Abstract
Plants can be considered an open system. Throughout their life cycle, plants need to exchange material, energy and information with the outside world. To improve their survival and complete their life cycle, plants have developed sophisticated mechanisms to maintain cellular homeostasis during development and in response to environmental changes. Autophagy is an evolutionarily conserved self-degradative process that occurs ubiquitously in all eukaryotic cells and plays many physiological roles in maintaining cellular homeostasis. In recent years, an increasing number of studies have shown that autophagy can be induced not only by starvation but also as a cellular response to various abiotic stresses, including oxidative, salt, drought, cold and heat stresses. This review focuses mainly on the role of autophagy in plant abiotic stress management.
1. Introduction
Due to their sessile nature, plants are constantly exposed to many transient but recurring stresses. To improve their survival and complete their life cycle, plants have developed sophisticated mechanisms to maintain cellular homeostasis during development and in response to environmental changes [1,2]. Autophagy (which means "eating itself") is one of the most important homeostasis regulation systems for degradation and recycling of proteins and cytoplasmic organelles in plants throughout their life cycle [3]. Under normal conditions, basal autophagy functions as a housekeeping process to clear unwanted cytoplasmic contents and remobilize nutrients during leaf senescence and seed germination, whereas under certain stresses (starvation, oxidative, salt, drought, heat, cold, pathogen or fungal infection, and programmed cell death (PCD)), autophagy is up-regulated and contributes to the recycling of damaged or unwanted cell materials [4].
Several types of autophagy have been characterized to date, including macroautophagy, microautophagy, chaperone-mediated autophagy and organelle-specific autophagy [3]. The best-studied type of autophagy in plants is macroautophagy, in which autophagosomes form and then fuse with lysosomes or vacuoles to degrade cellular components and organelles [5]. In microautophagy, tonoplast or lysosome membrane invagination packages cytoplasmic components to form single-membrane autophagic bodies, and the substrate components are then released into the vacuolar lumen or lysosome. Plants microautophagy eliminates damaged chloroplasts and degrades cellular components during starvation [6]. Chaperone-mediated autophagy, which is facilitated by chaperones and does not involve membrane reorganization, directly targets nonessential proteins, which are transported through the lysosomal membrane for degradation [7]. Autophagy can be either selective or nonselective, and organelle-specific autophagy is also considered as selective autophagy [8]. Though there are several types of autophagy, the core autophagy components are conserved. This review will focus mainly on the role of core autophagy components in plant abiotic stress management.
2. Comparison of Plants and Yeast Core Autophagy Components
Over the past decade, the molecular and physiological understanding of plant autophagy has greatly increased. Most of the essential machinery required for autophagy seems to be conserved from yeast to plants [9,10,11]. In yeast, more than 40 autophagy-related genes (Atg) have been identified, half of which constitute the core autophagy machinery, including the Atg1–Atg13 kinase complex, the Atg9 cycling system, the phosphatidylinositol 3-kinase (PtdIns3K) complex and two ubiquitin-like conjugation systems (Atg8 and Atg12) [12]. According to the various available plant genome databases, the orthologs and paralogs of the yeast core autophagy components were identified in plants, ranging from lower species, such as algae, to major agricultural plants, such as cereals and vegetables [13,14,15,16,17,18,19]. Although there are differences among plant species, some components were unidentified in a regular BLAST search, but most of the core autophagy components were conserved. Arabidopsis is the model plant and has been extensively studied. Oryza sativa (rice), the most important crop in the world, has also been well studied. To compare the core autophagy components of plants and yeast, the following discussion is primarily based on the well-studied monocot plant rice and the dicot plant Arabidopsis.
The Atg1–Atg13 kinase complex is negatively regulated by targeting of rapamycin (TOR) in a nutrient-dependent manner. This complex includes five core components: Atg1, Atg13, Atg17, Atg29 and Atg31. Atg17 forms a complex with Atg29 and Atg31 under normal conditions and further interacts with Atg1 and Atg13 upon starvation to mediate preautophagosomal structure (PAS, a specific structure for autophagosome formation in yeast) organization [20,21]. Atg11 can then participate in this process by forming a scaffold with Atg17 for selective autophagy [22]. The PAS is a punctate structure proximal to the endoplasmic reticulum, which is the site where the Atg machinery assembles upon autophagy induction. A recent study showed that the PAS is a transient structure, and consists of a liquid-like condensate of Atg proteins formed through liquid–liquid phase separation [23]. Homologs of yeast Atg1, Atg13 and Atg11 have been identified in plants. There are three Atg1 homologs in Arabidopsis, four in rice, and two Atg13 homologs both in Arabidopsis and rice through alignment searching. In Arabidopsis, ATG1 and ATG13 can form a kinase complex facilitated by ATG11 under starvation conditions [24,25]. Two sensor kinases, SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE1 (SnRK1) and TOR, have also been identified in Arabidopsis and rice. These two sensors can cooperatively perceive nutritional status to mediate ATG1–ATG13 kinase complex states and in turn activate or inhibit autophagy [26]. An ATG1-independent autophagy pathway exists in Arabidopsis under prolonged carbon starvation, in which the SnRK1 catalytic KIN10 subunit can directly phosphorylate the PtdIns3K complex ATG6 subunit to trigger autophagy [27]. These results showed that although the core autophagy mechanism is conserved in eukaryotic cells, plants need to engage multiple pathways to activate autophagy in response to the changeable ecological environment. A homolog of mammalian mATG101 has also been identified in the Arabidopsis genome through DELTA-BLAST analysis, which is conserved in various eukaryotes, but not in Saccharomyces cerevisiae. It has also been shown to assemble into an active complex to initiate autophagy, but homologous or similar function genes to Atg17, Atg19 and Atg31 have not yet been found in plants (Figure 1a) [28]. The PAS is a specific structure involved in autophagosome formation in yeast. Recently, studies have confirmed the PAS is formed from liquid–liquid phase separation. Mammalian aggregaphy, the selective degradation of protein aggregates, also forms through liquid–liquid phase separation [29]. Whether liquid–liquid phase separation occurs in plant autophagy remains to be determined.
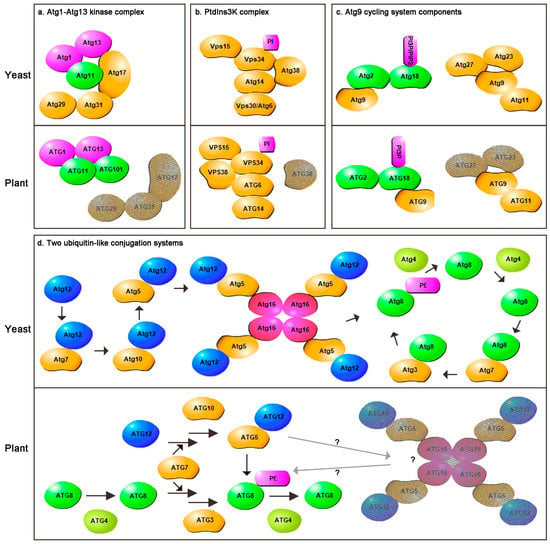
Figure 1.
