assay to cover two binding events simultaneously, which can be reported as one response. Assay setup utilized a Biacore™ instrument and commonly used CM5 chips, and followed a scheme of sequential additions of the CrossMab and then Ang-2 to immobilized VEGF. The second binding event (Ang-2 binding to the VEGF-bound CrossMab) included surface regeneration. As a result, Ang-2-binding response is dependent on the amount of VEGF-bound CrossMab molecules and therefore reflects the actual bridging signal. In this assay, SPR-detected bridging signal reflects the active concentration of Ang-2- and VEGF-binding molecules, where the loss of overall binding can be attributed to either the VEGF or the Ang-2 binding contribution, and therefore, covers both antigen interactions. A modified SPR-based dual-binding assay was developed by Meschendoerfer et al. [89] to address the pitfalls associated with the bridging assay—specifically the change of antigen activity upon immobilization. The main objective was to allow for the individual assessment of both targets in solution while avoiding the need for immobilization and regeneration of the target. They determined the individual VEGFA-121 and Ang2 activities of an anti-VEGFA-121/Ang2 BsAb where an anti-human-Fab capture system (for the Ang2 antigen) was used to measure different antibody concentrations with the same Biosensor (regeneration cycles included). The findings suggested that comparable binding signals can be read from individual injections, when compared to an approach with sequential antigen injection. Using this assay, they showed that simultaneous binding can be calculated based on both individual readouts: two binding events can be measured, and the third parameter can be accurately calculated based on these measurements.
Cell Surface Ligand Binding Assays
- Case Study: Flow cytometry-based binding and blocking assays for GPC3/CD47 BsAb [92]: To develop a potential immune-modulating therapeutic to treat hepatocellular carcinoma (HCC), the authors designed a novel BsAb directed against the HCC-associated antigen Glypican-3 (GPC3) and CD47, an inhibitory innate immune checkpoint that inhibits ADCP by binding to SIRPa on myeloid cells. Due to the fact that CD47 is widely expressed on both healthy and cancerous cells, treatment with anti-CD47 mAbs is associated with toxicity. Therefore, the authors sought to direct the ADCP-enhancing activity of targeting CD47 to GPC3+ tumor cells using a bispecific approach. Several flow cytometry-based binding assays were used to demonstrate selective targeting of GPC3+ cells. For example, wild-type Raji cells (GPC3-) were labeled with a fluorescent dye and mixed in a 1:1 ratio with unlabeled Raji cells engineered to express GPC3 (Raji-GPC3H) prior to incubation with GPC3/CD47 BsAb or anti-CD47 mAb. Following incubation with a FITC-labeled secondary antibody, labeled and unlabeled cells were separated by flow cytometry and the binding of the BsAb to each cell population was assessed. The results showed higher levels of BsAb binding to Raji-GPC3H cells compared to the wild-type cells. In contrast, no difference in binding was observed for an anti-CD47 mAb. The authors further tested the ability of the BsAb to block the interaction between CD47 and SIRPa in each cell type using a competitive flow cytometry assay. Wild-type and Raji-GPC3H cells were incubated with biotinylated SIRPa-mF in the presence of anti-CD47 mAb or GPC3/CD47 BsAb, followed by the addition of FITC-labeled streptavidin. The results showed that the BsAb prevented SIRPa binding more effectively in the Raji-GPC3H cells, while the anti-CD47 mAb showed similar blocking activity in both cell types. The results of the flow cytometry-based binding assays demonstrate preferential binding and blocking activities of the GPC3/CD47 BsAb in GPC3+/CD47+ compared to GPC3-/CD47+ cells in vitro. These results suggest that the bispecific targeting of GPC3+ and CD47 may preferentially induce killing of GPC3+ tumor cells by ADCP.
- Case Study: Flow cytometry-based assay to characterize binding activity of PD-L1xCSPG4 BsAb [93]: To improve antibody-therapy efficacy in patients with advanced melanoma, the authors developed a BsAb, PD-L1xCSPG4, to selectively reactivate T cells by directing PD-1/PD-L1 disrupting activity to chondroitin sulfate proteoglycan 4 (CSPG4)-expressing tumor cells. A flow cytometry-based assay was employed to evaluate the binding activities of the investigational BsAb. Wild-type ectopically hPD-L1-expressing CHO cells (CHO.PD-L1) were incubated with test antibodies, labeled with a fluorescent secondary antibody, and analyzed by flow cytometry. Dose-dependent binding specific to CHO.PD-L1 cells was observed for the BsAb. These binding activities were replicated in several representative cancer cells endogenously expressing both CSPG4 or PD-L1. In addition, a flow cytometry-based competitive binding assay was used to assess the overall binding strength (avidity) of PD-L1xCSPG4 BsAb to CSPG4+/PD-L1+ cancer cells. BsAb binding was strongly inhibited in the presence of competing parental anti-CSPG4 mAb and only weakly inhibited in the presence of competing PD-L1-blocking mAb. These experiments demonstrate that PD-L1xCSPG4 binds to both PD-L1 and CSPG4 and that the strength of the interaction between the BsAb and CSPG4+/PD-L1+ cancer cells is primarily dominated by binding to CSGA4. To further show that the enhanced binding of the BsAb to CSPG4+/PD-L1+ cells is driven by avidity, cells were pre-incubated with a fluorescent anti-PD-L1 mAb, before being exposed to the test BsAb and a control BsAb, capable of binding PD-L1 but not CSPG4. The EC50 of PD-L1xCSPG4 for displacing the probe was substantially lower compared to the control BsAb. Performing the experiment in the presence of an anti-GSPG4 mAb increased the EC50 of the PD-L1xCSPG4 BsAb to a level similar to the control BsAb. Together these flow cytometry-based binding assays demonstrated that the PDL1xCSPG4 BsAb has enhanced selectivity for CSPG4+/PD-L1+ cancer cells driven by avidity binding.
- Case Study: Cell-based reporter assay to measure cell surface binding of PDL1xCSPG4 BsAb [93]: The authors further evaluated the role of CSPG4 in mediating the PD-L1-blocking capacity of the PDL1xCSPG4 BsAb using a PD-1/PD-L1 blockade reporter bioassay. The assay relies on co-culturing of Jurkat.PD-1-NFAT-luc reporter T cells (Jurkat cells engineered to express luciferase under the control of a NFAT response element and PD-1) and CHO.PDL1/CD3 cells (CHO cells engineered to express PD-L1 and a membrane-linked agonistic anti-CD3 antibody). Upon successful interaction of PD-1 and PD-L1 between the two cell types, TCR signaling and downstream NFAT-mediated luciferase activity in the Jurkat cells is inhibited. In contrast, interrupting the PD-1/PD-L1 interaction leads to NFAT-mediated luciferase activity. Addition of the PDL1xCSPG4 BsAb to the co-culture disrupted the PD-1/PD-L1 interaction between the two cell types in a dose-dependent manner, as measured by luminescence detection. Next, they tested the role of CSPG4 mAb in PD-1/PD-L1 blocking capacity of PDL1xCSPG4 BsAb by replacing the CHO.PD-L1/CD3 cells with a CSPG4+/PD-L1+ cancer cell line (the CD3 stimulation of T cells was achieved by pre-treating the cells with BIS1; an EpCAM-directed CD3-agonistic bsAb). Stimulated reporter T cells were co-cultured with the double-positive cells in the presence of PDL1xCSPG4 BsAb or controls, with and without anti-CSPG4 mAb. The ability of the PDL1xCSPG4 BsAb to block PD-1/PD-L1 interaction was reduced in the presence of anti-CSPG4 mAb. These findings suggest that the BsAb’s PD-1/PD-L1-disrupting activity will be enhanced against CSPG4+/PD-L1+ cells compared to CSPG4-/PD-L1+ cells.