The comparison of yeast and Arabidopsis core autophagy components. (a) Atg1–Atg13 kinase complex. (b) The phosphatidylinositol 3-kinase (PtdIns3K) complex. (c) The Atg9 cycling system components. (d) The two ubiquitin-like conjugation systems. The unidentified components or complexes in plant are covered by grey.
The PtdIns3K complex produces PI3P in the PAS or the ER to recruit downstream proteins. Yeast contains PtdIns3K complex I and PtdIns3K complex II. Only PtdIns3K complex I participates in autophagy, while PtdIns3K complex II functions in vacuolar protein sorting. The PtdIns3K complex I includes a class III PtdIns3K, vacuolar protein sorting protein 34 (Vps34); a serine/threonine kinase (Vps15); Vps30/Atg6; Atg14 and Atg38. Vps15 is required for the membrane association of Vps34. Atg14 is thought to recruit Vps34 and Vps30/Atg6 to PAS localization [30]. Atg38 helps to maintain the integrity of the PtdIns3K complex. A previous study identified that Atg38 can also interact with Atg8 via an ATG8-interacting motif (AIM), which is responsible for the PAS accumulation of the PtdIns3K complex [31]. Sequence homologs of these genes, except ATG38, have been found in plants. All of these genes appear to have only one copy except for ATG14, which has two copies in Arabidopsis [32]. However, the sequence homologs or functional similarity of ATG14 was still missing in rice. In Arabidopsis, VPS34, VPS15 and ATG6 form the core complex, which can synthesize sufficient levels of PI3P [33,34,35]. This complex combines with ATG14 and VPS38 (an orthologue of the mammalian UV RADIATION RESISTANCE-ASSOCIATED GENE (UVRAG)) to better control the assembly of autophagosomes [32]. In yeast, Vps38 is responsible for endosome localization of the PtdIns3K complex II. In Arabidopsis, VSP38 is not only required for endosomal trafficking, but also promotes autophagy [36]. Whether VPS38 is involved in autophagy remains controversial [36,37], and the homolog of ATG38 remains to be identified (Figure 1b).
Atg9 is an integral membrane protein that has been proposed to deliver lipids to autophagosomes and plays a critical role in phagophore nucleation and expansion in yeast [38]. Atg9 cycles between the PAS and non-PAS structures, which requires the help of Atg2, Atg8, Atg11, Atg23, Atg27, the Atg1-Atg13 complex and the PtdIns3K complex (Figure 1c). Under starvation conditions, Atg9 localizes to the PAS in an Atg23- and Atg27-dependent manner. Retrograde Atg9 transport from the PAS to cytoplasmic compartments depends on the Atg1–Atg13, Atg2–Atg18, and PtdIns3K complexes. Defects in any of the components of these complexes leads to the accumulation of Atg9 at the PAS [39]. Atg18 is a peripheral membrane protein that not only interacts with Atg9 but also binds to PI3P and PI(3,5)P2. Atg2, a ∼200 kDa peripheral membrane protein, is important for the PAS localization of Atg18 [40]. The results of a recent study suggest that Atg2 facilitates Atg18 binding to PI3P, and these two proteins act cooperatively to target the Atg2–Atg18 complex to Atg9 vesicles [40,41]. Atg2 may mediate autophagosome membrane expansion through its endoplasmic reticulum (ER) and pre-autophagosomal membrane location [38,40]. This previous work also provides evidence for the involvement of the ER in autophagosome formation in yeast. In addition, Atg2 can transfer lipids for Atg8 lipidation, and Atg18 can protect Atg8-PE from unregulated cleavage by Atg4 [42]. In Arabidopsis, atg9 mutation causes the accumulation of abnormal autophagosome-related tubules upon autophagic induction. Immuno-electron microscopy (EM) and confocal microscopy analyses showed that these autophagosome-like structures were located near the rough ER. Three-dimensional tomographic reconstruction further revealed a direct connection between abnormal autophagosomal tubular structures and the ER in the atg9-3 mutant [43]. All of these data demonstrate that ATG9 is required for the efficient budding of autophagosomes and that the original autophagosome is formed from the ER membrane. ATG9 also cycles between the ER and the cytoplasmic pool and regulates the trafficking of ATG18 on the autophagosomal membrane in a PI3P-dependent manner [43,44]. Although there is no PAS in plants, they have large and complex membrane systems, and the ER may not be the only source of autophagosome formation. ATG2 and ATG8 present multiple functions in plant development and abiotic stress responses, which will be discussed in detail in the next section. The sequence homologs of Atg2, Atg9 and Atg18 were also identified in rice. There is a single ATG2 gene in rice like in Arabidopsis. In addition, there are two ATG9 homologs in rice versus one in Arabidopsis. There is also a large ATG18 family in rice including six numbers. Although the core ATG genes do exist in plants, plants have evolved either small or large families of specific ATG genes to perform specialized functions.
Two ubiquitin-like conjugation systems, Atg8-PE (phosphatidylethanolamine) and Atg12-Atg5-Atg16, are essential for autophagy progression. The lipidation of Atg8 requires the participation of a series of proteins. First, the C-terminus of Atg8 needs to be cleaved by a cysteine protease, Atg4, to expose a glycine residue. Then, it binds the E1-like enzyme Atg7 and is transferred to the E2-like enzyme Atg3. Finally, Atg8 is covalently linked to the membrane lipid phosphatidylethanolamine (PE) to form the Atg8-PE complex [45]. The conjugation between Atg8 and PE is reversible. Atg8 can also be released from the membrane lipid by the deconjugating enzyme function of Atg4. The N-terminal motif of Atg4, close to the catalytic motif, plays a key role in specific Atg8 deconjugation [46]. Atg12 and Atg8 are both ubiquitin-like proteins. The activity of Atg12 also requires the E1-like enzyme Atg7. Atg12 is then transferred to another E2-like enzyme, Atg10, and is finally conjugated to Atg5. The Atg12-Atg5 conjugate further interacts with a coiled-coil protein, Atg16, to form a tetrameric Atg12-Atg5-Atg16 complex. Atg16 interacts with Atg21 in a PI3P-dependent manner to recruit this complex to the PAS [47,48]. The two conjugation systems are closely related, as Atg12-Atg5-Atg16 is necessary for Atg8-PE formation and lipidation site determination [39,49]. In addition, Atg12 can interact with the Atg1 kinase complex, which serves as a scaffold for PAS organization [40]. These two ubiquitin-like conjugation systems have been well studied in plants, especially the ATG8 system, which also needs to be cleaved by ATG4 to finally form the ATG8-PE lipid complex [50,51,52]. The ATG12-ATG5 complex is essential for ATG8-mediated autophagy in plants by promoting ATG8 lipidation [53,54]. The homolog of ATG16 also exists in plants, but its specific function in autophagy remains to be determined (Figure 1d). ATG8 has been analyzed in more detail. There are nine homologs in Arabidopsis and five homologs in rice. Different members show distinct expression patterns and may have distinct functions during a plant’s development or its response to different stress conditions [55]. Almost all autophagy-related receptors/adaptors identified to date have ATG8 interaction domains (AIMs/UIMs) or can directly interact with ATG8. These results imply sophisticated roles of ATG8 in autophagy and plant development or stress response, which were illustrated in detail in recent reviews [55,56,57].