3.3. BsAb Bioassay: Potency Assays
- Case Study: Reporter gene T cell activation assay to measure potency of CD3e-binding TDB [96]: The authors developed a reporter gene potency assay that measures T-cell activation in the presence of a CD3e-binding TDB, using Jurkat T-cells engineered to express luciferase under the control of a T-cell activation-sensitive transcriptional response element. The assay was shown to be quantitative and stability indicating. Additionally, it is robust and relatively fast/easy to perform compared to a traditional cell-based cell killing assay, such as a Cr51 release or dye release assay, making it more amenable to a QC testing environment. The MoA of the TDB is complex as it consists of multiple factors—concurrent antigen binding, T-cell activation, and target-cell depletion. In order to show that T-cell activation in an engineered context is a suitable surrogate measure of the TDB’s overall bioactivity, the authors generated a characterization data package consisting of data from individual antigen binding assays (cell-based ELISA to measure binding to the target receptor, SPR to measure binding to CD3) and a flow cytometry-based cell killing assay that used human peripheral blood mononuclear cells as a source of cytolytic T-cells. By using the data generated from the characterization assays, the authors were able to show that changes in potency detected by the reporter gene assay agreed well with changes in affinity for either antigen and cell killing activity. The characterization results support the assertion that the reporter gene assay is sufficiently MoA-reflective to serve as a single potency assay on the control system without the need for additional assays. The authors’ overall strategy can be applied to justify potency for other TDBs with similar MoAs.
- Case Study: Cell-based potency assay to measure biological activity of HER3/EGFR DAF BsAb [69]: In order to measure the activity of a DAF molecule designed to simultaneously inhibit HER3 and EGFR, the authors developed a cell-based potency assay that measures cell proliferation using a cell-permeable redox dye, in which the fluorescence signal is proportional to the number of viable cells. This method was selected based on the molecule’s proposed MoA, which is characterized by blocking ligand binding to each receptor, prevention of receptor dimerization (hetero- and homo-), and inhibition of cell proliferation. A cell line that naturally expresses both receptors and their cognate ligands was selected in order to enable monitoring of the effects of inhibiting both receptors. As in the case study described above, the author’s generated a characterization data package using the potency assay and individual ELISA binding assays for HER3 and EGFR to show that the single potency assay is reflective of the DAF’s overall bioactivity. The fact that the potency assay was demonstrated to be sensitive to changes in affinity for either target and sensitive to inhibition of both receptors, with inhibition of both HER3 and EGFR by the DAF producing the most potent anti-proliferative activity, provides a strong justification that the potency assay sufficiently captures the molecule’s MoA. This, combined with the potency assay’s quantitative ability and stability indicating properties, provides a persuasive argument that it is suitable as a single control system assay for monitoring the impact of product quality on bioactivity.
3.4. BsAb Bioassay: Effector Function Assays
- Case Study: ADCC reporter assay and competitive ADCP assay to measure enhanced Fc-mediated effector function of GPC3/CD47 BsAb [92]: As mediated by the Fc domain function of therapeutic antibodies, ADCC and ADCP assays are among the appropriate ones to assess the enhanced Fc-mediated effector functions of investigational BsAbs. The authors employed a cell-based reporter system to evaluate the ability of BsAb in inducing ADCC against dual-antigen-expressing Raji-GPC3H cells. In this assay format, engineered Jurkat T lymphocyte cells were used as effector cells. Target cells, including wild-type Raji and Raji-GPC3H cells, were incubated with each mAb and BsAb test antibodies and effector cells. A luminescent substrate was used to measure the luciferase activity at the end of co-incubation that corresponds to the extent of the effector activities. This bioassay revealed that GPC3/CD47 BsAb could induce ADCC against dual-antigen-expressing Raji-GPC3H cells in a dose-dependent manner and to a greater extent compared to the wild-type Raji cells. The ability of BsAb to induce ADCP in vitro was also evaluated upon co-incubation of Raji-GPC3H cells with macrophages. In this assay setup, the effector cells [mouse hSIRPa expressing bone marrow-derived macrophages (BMDMs) harvested from humanized mouse bone marrow] were Alexa Fluor647-labeled and incubated with target cells (Raji or Raji-GPC3H cells) stained with a fluorescent proliferation dye and each antibody. Effector:target cell mixture was then evaluated for ADCP where the phagocytosis of fluorescent-labeled target cells by labeled BMDMs was recorded using a confocal microscope (unphagocytosed cells were washed away prior to microscopy). In this bioassay format, using fluorescence microscopy and quantification of phagocytosis, the authors showed a preferential phagocytosis of dual-antigen-expressing Raji-GPC3H cells specifically in the presence of GPC3/CD47 BsAb.
- Case Study: Use of a CDC assay to assess activity of a complement-regulator neutralizing BsAbs directed against CD20 and CD55/CD59 [75]: The authors designed BsAbs to increase complement-mediated killing of CD20-expressing B cells. By simultaneous targeting CD20 and CD55/CD59, the BsAbs are able to neutralize the C-regulating proteins on B cells, leading to more efficient killing by CDC. Various CD20-expressing cells were treated with the BsAbs, followed by incubation with human sera (source of complement). Following an incubation period, cells were assessed for viability using MTT (a dye that is reduced to form a purple dye in the presence of metabolically active cells). Cell killing was enhanced by treatment with the BsAbs compared to treatment with an effector-competent aCD20 mAb. Additionally, cell killing levels remained consistent in the presence of CD20-bystander cells expressing CD55 and CD59, suggesting that the BsAbs are selectively killing CD20+ B cells. Flow cytometry-based binding assays were used to confirm binding to cells expressing CD20, CD55, and CD59.
3.5. BsAb Bioassay: Impurities Assays
- Case Study: Luciferase reporter T cell activation assay to measure functional effects of impurities on CD3e-targeting TDB [95]: A CD3e-targeting TDB produced by knobs-into-holes technology and assembled in vitro contains a number of product-related impurities with the potential to activate T-cells in the absence of target cells. For example, aggregates and aCD3 homodimer, which result from the mispairing of aCD3 half antibody fragments during production, are characterized by multivalent binding to CD3 and can crosslink the TCR resulting in activation. These impurities are a safety concern because T-cell activation is linked to adverse events such as cytokine release syndrome. While aggregates and aCD3 HD can be measured using analytical methods, a bioassay is needed to assess their biological impact, such as target-independent T-cell activation. To address this need, the authors developed a reporter gene assay that measures T-cell activation in the absence of target cells using Jurkat T-cells engineered to express luciferase when activated. T-cell activation of product-related impurities present in the TDB formulation was quantified relative to T-cell activation by aCD3 HD standard. Using this assay, the authors were able to characterize the T-cell activating activities of aggregates and other product-related impurities, in order to get an idea of their potential impacts to safety and inform on the overall control strategy. Additionally, because the assay is a “catchall” assay that measures the combined T-cell activating activity of product-related impurities that may be present in a given sample, the method is able to provide reassurance that combinations of impurities are not leading to unexpected T-cell activation. Such combination effects would not be identified using physicochemical methods alone.
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lejeune, M.; Köse, M.C.; Duray, E.; Einsele, H.; Beguin, Y.; Caers, J. Bispecific, T-cell-recruiting antibodies in B-cell malignancies. Front. Immunol. 2020, 11, 762. [Google Scholar] [CrossRef] [PubMed]
- Perez, P.; Hoffman, R.W.; Shaw, S.; Bluestone, J.A.; Segal, D.M. Specific targeting of cytotoxic T cells by anti-T3 linked to anti-target cell antibody. Nature 1985, 316, 354–356. [Google Scholar] [CrossRef] [PubMed]
- Labrijn, A.F.; Janmaat, M.L.; Reichert, J.M.; Parren, P.W.H.I. Bispecific antibodies: A mechanistic review of the pipeline. Nat. Rev. Drug Discov. 2019, 18, 585–608. [Google Scholar] [CrossRef]
- Reichert, J. Bispecific Antibodies Come to the Fore. Available online: https://www.antibodysociety.org/antibody-therapeutics-pipeline/bispecific-antibodies-come-to-the-fore/ (accessed on 15 March 2021).
- Spiess, C.; Zhai, Q.; Carter, P.J. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol. Immunol. 2015, 67, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, U.; Kontermann, R.E. The making of bispecific antibodies. mAbs 2017, 9, 182–212. [Google Scholar] [CrossRef]
- Dickopf, S.; Georges, G.J.; Brinkmann, U. Format and geometries matter: Structure-based design defines the functionality of bispecific antibodies. Comput. Struct. Biotechnol. J. 2020, 18, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Fabozzi, G.; Pegu, A.; Koup, R.A.; Petrovas, C. Bispecific antibodies: Potential immunotherapies for HIV treatment. Methods 2019, 154, 118–124. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, Y.; Park, J.; Liu, X.; Hu, Y.; Wang, T.; McFarland, K.; Betenbaugh, M.J. Design and production of bispecific antibodies. Antibodies 2019, 8, 43. [Google Scholar] [CrossRef]
- Kontermann, R.E.; Brinkmann, U. Bispecific antibodies. Drug Discov. Today 2015, 20, 838–847. [Google Scholar] [CrossRef]
- Slaga, D.; Ellerman, D.; Lombana, T.N.; Vij, R.; Li, J.; Hristopoulos, M.; Clark, R.; Johnston, J.; Shelton, A.; Mai, E.; et al. Avidity-based binding to HER2 results in selective killing of HER2-overexpressing cells by anti-HER2/CD3. Sci. Transl. Med. 2018, 10, eaat5775. [Google Scholar] [CrossRef]
- Kontermann, R.E. Strategies to extend plasma half-lives of recombinant antibodies. BioDrugs 2009, 23, 93–109. [Google Scholar] [CrossRef]
- Huang, S.; Segués, A.; Hulsik, D.L.; Zaiss, D.M.; Sijts, A.J.A.M.; van Duijnhoven, S.M.J.; van Elsas, A. A novel efficient bispecific antibody format, combining a conventional antigen-binding fragment with a single domain antibody, avoids potential heavy-light chain mis-pairing. J. Immunol. Methods 2020, 483, 112811. [Google Scholar] [CrossRef] [PubMed]
- Atwell, S.; Ridgway, J.B.B.; Wells, J.A.; Carter, P. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J. Mol. Biol. 1997, 270, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, W.; Regula, J.T.; Bähner, M.; Schanzer, J.; Croasdale, R.; Dürr, H.; Gassner, C.; Georges, G.; Kettenberger, H.; Imhof-Jung, S.; et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc. Natl. Acad. Sci. USA 2011, 201019002. [Google Scholar] [CrossRef] [PubMed]
- Krah, S.; Schröter, C.; Eller, C.; Rhiel, L.; Rasche, N.; Beck, J.; Sellmann, C.; Günther, R.; Toleikis, L.; Hock, B.; et al. Generation of human bispecific common light chain antibodies by combining animal immunization and yeast display. Protein Eng. Des. Sel. 2017, 30, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Ahamadi-Fesharaki, R.; Fateh, A.; Vaziri, F.; Solgi, G.; Siadat, S.D.; Mahboudi, F.; Rahimi-Jamnani, F. Single-chain variable fragment-based bispecific antibodies: Hitting two targets with one sophisticated arrow. Mol. Ther. Oncolytics 2019, 14, 38–56. [Google Scholar] [CrossRef] [PubMed]
- Portell, C.A.; Wenzell, C.M.; Advani, A.S. Clinical and pharmacologic aspects of blinatumomab in the treatment of B-cell acute lymphoblastic leukemia. Clin. Pharm. 2013, 5, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.; Karle, A.; Meißburger, B.; Höfig, I.; Stork, R.; Kontermann, R.E. Improved pharmacokinetics of recombinant bispecific antibody molecules by fusion to human serum albumin. J. Biol. Chem. 2007, 282, 12650–12660. [Google Scholar] [CrossRef] [PubMed]
- Lum, L.; Davol, P.; Lee, R. The new face of bispecific antibodies: Targeting cancer and much more. Exp. Hematol. 2006, 34, 1–6. [Google Scholar] [CrossRef]
- Hoffmann, P.; Hofmeister, R.; Brischwein, K.; Brandl, C.; Crommer, S.; Bargou, R.; Itin, C.; Prang, N.; Baeuerle, P.A. Serial killing of tumor cells by cytotoxic T cells redirected with a CD19-/CD3-bispecific single-chain antibody construct. Int. J. Cancer 2005, 115, 98–104. [Google Scholar] [CrossRef]
- Ross, S.L.; Sherman, M.; McElroy, P.L.; Lofgren, J.A.; Moody, G.; Baeuerle, P.A.; Coxon, A.; Arvedson, T. Bispecific T cell engager (BiTE®) antibody constructs can mediate bystander tumor cell killing. PLoS ONE 2017, 12, e0183390. [Google Scholar] [CrossRef] [PubMed]
- Bacac, M.; Fauti, T.; Sam, J.; Colombetti, S.; Weinzierl, T.; Ouaret, D.; Bodmer, W.; Lehmann, S.; Hofer, T.; Hosse, R.J.; et al. A Novel carcinoembryonic antigen t-cell bispecific antibody (CEA TCB) for the treatment of solid tumors. Clin. Cancer Res. 2016, 22, 3286–3297. [Google Scholar] [CrossRef] [PubMed]
- Kruse, R.L.; Shum, T.; Legras, X.; Barzi, M.; Pankowicz, F.P.; Gottschalk, S.; Bissig, K.-D. In situ liver expression of HBsAg/CD3-bispecific antibodies for HBV immunotherapy. Mol. Ther. Methods Clin. Dev. 2017, 7, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Laszlo, G.S.; Gudgeon, C.J.; Harrington, K.H.; Dell’Aringa, J.; Newhall, K.J.; Means, G.D.; Sinclair, A.M.; Kischel, R.; Frankel, S.R.; Walter, R.B. Cellular determinants for preclinical activity of a novel CD33/CD3 bispecific T-cell engager (BiTE) antibody, AMG 330, against human AML. Blood 2014, 123, 554–561. [Google Scholar] [CrossRef]