Through the above steps, the autophagosome is filled with cargos. The mature autophagosome then fuses with and delivers its contents to the vacuole. The contents are then broken down by various hydrolases into carbohydrates, amino acids and lipids that are returned to the cytosol and recycled to synthesize new products or reused for other purposes. The study of yeast autophagy has provided a solid foundation for autophagy-related research in plants. Through homologous comparison and mutants analysis, many functional autophagy genes have been identified in plants. By screening autophagy-related interacting proteins, plant-specific genes required for autophagy were also identified, most of which are induced by biotic and abiotic stresses. Autophagy is induced by starvation and senescence, which is well established and is not included in this review. Abiotic stresses, including drought, salt, heat and cold, are inevitable environmental stresses encountered by plants. In recent years, an increasing number of studies have shown that autophagy plays an important role in plant stress management. In this review, we will focus on the role of autophagy in plant abiotic stress regulation in the order of autophagosome formation.
3. The Role of Core Autophagy Components in Plant Abiotic Stress
Adverse environmental conditions negatively affect agricultural production and reduce crop yield, both qualitatively and quantitatively, and accompany significant transcriptomic changes in plant cells [58,59,60,61,62,63,64,65]. Abiotic stresses, including oxidative, osmotic, drought, salt, cold and heat stresses, can not only influence ATG genes expression but also induce autophagosome formation in plant cells. Autophagy is an important stress response mechanism in plants. Homology-based analyses have identified conserved ATG genes in plants, some of which have been reported to participate in the plant abiotic stress response.
In plants, the homologs of the Atg1–Atg13 complex, the PtdIns3K complex, and the Atg2–Atg8 complex have been identified. Only ATG1, ATG2, ATG13 and ATG18 have been found to be needed for ATG9 cycles. ATG9, ATG11 and PI3K act upstream of ATG2 [25,43,66,67]. Their functions have been characterized in detail, and most are related to plant abiotic stress responses (not including starvation). There are four homologs of Atg1 in Arabidopsis. KIN10, an Arabidopsis ortholog of mammalian AMPK, can activate autophagy in response to drought and hypoxic stress by affecting ATG1 phosphorylation [68]. The PtdIns3K complex is also conserved in Arabidopsis. The homologs of Vps34, Atg6 and Vps15 are all present with only one copy, while Atg14 appears to have two homologs. The core components of the PtdIns3K complex are involved in plants abiotic stress regulation. Arabidopsis PI3K plays a positive role in salt tolerance. The pi3K mutants and wortmannin-treated plants exhibit an overly salt-sensitive phenotype [69]. PI3K can facilitate the internalization of plasma membrane intrinsic protein 2;1 (PIP2;1) from the plasma membrane (PM) into the vacuole under salt stress to decrease root water permeability [70]. In rice, there are three Atg6 homologs, which show differential expression when subjected to heat, cold and drought stress. OsATG6a is up-regulated by drought but down-regulated by HS. OsATG6b is up-regulated by drought but down-regulated by cold stress. OsATG6c is up-regulated by all examined stresses, including heat, drought and cold [71]. In barley, the expression of HvATG6 is also up-regulated by various abiotic stresses, including H2O2 treatment, high salinity, drought and low temperature. Knockdown of HvATG6 in barley leaves through barley strip mosaic virus (BSMV)-induced gene silencing leads to accelerated yellowing under H2O2 treatment [72]. These results show that the stress-specific response of ATG6 will help plants better deal with different stress conditions, but the biological mechanisms and action of these genes under abiotic stresses remain unclear. Although autophagy normally plays a positive role in plant abiotic stress regulation, not all of its components are up-regulated under stress conditions, and some can also be inhibited, which may be related to the extensive functions of autophagy and the extent of plant damage.
Arabidopsis has a single ATG2 gene that is expressed ubiquitously throughout the plant. The atg2 knockout mutants display typical autophagy-defective phenotypes during senescence and stress conditions [73,74]. The T-DNA insertion Arabidopsis mutant atg2-5 shows impaired low-CO2-induced stomatal opening. Plants can regulate stomatal opening and closure to cope with diverse environmental stresses [75]. In studying this mutant, the author found that autophagy controls guard cell reactive oxygen species (ROS) homeostasis by eliminating oxidized peroxisomes, thereby allowing stomatal opening. The disruption of other autophagy genes in Arabidopsis, including ATG5, ATG7, ATG10 and ATG12, causes similar stomatal defects and results in the constitutive accumulation of high ROS levels in guard cells [76]. These results illustrate that autophagy can alter ROS levels to regulate stomatal states. Moreover, ATG2 is induced by high temperatures in Arabidopsis. The fresh weight and chlorophyll retention of atg2-1 mutants are dramatically decreased compared with those of wild-type (WT) plants under high-temperature conditions. The mRNA levels of ATG5, ATG6, ATG12A and ATG18A also significantly increase in WT plants under high-temperature conditions [74]. In addition, autophagy plays a role in resetting the cellular memory of heat stress (HS). Autophagy is induced by thermopriming and remains at a high level long after stress termination. The autophagy-deficient mutants atg2-1, atg5-1, atg12ab and atg18a-2 show significantly better survival under postmemory HS than WT plants, suggesting that autophagy negatively controls thermomemory in Arabidopsis. Upon further investigation, the author reported that the autophagy mutants retain heat shock proteins longer than the WT and concomitantly display improved thermomemory. These results show that autophagy mediates the specific degradation of heat shock proteins at later stages of the thermorecovery phase, leading to compromised heat tolerance after the second HS [77]. CaATG6 was predicted to interact with CaHSP90 family members and is related to heat stress tolerance in pepper [78]. These link autophagy to heat stress through regulation heat shock protein levels. In the presence of low NaCl concentrations, Arabidopsis atg9 and atg2 mutants germinated faster than the WT, while the atg5 and atg7 mutants showed the opposite behavior. In the presence of higher NaCl concentrations, germination slowed down in all lines [79]. These results demonstrate that under different degrees of stress, autophagy has different manifestations. The ATG18 proteins are a large protein family in plants, similar to that found in mammals but not in yeast. There are eight Atg18 homologs in Arabidopsis and six in rice. In Arabidopsis, only ATG18A is up-regulated by both salt and osmotic stresses. RNAi-ATG18A Arabidopsis plants are more sensitive to salt and osmotic treatment. All of these results show that ATG18A plays a positive role in salt and osmotic stress regulation in Arabidopsis. Furthermore, RNAi-ATG18A Arabidopsis seedlings are hypersensitive to oxidative stress and exhibit delayed growth and severe bleaching relative to WT Arabidopsis. Following treatment with methyl viologen (MV), an inducer of ROS, autophagosome accumulation is observed in WT Arabidopsis plants but not in RNAi-ATG18A Arabidopsis plants, and RNAi-ATG18A Arabidopsis plants accumulate higher levels of oxidized proteins than WT Arabidopsis plants [80,81]. These results show that autophagy plays a role in the clearance of oxidized proteins following oxidative stress in Arabidopsis. Inhibiting ROS production also has an effect on autophagy induction under salt stress conditions but has no effect under osmotic stress. These results illustrate that ROS can link autophagy and abiotic stress by acting as signaling molecules. In addition, ATG18A overexpression can improve transgenic plants resistance to drought stress in both tomato and apple plants [82,83]. Large numbers of ATG18 homologs exist in plants, and different numbers of these homologs respond to different stresses, demonstrating that plants have evolved complex autophagy mechanisms to improve their survival in extreme environments.