- Junttila, T.T.; Li, J.; Johnston, J.; Hristopoulos, M.; Clark, R.; Ellerman, D.; Wang, B.-E.; Li, Y.; Mathieu, M.; Li, G.; et al. Antitumor efficacy of a bispecific antibody that targets HER2 and activates T cells. Cancer Res. 2014, 74, 5561–5571. [Google Scholar] [CrossRef]
- Li, J.; Ybarra, R.; Mak, J.; Herault, A.; De Almeida, P.; Arrazate, A.; Ziai, J.; Totpal, K.; Junttila, M.R.; Walsh, K.B.; et al. IFNγ-induced chemokines are required for CXCR3-mediated T-cell recruitment and antitumor efficacy of anti-HER2/CD3 bispecific antibody. Clin. Cancer Res. 2018, 24, 6447–6458. [Google Scholar] [CrossRef]
- Meng, W.; Tang, A.; Ye, X.; Gui, X.; Li, L.; Fan, X.; Schultz, R.D.; Freed, D.C.; Ha, S.; Wang, D.; et al. Targeting human-cytomegalovirus-infected cells by redirecting T Cells using an anti-CD3/anti-glycoprotein B bispecific antibody. Antimicrob. Agents Chemother. 2018, 62, e01717–e01719. [Google Scholar] [CrossRef]
- Osada, T.; Patel, S.P.; Hammond, S.A.; Osada, K.; Morse, M.A.; Lyerly, H.K. CEA/CD3-bispecific T cell-engaging (BiTE) antibody-mediated T lymphocyte cytotoxicity maximized by inhibition of both PD1 and PD-L1. Cancer Immunol. Immunother. 2015, 64, 677–688. [Google Scholar] [CrossRef]
- Reusch, U.; Harrington, K.H.; Gudgeon, C.J.; Fucek, I.; Ellwanger, K.; Weichel, M.; Knackmuss, S.H.; Zhukovsky, E.A.; Fox, J.A.; Kunkel, L.A.; et al. Characterization of CD33/CD3 Tetravalent bispecific tandem diabodies (TandAbs) for the treatment of acute myeloid leukemia. Clin. Cancer Res. 2016, 22, 5829–5838. [Google Scholar] [CrossRef]
- Schlereth, B.; Kleindienst, P.; Fichtner, I.; Lorenczewski, G.; Brischwein, K.; Lippold, S.; Silva, A.D.; Locher, M.; Kischel, R.; Lutterbüse, R.; et al. Potent inhibition of local and disseminated tumor growth in immunocompetent mouse models by a bispecific antibody construct specific for Murine CD3. Cancer Immunol. Immunother. 2006, 55, 785–796. [Google Scholar] [CrossRef]
- Shiraiwa, H.; Narita, A.; Kamata-Sakurai, M.; Ishiguro, T.; Sano, Y.; Hironiwa, N.; Tsushima, T.; Segawa, H.; Tsunenari, T.; Ikeda, Y.; et al. Engineering a bispecific antibody with a common light chain: Identification and optimization of an anti-CD3 epsilon and anti-GPC3 bispecific antibody, ERY974. Methods 2019, 154, 10–20. [Google Scholar] [CrossRef]
- Sung, J.A.M.; Pickeral, J.; Liu, L.; Stanfield-Oakley, S.A.; Lam, C.-Y.K.; Garrido, C.; Pollara, J.; LaBranche, C.; Bonsignori, M.; Moody, M.A.; et al. Dual-affinity re-targeting proteins direct T cell–mediated cytolysis of latently HIV-infected cells. J. Clin. Investig. 2015, 125, 4077–4090. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.L.; Ellerman, D.; Mathieu, M.; Hristopoulos, M.; Chen, X.; Li, Y.; Yan, X.; Clark, R.; Reyes, A.; Stefanich, E.; et al. Anti-CD20/CD3 T cell–dependent bispecific antibody for the treatment of B cell malignancies. Sci. Transl. Med. 2015, 7, 287ra270. [Google Scholar] [CrossRef] [PubMed]
- Grosse-Hovest, L.; Hartlapp, I.; Marwan, W.; Brem, G.; Rammensee, H.-G.; Jung, G. A recombinant bispecific single-chain antibody induces targeted, supra-agonistic CD28-stimulation and tumor cell killing. Eur. J. Immunol. 2003, 33, 1334–1340. [Google Scholar] [CrossRef]
- Kroesen, B.J.; Bakker, A.; van Lier, R.A.; The, H.T.; de Leij, L. Bispecific antibody-mediated target cell-specific costimulation of resting T cells via CD5 and CD28. Cancer Res. 1995, 55, 4409–4415. [Google Scholar] [PubMed]
- Li, J.; Stagg, N.J.; Johnston, J.; Harris, M.J.; Menzies, S.A.; DiCara, D.; Clark, V.; Hristopoulos, M.; Cook, R.; Slaga, D.; et al. Membrane-proximal epitope facilitates efficient T cell synapse formation by anti-FcRH5/CD3 and is a requirement for myeloma cell killing. Cancer Cell 2017, 31, 383–395. [Google Scholar] [CrossRef]
- Strohl, W.R.; Naso, M. Bispecific T-cell redirection versus chimeric antigen receptor (CAR)-T cells as approaches to kill cancer cells. Antibodies 2019, 8, 41. [Google Scholar] [CrossRef]
- Piscopo, N.J.; Mueller, K.P.; Das, A.; Hematti, P.; Murphy, W.L.; Palecek, S.P.; Capitini, C.M.; Saha, K. Bioengineering solutions for manufacturing challenges in CAR T cells. Biotechnol. J. 2018, 13, 1700095. [Google Scholar] [CrossRef]
- Shimabukuro-Vornhagen, A.; Gödel, P.; Subklewe, M.; Stemmler, H.J.; Schlößer, H.A.; Schlaak, M.; Kochanek, M.; Böll, B.; von Bergwelt-Baildon, M.S. Cytokine release syndrome. J. Immunother. Cancer 2018, 6, 56. [Google Scholar] [CrossRef]
- Subklewe, M. BiTEs better than CAR T cells. Blood Adv. 2021, 5, 607–612. [Google Scholar] [CrossRef]
- Pahl, J.H.W.; Koch, J.; Götz, J.J.; Arnold, A.; Reusch, U.; Gantke, T.; Rajkovic, E.; Treder, M.; Cerwenka, A. CD16A Activation of NK cells promotes NK cell proliferation and memory-like cytotoxicity against cancer cells. Cancer Immunol. Res. 2018, 6, 517–527. [Google Scholar] [CrossRef]
- Reusch, U.; Burkhardt, C.; Fucek, I.; Le Gall, F.; Le Gall, M.; Hoffmann, K.; Knackmuss, S.H.J.; Kiprijanov, S.; Little, M.; Zhukovsky, E.A. A novel tetravalent bispecific TandAb (CD30/CD16A) efficiently recruits NK cells for the lysis of CD30+ tumor cells. mAbs 2014, 6, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Oberg, H.H.; Kellner, C.; Gonnermann, D.; Sebens, S.; Bauerschlag, D.; Gramatzki, M.; Kabelitz, D.; Peipp, M.; Wesch, D. Tribody [(HER2)2xCD16] is more effective than trastuzumab in enhancing γδ T cell and natural killer cell cytotoxicity against HER2-expressing cancer cells. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Schmohl, J.U.; Felices, M.; Taras, E.; Miller, J.S.; Vallera, D.A. Enhanced ADCC and NK cell activation of an anticarcinoma bispecific antibody by genetic insertion of a modified IL-15 cross-linker. Mol. Ther. 2016, 24, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Xu, L.; Pi, C.; Yin, Y.; Xie, K.; Tao, F.; Li, R.; Gu, H.; Fang, J. CD89-mediated recruitment of macrophages via a bispecific antibody enhances anti-tumor efficacy. OncoImmunology 2018, 7, e1380142. [Google Scholar] [CrossRef] [PubMed]
- Kolumam, G.; Chen, M.Z.; Tong, R.; Zavala-Solorio, J.; Kates, L.; van Bruggen, N.; Ross, J.; Wyatt, S.K.; Gandham, V.D.; Carano, R.A.D.; et al. Sustained brown fat stimulation and insulin sensitization by a humanized bispecific antibody agonist for fibroblast growth factor receptor 1/βKlotho complex. EBioMedicine 2015, 2, 730–743. [Google Scholar] [CrossRef]
- Yu, S.; Liu, Q.; Han, X.; Qin, S.; Zhao, W.; Li, A.; Wu, K. Development and clinical application of anti-HER2 monoclonal and bispecific antibodies for cancer treatment. Exp. Hematol. Oncol. 2017, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Perry, S.R.; Muniz-Medina, V.; Wang, X.; Wetzel, L.K.; Rebelatto, M.C.; Hinrichs, M.J.M.; Bezabeh, B.Z.; Fleming, R.L.; Dimasi, N.; et al. A biparatopic HER2-targeting antibody-drug conjugate induces tumor regression in primary models refractory to or ineligible for HER2-targeted therapy. Cancer Cell 2016, 29, 117–129. [Google Scholar] [CrossRef]
- Veri, M.-C.; Burke, S.; Huang, L.; Li, H.; Gorlatov, S.; Tuaillon, N.; Rainey, G.J.; Ciccarone, V.; Zhang, T.; Shah, K.; et al. Therapeutic control of B cell activation via recruitment of Fcγ receptor IIb (CD32B) inhibitory function with a novel bispecific antibody scaffold. Arthritis Rheum. 2010, 62, 1933–1943. [Google Scholar] [CrossRef]
- DiGiandomenico, A.; Keller, A.E.; Gao, C.; Rainey, G.J.; Warrener, P.; Camara, M.M.; Bonnell, J.; Fleming, R.; Bezabeh, B.; Dimasi, N.; et al. A multifunctional bispecific antibody protects against Pseudomonas aeruginosa. Sci. Transl. Med. 2014, 6, 262ra155. [Google Scholar] [CrossRef]
- Jimeno, A.; Moore, K.N.; Gordon, M.; Chugh, R.; Diamond, J.R.; Aljumaily, R.; Mendelson, D.; Kapoun, A.M.; Xu, L.; Stagg, R.; et al. A first-in-human phase 1a study of the bispecific anti-DLL4/anti-VEGF antibody navicixizumab (OMP-305B83) in patients with previously treated solid tumors. Investig. New Drugs 2019, 37, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Moores, S.; Jarantow, S.; Pardinas, J.; Chiu, M.; Zhou, H.; Wang, W. Cross-arm binding efficiency of an EGFR x c-Met bispecific antibody. mAbs 2016, 8, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Grugan, K.D.; Dorn, K.; Jarantow, S.W.; Bushey, B.S.; Pardinas, J.R.; Laquerre, S.; Moores, S.L.; Chiu, M.L. Fc-mediated activity of EGFR x c-Met bispecific antibody JNJ-61186372 enhanced killing of lung cancer cells. mAbs 2017, 9, 114–126. [Google Scholar] [CrossRef]
- Patel, A.; DiGiandomenico, A.; Keller, A.E.; Smith, T.R.F.; Park, D.H.; Ramos, S.; Schultheis, K.; Elliott, S.T.C.; Mendoza, J.; Broderick, K.E.; et al. An engineered bispecific DNA-encoded IgG antibody protects against Pseudomonas aeruginosa in a pneumonia challenge model. Nat. Commun. 2017, 8, 637. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Ying, H.; Grinnell, C.; Bryant, S.; Miller, R.; Clabbers, A.; Bose, S.; McCarthy, D.; Zhu, R.-R.; Santora, L.; et al. Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat. Biotechnol. 2007, 25, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Schanzer, J.M.; Wartha, K.; Croasdale, R.; Moser, S.; Künkele, K.P.; Ries, C.; Scheuer, W.; Duerr, H.; Pompiati, S.; Pollman, J.; et al. A novel glycoengineered bispecific antibody format for targeted inhibition of epidermal growth factor receptor (EGFR) and insulin-like growth factor receptor type I (IGF-1R) demonstrating unique molecular properties. J. Biol. Chem. 2014, 289, 18693–18706. [Google Scholar] [CrossRef] [PubMed]
- Godar, M.; Deswarte, K.; Vergote, K.; Saunders, M.; de Haard, H.; Hammad, H.; Blanchetot, C.; Lambrecht, B.N. A bispecific antibody strategy to target multiple type 2 cytokines in asthma. J. Allergy Clin. Immunol. 2018, 142, 1185–1193.e1184. [Google Scholar] [CrossRef]
- Lyman, M.; Lieuw, V.; Richardson, R.; Timmer, A.; Stewart, C.; Granger, S.; Woods, R.; Silacci, M.; Grabulovski, D.; Newman, R. A bispecific antibody that targets IL-6 receptor and IL-17A for the potential therapy of patients with autoimmune and inflammatory diseases. J. Biol. Chem. 2018, 293, 9326–9334. [Google Scholar] [CrossRef]
- Dovedi, S.J.; Elder, M.J.; Yang, C.; Sitnikova, S.I.; Irving, L.; Hansen, A.; Hair, J.; Jones, D.C.; Hasani, S.; Wang, B.; et al. Design and efficacy of a monovalent bispecific PD-1/CTLA-4 antibody that enhances CTLA-4 blockade on PD-1+ activated T cells. Cancer Discov. 2021. [Google Scholar] [CrossRef]
- Kitazawa, T.; Igawa, T.; Sampei, Z.; Muto, A.; Kojima, T.; Soeda, T.; Yoshihashi, K.; Okuyama-Nishida, Y.; Saito, H.; Tsunoda, H.; et al. A bispecific antibody to factors IXa and X restores factor VIII hemostatic activity in a hemophilia A model. Nat. Med. 2012, 18, 1570–1574. [Google Scholar] [CrossRef]