The two ubiquitin-like conjugation systems are also well conserved in plants. Plants contains more members of the protein family involved in these systems. There are nine ATG8 family numbers in Arabidopsis and five ATG8 family numbers in rice versus one in yeast. In Arabidopsis, all nine ATG8s are expressed throughout the plant in distinct expression patterns, implying that each member may have a distinct function during development or under various stress conditions; however, only some of these genes have been reported to be involved in abiotic stress responses. The overexpression of a GFP-ATG8F-HA fusion protein was found to exert a considerably stronger negative effect on the growth of Arabidopsis plants under mild NaCl treatment with increased electrolyte leakage, indicating greater damage to the cell membrane system than was observed in the control seedlings. The effect of the expression of the GFP-ATG8F-HA construct on the response of the plants to relatively mild osmotic stresses is the same as that of salt [84]; however, Arabidopsis overexpressing ATG8A performed better than WT plants in germination assays on NaCl-containing plates. In addition, ATG8A and ATG8E can serve as autophagy markers. When plants were subjected to different abiotic stresses, GFP-ATG8A or ATG8E-GFP accumulated in autophagosomes, demonstrating that ATG8 participates in plant abiotic stress regulation [77,79,81]. The different ATG8 family members associated with the responses to different stresses allow fine-tuning of the regulatory mechanism of autophagy [85]. In wild emmer wheat, the expression of TdATG8 is also strongly induced by drought and osmotic stress, especially in the roots relative to the leaves [86]. There are also two/nine members, respectively, of the ATG4/ATG8 families in common wheat (Triticum aestivum L.). The expression profiles of TaATG4a, 4b, 8a, 8g and 8h all show up-regulated expression upon exposure to high salinity, drought and low temperature [87]. Two other ubiquitin-like conjugation system components, ATG5 and ATG7, have been reported to respond to salt and oxidative stress in Arabidopsis. ATG5- or ATG7-overexpressing plants exhibited increased resistance to oxidative stress, delayed ageing and enhanced growth, and plants with mutations in these genes showed the opposite behavior [88]. In addition, both Arabidopsis and tomato plants with mutations in these two genes are more sensitive to HS than WT plants [77,89]. Atg10, an E2-like enzyme, is another important component of the Atg12-Atg5-Atg16 ubiquitin-like conjugation systems. There is one Atg10 homologue in Arabidopsis, but there are two Atg10 homologs in rice, OsATG10a and OsATG10b. Only osatg10b mutants are sensitive to high salt and methyl viologen treatment, resulting in the formation of fewer autophagosomes than that observed in WT [90]. In addition, overexpression of the autophagy-related gene MdATG10 in apples can increase autophagic activity in the roots and enhance transgenic plants salt tolerance [91]. These results demonstrate that autophagy plays an important role in the survival of plant cells under different abiotic stresses. Importantly, ATG genes can quickly respond to different abiotic stress conditions in the initiation phase of autophagy.
Autophagy is also involved in the regulation of abiotic stress in other plants. Autophagy has been reported to be involved in pepper (Capsicum annuum L.) tolerance to abiotic stresses. The author identified 15 core ATG members, including 29 ATG proteins with corresponding conserved functional domains, in the whole genome of pepper via the HMM method. Under salt, drought, heat and cold stresses, the expression levels of CaATG genes changed in a stress type-dependent pattern, and the accumulation of autophagosome puncta increased. These results indicate the linkage of autophagy in the response of pepper to abiotic stresses [78]. Camellia sinensis autophagy-related genes (CsARGs) also respond to abiotic stress and most CsARGs were upregulated at different time points during the abiotic stress treatment [17]. Autophagy-related genes were also identified in Citrus sinensis. Most of the CsATGs were significantly changed in response to drought, cold, heat, salt and mannitol treatment. In addition, ectopically expressed CsATG18a and CsATG18b in Arabidopsis showed enhanced tolerance to osmotic, salt, drought (CsATG18a) or cold (CsATG18b) stress, compared to wild-type plants [14]. Drought stress also up-regulates the expression of the autophagy-related genes ATG1, ATG8, ATG9 and ATG12 in Caragana korshinskii [15]. Homologs of Atg3 from apples are reported to improve transgenic Arabidopsis salt and osmotic stress resistance [92]. The above findings demonstrate that the core autophagy components are conserved among plants and play a wide range of roles in plant abiotic stress management.
4. The Role of Selected Autophagy in Plant Abiotic Stress
Autophagy was initially defined as a bulk degradation process that causes massive degradation of cellular components; however, in recent years, cumulative evidence has indicated that the recruitment of cargo to autophagosomes is highly selective [56,93]. Several selective autophagy receptors have been characterized in plants. These selective autophagy receptors link organelles, protein aggregates or other cargo to the autophagy machinery by binding to both the fated cargo and ATG8 through conserved ATG8-interacting motif (AIM) or ubiquitin-interacting motif (UIM). Several characterized autophagy receptors function in the plant abiotic stress response (Table 1).