- Kitazawa, T.; Esaki, K.; Tachibana, T.; Ishii, S.; Soeda, T.; Muto, A.; Kawabe, Y.; Igawa, T.; Tsunoda, H.; Nogami, K.; et al. Factor VIIIa-mimetic cofactor activity of a bispecific antibody to factors IX/IXa and X/Xa, emicizumab, depends on its ability to bridge the antigens. Thromb. Haemost. 2017, 117, 1348–1357. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.J.; Atwal, J.K.; Zhang, Y.; Tong, R.K.; Wildsmith, K.R.; Tan, C.; Bien-Ly, N.; Hersom, M.; Maloney, J.A.; Meilandt, W.J.; et al. Therapeutic bispecific antibodies cross the blood-brain barrier in nonhuman primates. Sci. Transl. Med. 2014, 6, 261ra154. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.J.; Zhang, Y.; Kenrick, M.; Hoyte, K.; Luk, W.; Lu, Y.; Atwal, J.; Elliott, J.M.; Prabhu, S.; Watts, R.J.; et al. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci. Transl. Med. 2011, 3, 84ra44. [Google Scholar] [CrossRef]
- Niewoehner, J.; Bohrmann, B.; Collin, L.; Urich, E.; Sade, H.; Maier, P.; Rueger, P.; Stracke, J.O.; Lau, W.; Tissot, A.C.; et al. Increased brain penetration and potency of a therapeutic antibody using a monovalent molecular shuttle. Neuron 2014, 81, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, M.; Wang, X.; Lim, B.; Leitner, E.; Klingberg, F.; Ching, V.; Yao, Y.; Huang, D.; Gao, X.-M.; Kiriazis, H.; et al. Platelet-targeted delivery of peripheral blood mononuclear cells to the ischemic heart restores cardiac function after ischemia-reperfusion injury. Theranostics 2017, 7, 3192–3206. [Google Scholar] [CrossRef] [PubMed]
- De Goeij, B.E.; Vink, T.; Ten Napel, H.; Breij, E.C.; Satijn, D.; Wubbolts, R.; Miao, D.; Parren, P.W. Efficient payload delivery by a bispecific antibody-drug conjugate targeting HER2 and CD63. Mol. Cancer Ther. 2016, 15, 2688–2697. [Google Scholar] [CrossRef]
- Wec, A.Z.; Nyakatura, E.K.; Herbert, A.S.; Howell, K.A.; Holtsberg, F.W.; Bakken, R.R.; Mittler, E.; Christin, J.R.; Shulenin, S.; Jangra, R.K.; et al. A “Trojan horse” bispecific-antibody strategy for broad protection against ebolaviruses. Science 2016, 354, 350–354. [Google Scholar] [CrossRef]
- Lee, H.Y.; Schaefer, G.; Lesaca, I.; Lee, C.V.; Wong, P.Y.; Jiang, G. “Two-in-One” approach for bioassay selection for dual specificity antibodies. J. Immunol. Methods 2017, 448, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, S.; Li, Y.; Ngoh, E.; Wong, H.Y.; Cheng, M.S.; Wang, C.-I.; Schwarz, H. Development of a bispecific antibody targeting CD30 and CD137 on Hodgkin and Reed-Sternberg cells. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Karlsson, R.; Fridh, V.; Frostell, Å. Surrogate potency assays: Comparison of binding profiles complements dose response curves for unambiguous assessment of relative potencies. J. Pharm. Anal. 2018, 8, 138–146. [Google Scholar] [CrossRef]
- Ritter, N.; Russell, R.; Schofield, T.; Graham, L.; Dillon, P.; Maggio, F.; Bhattacharyya, L.; Schmalzing, D.; Zhou, W.-M.; Miller, K.; et al. Bridging analytical methods for release and stability testing: Technical, quality and regulatory considerations. BioProcess Int. 2016, 14, 12–23. [Google Scholar]
- Brünker, P.; Wartha, K.; Friess, T.; Grau-Richards, S.; Waldhauer, I.; Koller, C.F.; Weiser, B.; Majety, M.; Runza, V.; Niu, H.; et al. RG7386, a novel tetravalent FAP-DR5 antibody, effectively triggers FAP-dependent, avidity-driven DR5 hyperclustering and tumor cell apoptosis. Mol. Cancer Ther. 2016, 15, 946–957. [Google Scholar] [CrossRef]
- Dong, J.; Sereno, A.; Snyder, W.B.; Miller, B.R.; Tamraz, S.; Doern, A.; Favis, M.; Wu, X.; Tran, H.; Langley, E.; et al. Stable IgG-like bispecific antibodies directed toward the type I insulin-like growth factor receptor demonstrate enhanced ligand blockade and anti-tumor activity. J. Biol. Chem. 2011, 286, 4703–4717. [Google Scholar] [CrossRef] [PubMed]
- Macor, P.; Secco, E.; Mezzaroba, N.; Zorzet, S.; Durigutto, P.; Gaiotto, T.; De Maso, L.; Biffi, S.; Garrovo, C.; Capolla, S.; et al. Bispecific antibodies targeting tumor-associated antigens and neutralizing complement regulators increase the efficacy of antibody-based immunotherapy in mice. Leukemia 2015, 29, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-J.; Kim, Y.-J.; Lee, S.; Kim, Y.-S. A heterodimeric Fc-based bispecific antibody simultaneously targeting VEGFR-2 and Met exhibits potent antitumor activity. Mol. Cancer Ther. 2013, 12, 2748–2759. [Google Scholar] [CrossRef]
- FDA. Bispecific Antibody Development Programs: Guidance for Industry: Draft Guidance. Available online: https://r.search.yahoo.com/_ylt=AwrJ7Jz7h1NgnVQA4ltXNyoA;_ylu=Y29sbwNiZjEEcG9zAzEEdnRpZANDMTc1NF8xBHNlYwNzcg--/RV=2/RE=1616115836/RO=10/RU=https%3a%2f%2fwww.fda.gov%2fmedia%2f123313%2fdownload/RK=2/RS=I6KRkNsu63Y7IxvL96a96IT0LUs- (accessed on 15 March 2021).
- Trivedi, A.; Stienen, S.; Zhu, M.; Li, H.; Yuraszeck, T.; Gibbs, J.; Heath, T.; Loberg, R.; Kasichayanula, S. Clinical pharmacology and translational aspects of bispecific antibodies. Clin. Transl. Sci. 2017, 10, 147–162. [Google Scholar] [CrossRef]
- Jain, T.; Sun, T.; Durand, S.; Hall, A.; Houston, N.R.; Nett, J.H.; Sharkey, B.; Bobrowicz, B.; Caffry, I.; Yu, Y.; et al. Biophysical properties of the clinical-stage antibody landscape. Proc. Natl. Acad. Sci. USA 2017, 114, 944–949. [Google Scholar] [CrossRef]
- Davda, J.; Declerck, P.; Hu-Lieskovan, S.; Hickling, T.P.; Jacobs, I.A.; Chou, J.; Salek-Ardakani, S.; Kraynov, E. Immunogenicity of immunomodulatory, antibody-based, oncology therapeutics. J. Immunother. Cancer 2019, 7, 105. [Google Scholar] [CrossRef]
- FDA. Immunogenicity Testing of Therapeutic Protein Products—Developing and Validating Assays for Anti-Drug Antibody Detection: Guidance for Industry. Available online: https://r.search.yahoo.com/_ylt=A0geK.L9iVNgLn8A6y5XNyoA;_ylu=Y29sbwNiZjEEcG9zAzEEdnRpZANDMTc1NF8xBHNlYwNzcg--/RV=2/RE=1616116349/RO=10/RU=https%3a%2f%2fwww.fda.gov%2fmedia%2f119788%2fdownload/RK=2/RS=bmD0ZLN67iFU.k2iXNU7prdoxY8- (accessed on 15 March 2021).