Table 1.
The cargo receptors/adapters in plant abiotic stress responses.
The next to BRCA1 gene 1 (NBR1) is a functional hybrid protein of the mammalian autophagy receptor p62 (also known as Sequestosome1/SQSTM1) and a neighbor of BRCA1 (NBR1) that specifically targets stress-induced, ubiquitinated protein aggregates [94,95]. Both p62 and NBR1 preferentially target K63-linked polyubiquitylated proteins and mediate their aggregation and autophagic clearance in an LC3-interacting region (LIR)- and UBA-dependent manner. NBR1 and p62 oligomerize but can also function independently [96]. Arabidopsis AtNBR1 can bind ATG8 via the AIM motif and ubiquitinate proteins via the ubiquitin-associated domain [97]. The expression of AtNBR1 is upregulated under HS in Arabidopsis [98]. Moreover, under HS conditions, atnbr1 mutants accumulate more puncta in the cytoplasm compared with WT. GFP-NBR1 puncta accumulate in WT plants but not in atg7 mutants under HS conditions. During the HS recovery phase, more NBR1 puncta accumulate in WT plants, and the NBR1 protein accumulates at substantially higher levels in atg5-1 and atg18a-2 mutants than in WT plants [98,99]. These findings demonstrate that NBR1 puncta formation is autophagy dependent and that NBR1 is required not only for the heat-induced formation of autophagosomes but also for the degradation of substrates in an autophagy-dependent pathway throughout the HS stage. Further analysis showed that NBR1 plays a crucial role as a receptor for the selective autophagy-mediated degradation of heat shock protein 90.1 (HSP90.1) and rotamase FKBP1 (ROF1) during recovery from HS to regulate HS memory in Arabidopsis [100]. In addition, nbr1 mutants are hypersensitive to oxidative, drought and salt stress relative to WT plants [78,95,97,98]. Similar results were observed in tomato plants in which NBR1 was silenced by VIGS. ATGs and NBR1 gene silencing triggered the accumulation of ubiquitinated insoluble proteins and decreased the number of autophagosomes under cold and heat stress [88,101]. In poplars, PagNBR1 is also induced by salt stress. PagNBR1 overexpressing poplars displayed more salt stress tolerance by accelerating antioxidant system activity and autophagy activity [102]. These results demonstrate the important roles of NBR1 in resisting stress conditions via the autophagy pathway.
Atg8-Interacting Proteins 1/2/3 (ATI1/2/3) are AIM-motif-containing proteins identified through a yeast two-hybrid screen for proteins interacting with ATG8. ATI1 and ATI2 are homologous in that each contain two AIM motifs and a transmembrane domain. These two proteins define a newly identified stress-induced compartment that moves along the ER network and is subsequently transported to the vacuole in Arabidopsis plants [103,104]. Salt stress promotes ATI1 protein accumulation. Arabidopsis that are deficient in both homologs (ATI-KD) display increased sensitivity to salt treatment both at the seedling stage and in older plants but no effect was observed on germination. The authors found that ATI1 may play a role in the elimination of damaged plastid and ER proteins produced during salt stress [105]. ATI3 proteins contain a WxxL motif at the C-terminus required for ATG8 interaction. ATI3 homologs are found in dicots but not in other organisms, including monocots. The ati3 mutant plants display hypersensitivity to HS, and the interaction of ATI3 and ATG8 is increased under HS [106].
Dominant suppressor of Kar 2 (DSK2), a ubiquitin-binding receptor, is another ATG8-interacting protein with an AIM motif. DSK2-RNAi Arabidopsis plants display increased sensitivity to drought stress and increased levels of BES1 (BRI1-EMS SUPPRESSOR 1) relative to the WT. BES1 is a master regulator of the brassinosteroid (BR) pathway. DSK2 can be phosphorylated by another negative regulator of the BR pathway, BIN2, which promotes DSK2-ATG8 interaction [107]. These results suggest that DSK2, acting as an autophagy receptor, specifically directs BES1 degradation through the autophagy pathway under drought stress conditions. This link between BR signaling and autophagy means that these processes regulate the plant stress response together. Phytohormones play important roles in plant growth, development, abiotic and biotic stress responses. Phytohormone signals and the autophagy pathway jointly regulate plant responses to abiotic stress [108]; however, the relationship between phytohormones and autophagy in plant abiotic stress regulation remains unclear.
Tryptophan-rich sensory protein/translocator (TSPO) is a type of tryptophan-rich sensory protein/peripheral-type benzodiazepine receptor (TspO/MBR) domain-containing membrane protein, which also has an AIM motif that interacts with ATG8 [109]. The expression of AtTSPO is induced by osmotic and salt stress [110,111]. AtTSPO overexpression makes plants hypersensitive to salt stress, possibly because AtTSPO can bind the plasma membrane aquaporin AtPIP2;7 and regulate its degradation through the autophagy pathway, and the overdegradation of aquaporins impairs cell water status [112]. In addition, similar to NBR1, TSPO is degraded via the autophagy pathway in a manner dependent on ATG5 and the PI3K complex [109]. Another protein from Medicago, cold acclimation-specific 31 (MtCas31), can also regulate MtPIP2;7 stability through the autophagy pathway and participate in drought stress regulation. MtCAS31 directly interacts with MtATG8a in the AIM-like motifs YXXXI and MtPIP2;7, supporting its function in autophagic degradation. The overexpression of MtCAS31 promotes autophagy and MtPIP2;7 degradation under drought stress. These results demonstrate that MtCAS31 functions as a positive regulator of drought stress in Medicago and participates in the drought-induced autophagic degradation of MtPIP2;7 as a cargo receptor [113]. The selected substrates of AtTSPO and MtCAS31 are from the same family of proteins but do not overlap, which may be related to the meticulous regulation of autophagy.
Constitutively stressed 1 (COST1), a plant-specific gene, can also interact with ATG8E but does not have a typical AIM or UIM motif. The cost1 mutant exhibits decreased growth and increased drought tolerance, together with constitutive autophagy and increased expression of drought response genes. COST1 overexpression confers drought hypersensitivity and reduces autophagy. COST1 co-localizes with ATG8E and NBR1 in autophagosomes and directly affects the ATG8E protein level, indicating that it plays a pivotal role in the direct regulation of autophagy [114,115]. The above results illustrate that COST1 is a negative regulator of both drought resistance and autophagy. The increased drought tolerance of the cost1 mutant is due to autophagy activation [114,115,116]. The majority of the autophagy receptors/adaptors identified to date are ATG8-interacting proteins and thus ATG8 is an essential regulator of autophagy; however, the regulation of autophagy is complicated, and autophagy plays a role in almost every part of the plant life cycle. ATG8-independent receptors/adaptors must exist and require further exploration.
5. Conclusions and Prospects
Due to their sessile nature, plants are more vulnerable to abiotic stress than mobile organisms. These stresses challenge agriculture and food security globally. Plants have evolved many systems to alleviate abiotic stress. Autophagy is one of the most important and effective ways to degrade harmful proteins/damaged organs and recycle products/reuse these materials; however, most research on plant autophagy is based on yeast and mammals. Although the core autophagy pathway is conserved in plants, a few genes have yet to be identified in plant genomes, and some genes have expanded into gene families. Do other novel autophagy genes remain to be discovered in plants? Do the genes within families show functional differentiation? These questions warrant future investigation. In recent years, many studies on selected autophagy pathways have emerged, and some plant-specific cargo receptors and conserved cargo receptors have been identified. However, the molecular mechanisms of selective autophagy pathways still need further elucidation. Autophagy can respond to many abiotic stresses, and the same ATG gene can respond to different abiotic stresses. Does abiotic stress exhibit crosstalk in an autophagy-dependent pathway? Reported studies have increased the complexity of the known plant autophagy system. How these studies can be applied to cultivate more stress-resistant and high-yield crops must be given more attention.
Author Contributions
H.C. wrote the article. T.W. and J.D. reviewed and determined the design and structure of the article. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the China Postdoctoral Science Foundation (2019M660876), the China National Postdoctoral Program for Innovative Talents (BX20190371) and the Project for Extramural Scientists of the State Key Laboratory of Agrobiotechnology (2020SKLAB6-15).
Data Availability Statement
Please refer to suggested Data Availability Statements in section “MDPI Research Data Policies” at https://www.mdpi.com/ethics.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Signorelli, S.; Tarkowski, L.P.; Van den Ende, W.; Bassham, D.C. Linking Autophagy to Abiotic and Biotic Stress Responses. Trends Plant Sci. 2019, 24, 413–430. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for Self-Eating in Plant Cells. Annu. Rev. Plant Biol. 2012, 63, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Bao, Y.; Yu, X.; Xia, X.; Liu, C.; Yin, W. Autophagy and Its Regulators in Response to Stress in Plants. Int. J. Mol. Sci. 2020, 21, 8889. [Google Scholar] [CrossRef]
- Feng, Y.C.; He, D.; Yao, Z.Y.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Sienko, K.; Poormassalehgoo, A.; Yamada, K.; Goto-Yamada, S. Microautophagy in Plants: Consideration of Its Molecular Mechanism. Cells 2020, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Orenstein, S.J.; Cuervo, A.M. Chaperone-mediated autophagy: Molecular mechanisms and physiological relevance. Semin Cell Dev. Biol. 2010, 21, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.L.; Li, J.Y.; Wang, M.J.; Song, Z.T.; Liu, J.X. Protein Quality Control in Plant Organelles: Current Progress and Future Perspectives. Mol. Plant 2021, 14, 95–114. [Google Scholar] [CrossRef]
- Kroemer, G.; Marino, G.; Levine, B. Autophagy and the Integrated Stress Response. Mol. Cell. 2010, 40, 280–293. [Google Scholar] [CrossRef]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef]
- Üstün, S.; Hafrén, A.; Hofius, D. Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 2017, 40, 122–130. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T.; Honig, A.; Galili, G. Variations on a theme: Plant autophagy in comparison to yeast and mammals. Protoplasma 2012, 249, 285–299. [Google Scholar] [CrossRef]
- Fu, X.Z.; Zhou, X.; Xu, Y.Y.; Hui, Q.L.; Chun, C.P.; Ling, L.L.; Peng, L.Z. Comprehensive Analysis of Autophagy-Related Genes in Sweet Orange (Citrus sinensis) Highlights Their Roles in Response to Abiotic Stresses. Int. J. Mol. Sci. 2020, 21, 2699. [Google Scholar] [CrossRef]
- Luo, X.; Zhang, Y.; Wu, H.; Bai, J. Drought stress-induced autophagy gene expression is correlated with carbohydrate concentrations in Caragana korshinskii. Protoplasma 2020, 257, 1211–1220. [Google Scholar] [CrossRef]
- Shemi, A.; Ben-Dor, S.; Vardi, A. Elucidating the composition and conservation of the autophagy pathway in photosynthetic eukaryotes. Autophagy 2015, 11, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ding, Z.; Gou, M.; Hu, J.; Wang, Y.; Wang, L.; Wang, Y.; Di, T.; Zhang, X.; Hao, X.; et al. Genome-wide identification, characterization, and expression analysis of tea plant autophagy-related genes (CsARGs) demonstrates that they play diverse roles during development and under abiotic stress. BMC Genom. 2021, 22, 121. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, M.; Wang, E.; Hu, L.; Hawkesford, M.J.; Zhong, L.; Chen, Z.; Xu, Z.; Li, L.; Zhou, Y.; et al. Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. BMC Genom. 2016, 17, 797. [Google Scholar] [CrossRef] [PubMed]
- Stjepanovic, G.; Davies, C.W.; Stanley, R.E.; Ragusa, M.J.; Kim, D.J.; Hurley, J.H. Assembly and dynamics of the autophagy-initiating Atg1 complex. Proc. Natl. Acad. Sci. USA 2014, 111, 12793–12798. [Google Scholar] [CrossRef] [PubMed]
- Chew, L.H.; Lu, S.; Liu, X.; Li, F.K.; Yu, A.Y.; Klionsky, D.J.; Dong, M.Q.; Yip, C.K. Molecular interactions of the Saccharomyces cerevisiae Atg1 complex provide insights into assembly and regulatory mechanisms. Autophagy 2015, 11, 891–905. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Subramani, S. Mechanistic Insights into the Role of Atg11 in Selective Autophagy. J. Mol. Biol. 2020, 432, 104–122. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305. [Google Scholar] [CrossRef]