- Steiner, G.; Marban-Doran, C.; Langer, J.; Pimenova, T.; Duran-Pacheco, G.; Sauter, D.; Langenkamp, A.; Solier, C.; Singer, T.; Bray-French, K.; et al. Enabling Routine MHC-II-Associated Peptide Proteomics for Risk Assessment of Drug-Induced Immunogenicity. J. Proteome Res. 2020, 19, 3792–3806. [Google Scholar] [CrossRef]
- Tourdot, S.; Hickling, T.P. Nonclinical immunogenicity risk assessment of therapeutic proteins. Bioanalysis 2019, 11. [Google Scholar] [CrossRef]
- Stracke, J.; Emrich, T.; Rueger, P.; Schlothauer, T.; Kling, L.; Knaupp, A.; Hertenberger, H.; Wolfert, A.; Spick, C.; Lau, W.; et al. A novel approach to investigate the effect of methionine oxidation on pharmacokinetic properties of therapeutic antibodies. mAbs 2014, 6, 1229–1242. [Google Scholar] [CrossRef] [PubMed]
- Schlothauer, T.; Rueger, P.; Stracke, J.O.; Hertenberger, H.; Fingas, F.; Kling, L.; Emrich, T.; Drabner, G.; Seeber, S.; Auer, J.; et al. Analytical FcRn affinity chromatography for functional characterization of monoclonal antibodies. mAbs 2013, 5, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Ljungars, A.; Schiött, T.; Mattson, U.; Steppa, J.; Hambe, B.; Semmrich, M.; Ohlin, M.; Tornberg, U.-C.; Mattsson, M. A bispecific IgG format containing four independent antigen binding sites. Sci. Rep. 2020, 10, 1546. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.; Chen, Y.; Huang, A.; Moffat, B.; Scheer, J.M.; Leong, S.R.; Lee, W.P.; Zhang, J.; Sharma, N.; Lu, Y.; et al. Development of a two-part strategy to identify a therapeutic human bispecific antibody that inhibits IgE receptor signaling. J. Biol. Chem. 2010, 285, 20850–20859. [Google Scholar] [CrossRef] [PubMed]
- Gassner, C.; Lipsmeier, F.; Metzger, P.; Beck, H.; Schnueriger, A.; Regula, J.T.; Moelleken, J. Development and validation of a novel SPR-based assay principle for bispecific molecules. J. Pharm. Biomed. Anal. 2015, 102, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Meschendoerfer, W.; Gassner, C.; Lipsmeier, F.; Regula, J.T.; Moelleken, J. SPR-based assays enable the full functional analysis of bispecific molecules. J. Pharm. Biomed. Anal. 2017, 132, 141–147. [Google Scholar] [CrossRef]
- Kiesgen, S.; Messinger, J.C.; Chintala, N.K.; Tano, Z.; Adusumilli, P.S. Comparative analysis of assays to measure CAR T-cell-mediated cytotoxicity. Nat. Protoc. 2021, 16, 1331–1342. [Google Scholar] [CrossRef]
- Broussas, M.; Broyer, L.; Goetsch, L. Evaluation of antibody-dependent cell cytotoxicity using lactate dehydrogenase (LDH) measurement. In Glycosylation Engineering of Biopharmaceuticals: Methods and Protocols; Beck, A., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 305–317. [Google Scholar]
- Du, K.; Li, Y.; Liu, J.; Chen, W.; Wei, Z.; Luo, Y.; Liu, H.; Qi, Y.; Wang, F.; Sui, J. A bispecific antibody targeting GPC3 and CD47 induced enhanced antitumor efficacy against dual antigen-expressing HCC. Mol. Ther. 2021. [Google Scholar] [CrossRef]
- Koopmans, I.; Hendriks, M.A.J.M.; van Ginkel, R.J.; Samplonius, D.F.; Bremer, E.; Helfrich, W. Bispecific antibody approach for improved melanoma-selective PD-L1 immune checkpoint blockade. J. Investig. Dermatol. 2019, 139, 2343–2351. [Google Scholar] [CrossRef]
- Staflin, K.; Zuch de Zafra, C.L.; Schutt, L.K.; Clark, V.; Zhong, F.; Hristopoulos, M.; Clark, R.; Li, J.; Mathieu, M.; Chen, X.; et al. Target arm affinities determine preclinical efficacy and safety of anti-HER2/CD3 bispecific antibody. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Lee, H.Y.; Contreras, E.; Register, A.C.; Wu, Q.; Abadie, K.; Garcia, K.; Wong, P.Y.; Jiang, G. Development of a bioassay to detect T-cell-activating impurities for T-cell-dependent bispecific antibodies. Sci. Rep. 2019, 9, 3900. [Google Scholar] [CrossRef]
- Lee, H.Y.; Register, A.; Shim, J.; Contreras, E.; Wu, Q.; Jiang, G. Characterization of a single reporter-gene potency assay for T-cell-dependent bispecific molecules. mAbs 2019, 11, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Wallace, P.K.; Keler, T.; Deo, Y.M.; Akewanlop, C.; Hayes, D.F. Antibody dependent cellular phagocytosis (ADCP) and antibody dependent cellular cytotoxicity (ADCC) of breast cancer cells mediated by bispecific antibody, MDX-210. Breast Cancer Res. Treat. 1999, 53, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Syedbasha, M.; Linnik, J.; Santer, D.; O’Shea, D.; Barakat, K.; Joyce, M.; Khanna, N.; Tyrrell, D.L.; Houghton, M.; Egli, A. An ELISA based binding and competition method to rapidly determine ligand-receptor interactions. J. Vis. Exp. 2016, e53575. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018, 72, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, L.; Tissot, N.; Hartmann, D.J.; Cohen, R. Comparison of the results obtained by ELISA and surface plasmon resonance for the determination of antibody affinity. J. Immunol. Methods 2010, 352, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, R. Applications of surface plasmon resonance for detection of bispecific antibody activity. Biopharm. Int. 2015, 28, 38–44. [Google Scholar]
- Hadzhieva, M.; Pashov, A.D.; Kaveri, S.; Lacroix-Desmazes, S.; Mouquet, H.; Dimitrov, J.D. Impact of antigen density on the binding mechanism of IgG antibodies. Sci. Rep. 2017, 7, 3767. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed]
- Helmerhorst, E.; Chandler, D.; Nussio, M.; Mamotte, C. Real-time and label-free bio-sensing of molecular interactions by surface plasmon resonance: A laboratory medicine perspective. Clin. Biochem. Rev. Aust. Assoc. Clin. Biochem. 2012, 33, 161–173. [Google Scholar]
- Staffler, R.; Pasternack, R.; Hils, M.; Kaiser, W.; Möller, F.M. Nucleotide binding kinetics and conformational change analysis of tissue transglutaminase with switchSENSE. Anal. Biochem. 2020, 605, 113719. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, J.; Langer, A.; Hampel, P.A.; Kaiser, W.; Strasser, R.; Rant, U. Quantitation of Affinity, Avidity, and Binding Kinetics of Protein Analytes with a Dynamically Switchable Biosurface. J. Am. Chem. Soc. 2012, 134, 15225–15228. [Google Scholar] [CrossRef]
- Agez, M.; Mandon, E.D.; Iwema, T.; Gianotti, R.; Limani, F.; Herter, S.; Mossner, E.; Kusznir, E.A.; Huber, S.; Lauer, M.; et al. Biochemical and biophysical characterization of purified native CD20 alone and in complex with rituximab and obinutuzumab. Sci. Rep. 2019, 9, 13675. [Google Scholar] [CrossRef] [PubMed]
- Bjorkelund, H.; Gedda, L.; Barta, P.; Malmqvist, M.; Andersson, K. Gefitinib Induces Epidermal Growth Factor Receptor Dimers Which Alters the Interaction Characteristics with I-125-EGF. PLoS ONE 2011, 6, e24739. [Google Scholar] [CrossRef] [PubMed]