- Li, F.; Vierstra, R.D. Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy 2014, 10, 1466–1467. [Google Scholar] [CrossRef] [PubMed]
- Suttangkakul, A.; Li, F.; Chung, T.; Vierstra, R.D. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 2011, 23, 3761–3779. [Google Scholar] [CrossRef]
- Augustine, R.C. You Are What You Eat. An ATG1-Independent Path to Autophagy. Plant Cell 2019, 31, 2821–2822. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zheng, C.; Liu, F.; Yang, C.; Zheng, P.; Lu, X.; Tian, J.; Chung, T.; Otegui, M.S.; Xiao, S.; et al. Genetic Analyses of the Arabidopsis ATG1 Kinase Complex Reveal Both Kinase-Dependent and Independent Autophagic Routes during Fixed-Carbon Starvation. Plant Cell 2019, 31, 2973–2995. [Google Scholar] [CrossRef]
- Li, F.; Chung, T.; Vierstra, R.D. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 2014, 26, 788–807. [Google Scholar] [CrossRef]
- Sun, D.X.; Wu, R.B.; Li, P.L.; Yu, L. Phase Separation in Regulation of Aggrephagy. J. Mol. Biol. 2020, 432, 160–169. [Google Scholar] [CrossRef]
- Obara, K.; Sekito, T.; Ohsumi, Y. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006, 17, 1527–1539. [Google Scholar] [CrossRef]
- Yu, Z.Q.; Sun, L.L.; Jiang, Z.D.; Liu, X.M.; Zhao, D.; Wang, H.T.; He, W.Z.; Dong, M.Q.; Du, L.L. Atg38-Atg8 interaction in fission yeast establishes a positive feedback loop to promote autophagy. Autophagy 2020, 16, 2036–2051. [Google Scholar] [CrossRef]
- Liu, F.; Hu, W.; Li, F.; Marshall, R.S.; Zarza, X.; Munnik, T.; Vierstra, R.D. AUTOPHAGY-RELATED14 and Its Associated Phosphatidylinositol 3-Kinase Complex Promote Autophagy in Arabidopsis. Plant Cell 2020, 32, 3939–3960. [Google Scholar] [CrossRef]
- Xu, N.; Gao, X.Q.; Zhao, X.Y.; Zhu, D.Z.; Zhou, L.Z.; Zhang, X.S. Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation. Plant Mol. Biol. 2011, 77, 251–260. [Google Scholar] [CrossRef]
- Wang, W.Y.; Zhang, L.; Xing, S.; Ma, Z.; Liu, J.; Gu, H.; Qin, G.; Qu, L.J. Arabidopsis AtVPS15 plays essential roles in pollen germination possibly by interacting with AtVPS34. J. Genet. Genom. 2012, 39, 81–92. [Google Scholar] [CrossRef]
- Fujiki, Y.; Yoshimoto, K.; Ohsumi, Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007, 143, 1132–1139. [Google Scholar] [CrossRef]
- Liu, F.; Hu, W.; Vierstra, R.D. The Vacuolar Protein Sorting-38 Subunit of the Arabidopsis Phosphatidylinositol-3-Kinase Complex Plays Critical Roles in Autophagy, Endosome Sorting, and Gravitropism. Front. Plant Sci. 2018, 9, 781. [Google Scholar] [CrossRef]
- Lee, H.N.; Zarza, X.; Kim, J.H.; Yoon, M.J.; Kim, S.H.; Lee, J.H.; Paris, N.; Munnik, T.; Otegui, M.S.; Chung, T. Vacuolar Trafficking Protein VPS38 Is Dispensable for Autophagy. Plant Physiol. 2018, 176, 1559–1572. [Google Scholar] [CrossRef]
- Matoba, K.; Kotani, T.; Tsutsumi, A.; Tsuji, T.; Mori, T.; Noshiro, D.; Sugita, Y.; Nomura, N.; Iwata, S.; Ohsumi, Y.; et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 2020, 27, 1185–1224. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Young, L.N. Mechanisms of Autophagy Initiation. Annu. Rev. Biochem. 2017, 86, 225–244. [Google Scholar] [CrossRef]
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368. [Google Scholar] [CrossRef]
- Kobayashi, T.; Suzuki, K.; Ohsumi, Y. Autophagosome formation can be achieved in the absence of Atg18 by expressing engineered PAS-targeted Atg2. FEBS Lett. 2012, 586, 2473–2478. [Google Scholar] [CrossRef]
- Sawa-Makarska, J.; Baumann, V.; Coudevylle, N.; von Bulow, S.; Nogellova, V.; Abert, C.; Schuschnig, M.; Graef, M.; Hummer, G.; Martens, S. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science 2020, 369, 1206. [Google Scholar] [CrossRef]
- Zhuang, X.; Chung, K.P.; Cui, Y.; Lin, W.; Gao, C.; Kang, B.H.; Jiang, L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, e426–e435. [Google Scholar] [CrossRef]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005, 42, 535–546. [Google Scholar] [CrossRef]
- Noda, N.N.; Inagaki, F. Mechanisms of Autophagy. Annu. Rev. Biophys. 2015, 44, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Abreu, S.; Kriegenburg, F.; Gomez-Sanchez, R.; Mari, M.; Sanchez-Wandelmer, J.; Rasmussen, M.S.; Guimaraes, R.S.; Zens, B.; Schuschnig, M.; Hardenberg, R.; et al. Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation. EMBO Rep. 2017, 18, 765–780. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Kotani, T.; Kirisako, H.; Sakoh-Nakatogawa, M.; Oikawa, Y.; Kimura, Y.; Hirano, H.; Yamamoto, H.; Ohsumi, Y.; Nakatogawa, H. Two distinct mechanisms target the autophagy-related E3 complex to the pre-autophagosomal structure. eLife 2019, 8, 17. [Google Scholar] [CrossRef]
- Juris, L.; Montino, M.; Rube, P.; Schlotterhose, P.; Thumm, M.; Krick, R. PI3P binding by Atg21 organises Atg8 lipidation. EMBO J. 2015, 34, 955–973. [Google Scholar] [CrossRef] [PubMed]
- Martens, S.; Fracchiolla, D. Activation and targeting of ATG8 protein lipidation. Cell Discov. 2020, 6, 11. [Google Scholar] [CrossRef]
- Seo, E.; Woo, J.; Park, E.; Bertolani, S.J.; Siegel, J.B.; Choi, D.; Dinesh-Kumar, S.P. Comparative analyses of ubiquitin-like ATG8 and cysteine protease ATG4 autophagy genes in the plant lineage and cross-kingdom processing of ATG8 by ATG4. Autophagy 2016, 12, 2054–2068. [Google Scholar] [CrossRef]
- Zhen, X.; Li, X.; Yu, J.; Xu, F. OsATG8c-Mediated Increased Autophagy Regulates the Yield and Nitrogen Use Efficiency in Rice. Int. J. Mol. Sci. 2019, 20, 4956. [Google Scholar] [CrossRef]