- Peess, C.; von Proff, L.; Goller, S.; Andersson, K.; Gerg, M.; Malmqvist, M.; Bossenmaier, B.; Schraml, M. Deciphering the Stepwise Binding Mode of HRG1 beta to HER3 by Surface Plasmon Resonance and Interaction Map. PLoS ONE 2015, 10, e0116870. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.; Kim, H.S.; Tong, R.K.; Bainbridge, T.W.; Vernes, J.-M.; Zhang, Y.; Lin, Y.L.; Chung, S.; Dennis, M.S.; Zuchero, Y.J.Y.; et al. Effector-attenuating substitutions that maintain antibody stability and reduce toxicity in mice. J. Biol. Chem. 2017, 292, 3900–3908. [Google Scholar] [CrossRef] [PubMed]
- Couch, J.A.; Yu, Y.J.; Zhang, Y.; Tarrant, J.M.; Fuji, R.N.; Meilandt, W.J.; Solanoy, H.; Tong, R.K.; Hoyte, K.; Luk, W.; et al. Addressing safety liabilities of TfR bispecific antibodies that cross the blood-brain barrier. Sci. Transl. Med. 2013, 5, 183ra157. [Google Scholar] [CrossRef]
- Zhang, H.M.; Li, C.; Lei, M.; Lundin, V.; Lee, H.Y.; Ninonuevo, M.; Lin, K.; Han, G.; Sandoval, W.; Lei, D.; et al. Structural and Functional Characterization of a Hole-Hole Homodimer Variant in a “Knob-Into-Hole” Bispecific Antibody. Anal. Chem. 2017, 89, 13494–13501. [Google Scholar] [CrossRef]

| Bioassay | Method Principle | Examples |
|---|---|---|
| Bridging ELISA | To assess the ability of each arm of the BsAbs to bind two antigens simultaneously. The assay follows a sequential capture method, where antibody is allowed to bind the first coated antigen, followed by a wash step and addition of a biotinylated version of the second antigen. The bound biotinylated antigen can be detected using HRP-labeled streptavidin and luminescent substrate. | tetra-VH IgG bispecific tetravalent [86] |
| Sandwich ELISA | To assess binding specificity, including dual-specificity detection, of BsAbs. The assay format consists of antigen incubation with an immobilized capture antibody in the plate, followed by wash step to remove non-bound components. The antibody-antigen complex is then detected using a labeled antibody. | IgE receptor signaling blocking BsAb, FcεRI/FcγRIIb cross-link [87] |
| Bridging SPR | To measure the binding affinities of antibodies to their respective antigens. The assay follows Biacore™ SPR-based format, where two sequential binding events to a ligand immobilized on a chip and surface regeneration is used to measure a bridging signal and, as a result, the simultaneous binding of the assay to both antigens. | Ang-2/VEGF BsAb [88] |
| Dual-Binding SPR | A solution binding SPR-based assay for individual assessment of both targets in solution without the need for immobilization and regeneration of the target. | anti-VEGFA-121/Ang2 BsAb [89] |
| Direct Cell Killing | To evaluate cell killing potential by co-culturing the target and effector cells in the presence or absence of BsAb. Assay readout can be accomplished through luciferase reporter system or by flow cytometry-based methods to measure percent apoptotic cells or percent cytolysis of pre-labeled target cells by proliferation dye dilution analysis. Additional assays include labeling target cells with 51Cr or measuring the presence of extracellular LDH, where the release of the label or LDH by lysed target cells is used as surrogate for cell-killing activity [90,91]. | CD47 blocking BsAb specifically targeted to GPC3 expressing target cells [92]) PD-L1 blocking BsAb specifically directed to CSPG4-expressing target cells [93] CD3-bispecific (anti-HER2/CD3) TDB [94] |
| T Cell Activation | To assess BsAb effects on T-cell activation and proliferation potential. Assay readout mainly includes luciferase bioluminescence reporter signal using Jurkat T cells engineered with an NFAT-response-element driving luciferase expression. Depending on the MoA of the molecule, T-cell activation can be triggered either only by T cells expressing relevant receptors or in the presence of antigen-presenting target cells. In addition, activation and proliferation of T cells can be evaluated using an in-vitro mixed lymphocyte reaction followed by flow cytometry (proliferation dye dilution analysis or measurement of T-cell activation markers) and ELISA (for IFN-g and granzyme B secretion measurements). | CD3e-targeting TDB [95] PD-L1 blocking BsAb specifically directed to CSPG4-expressing target cells [93] CD3-bispecific (anti-HER2/CD3) TDB [94] |
| Type 1 Cell Bridging | Type 2 Receptor Blocking or Activating (Cis/Trans) | Type 3 Cofactor Mimicking | Type 4 Spatial Targeting (“Homing” BsAbs) | |
|---|---|---|---|---|
| Binding | ELISA (binding to single target) [96], SPR (binding to single target) [96] | ELISA (binding to either target), SPR (affinity for either target) [50], bridging ELISA (dual target recognition) [50], bridging SPR [92], ELISA (ligand blocking) [74], SPR (stoichiometry of binding) [74] | SPR (characterize affinity for FIX, FIXa, FX, FXa) [61,62], ELISA (confirm specificity for FIX and FX) [62] | BLI (measure affinity of each arm, and support 1:1 binding) [68], Competition ELISA [63] |
| Bioactivity (Major MoA) | Reporter gene effector cell activation assay [96], direct cell killing assay [21,22,42] | Cell Proliferation [50,69,76], Apoptosis [73], cytokine neutralization [56] | Enzymatic assays (FXa activity) [61] | Viral Inactivation [68], TR-FRET AB assay [63] |
| Functional (other supporting MoA as characterization) | Cell depletion by flow cytometery [21,96], Cytokine release [42], cell surface marker expression (per MoA) [22,42,45,96] | Tyrosine phosphorylation [74], inhibition of antibody production/secretion [50], Calcium flux assay [50] | Thrombin generation assay [61,62] | Fluorescence microscopy to assess subcellular localization [63,65,68], transcytosis assay [65] |
| Effector Function | ADCC [97], ADCP [97] | CDC assay [75], ADCC/ADCP bioassay [92], binding to FcgRs [92] | NA | NA |
| Impurity Bioassay | T-cell activating impurities [95] | NA | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Register, A.C.; Tarighat, S.S.; Lee, H.Y. Bioassay Development for Bispecific Antibodies—Challenges and Opportunities. Int. J. Mol. Sci. 2021, 22, 5350. https://doi.org/10.3390/ijms22105350
Register AC, Tarighat SS, Lee HY. Bioassay Development for Bispecific Antibodies—Challenges and Opportunities. International Journal of Molecular Sciences. 2021; 22(10):5350. https://doi.org/10.3390/ijms22105350
Chicago/Turabian StyleRegister, Ames C., Somayeh S. Tarighat, and Ho Young Lee. 2021. "Bioassay Development for Bispecific Antibodies—Challenges and Opportunities" International Journal of Molecular Sciences 22, no. 10: 5350. https://doi.org/10.3390/ijms22105350
APA StyleRegister, A. C., Tarighat, S. S., & Lee, H. Y. (2021). Bioassay Development for Bispecific Antibodies—Challenges and Opportunities. International Journal of Molecular Sciences, 22(10), 5350. https://doi.org/10.3390/ijms22105350