- Fujioka, Y.; Noda, N.N.; Fujii, K.; Yoshimoto, K.; Ohsumi, Y.; Inagaki, F. In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J Biol Chem. 2008, 283, 1921–1928. [Google Scholar] [CrossRef]
- Le Bars, R.; Marion, J.; Satiat-Jeunemaitre, B.; Bianchi, M.W. Folding into an autophagosome ATG5 sheds light on how plants do it. Autophagy 2014, 10, 1861–1863. [Google Scholar] [CrossRef]
- Chung, T.; Phillips, A.R.; Vierstra, R.D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010, 62, 483–493. [Google Scholar] [CrossRef]
- Bu, F.; Yang, M.K.; Guo, X.; Huang, W.; Chen, L. Multiple Functions of ATG8 Family Proteins in Plant Autophagy. Front. Cell Dev. Biol. 2020, 8, 13. [Google Scholar] [CrossRef]
- Luo, S.; Li, X.; Zhang, Y.; Fu, Y.; Fan, B.; Zhu, C.; Chen, Z. Cargo Recognition and Function of Selective Autophagy Receptors in Plants. Int. J. Mol. Sci. 2021, 22, 1013. [Google Scholar] [CrossRef]
- Woltering, S.B.; Isono, E. Knowing When to Self-Eat-Fine-Tuning Autophagy Through ATG8 Iso-forms in Plants. Front. Plant Sci. 2020, 11, 8. [Google Scholar]
- Yan, F.; Zhu, Y.; Zhao, Y.; Wang, Y.; Li, J.; Wang, Q.; Liu, Y. De novo transcriptome sequencing and analysis of salt-, alkali-, and drought-responsive genes in Sophora alopecuroides. BMC Genom. Genom. 2020, 21, 423. [Google Scholar] [CrossRef] [PubMed]
- Arisha, M.H.; Aboelnasr, H.; Ahmad, M.Q.; Liu, Y.; Tang, W.; Gao, R.; Yan, H.; Kou, M.; Wang, X.; Zhang, Y.; et al. Transcriptome sequencing and whole genome expression profiling of hexaploid sweetpotato under salt stress. BMC Genom. Genom. 2020, 21, 197. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Yu, X.; Shao, J.; Liu, Z.; Gao, T.; Zheng, Y.; Zeng, C.; Liang, C.; Chen, C. Transcriptomic profiling and analysis of differentially expressed genes in asparagus bean (Vigna unguiculata ssp. sesquipedalis) under salt stress. PLoS ONE. 2019, 14, e0219799. [Google Scholar] [CrossRef]
- Sun, M.; Huang, D.; Zhang, A.; Khan, I.; Yan, H.; Wang, X.; Zhang, X.; Zhang, J.; Huang, L. Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full-length transcriptome sequencing. BMC Plant Biol. 2020, 20, 323. [Google Scholar] [CrossRef]
- Kumar, R.R.; Goswami, S.; Sharma, S.K.; Kala, Y.K.; Rai, G.K.; Mishra, D.C.; Grover, M.; Singh, G.P.; Pathak, H.; Rai, A.; et al. Harnessing Next Generation Sequencing in Climate Change: RNA-Seq Analysis of Heat Stress-Responsive Genes in Wheat (Triticum aestivum L.). OMICS. 2015, 19, 632–647. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Jogaiah, S.; Burritt, D.J.; Tran, L.P. Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant Cell Environ. 2018, 41, 1972–1983. [Google Scholar] [CrossRef]
- He, J.; Jiang, Z.; Gao, L.; You, C.; Ma, X.; Wang, X.; Xu, X.; Mo, B.; Chen, X.; Liu, L. Genome-Wide Transcript and Small RNA Profiling Reveals Transcriptomic Responses to Heat Stress. Plant Physiol. 2019, 181, 609–629. [Google Scholar] [CrossRef]
- Bashir, K.; Matsui, A.; Rasheed, S.; Seki, M. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Research 2019, 8, F1000. [Google Scholar] [CrossRef]
- Papinski, D.; Schuschnig, M.; Reiter, W.; Wilhelm, L.; Barnes, C.A.; Maiolica, A.; Hansmann, I.; Pfaffenwimmer, T.; Kijanska, M.; Stoffel, I.; et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol. Cell. 2014, 53, 471–483. [Google Scholar] [CrossRef]
- Kang, S.; Shin, K.D.; Kim, J.H.; Chung, T. Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 2018, 37, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Su, Z.Z.; Huang, L.; Xia, F.N.; Qi, H.; Xie, L.J.; Xiao, S.; Chen, Q.F. The AMP-Activated Protein Kinase KIN10 Is Involved in the Regulation of Autophagy in Arabidopsis. Front. Plant Sci. 2017, 8, 11. [Google Scholar] [CrossRef]
- Leshem, Y.; Seri, L.; Levine, A. Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J. 2007, 51, 185–197. [Google Scholar] [CrossRef]
- Ueda, M.; Tsutsumi, N.; Fujimoto, M. Salt stress induces internalization of plasma membrane aquaporin into the vacuole in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2016, 474, 742–746. [Google Scholar] [CrossRef]
- Rana, R.M.; Dong, S.; Ali, Z.; Huang, J.; Zhang, H.S. Regulation of ATG6/Beclin-1 homologs by abiotic stresses and hormones in rice (Oryza sativa L.). Genet. Mol. Res. 2012, 11, 3676–3687. [Google Scholar] [CrossRef]
- Zeng, X.; Zeng, Z.; Liu, C.; Yuan, W.; Hou, N.; Bian, H.; Zhu, M.; Han, N. A barley homolog of yeast ATG6 is involved in multiple abiotic stress responses and stress resistance regulation. Plant Physiol Biochem. 2017, 115, 97–106. [Google Scholar] [CrossRef]
- Wang, Y.; Nishimura, M.T.; Zhao, T.; Tang, D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011, 68, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhu, L.; Wang, Q.; Hou, X. Autophagy-Related 2 Regulates Chlorophyll Degradation under Abiotic Stress Conditions in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 4515. [Google Scholar] [CrossRef]
- Postiglione, A.E.; Muday, G.K. The Role of ROS Homeostasis in ABA-Induced Guard Cell Signaling. Front. Plant Sci. 2020, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, S.; Mano, S.; Oikawa, K.; Hikino, K.; Teshima, K.M.; Kimori, Y.; Nishimura, M.; Shimazaki, K.I.; Takemiya, A. Autophagy controls reactive oxygen species homeostasis in guard cells that is essential for stomatal opening. Proc. Natl. Acad. Sci. USA 2019, 116, 19187–19192. [Google Scholar] [CrossRef]
- Sedaghatmehr, M.; Thirumalaikumar, V.P.; Kamranfar, I.; Marmagne, A.; Masclaux-Daubresse, C.; Balazadeh, S. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019, 42, 1054–1064. [Google Scholar] [CrossRef]
- Zhai, Y.; Guo, M.; Wang, H.; Lu, J.; Liu, J.; Zhang, C.; Gong, Z.; Lu, M. Autophagy, a Conserved Mechanism for Protein Degradation, Responds to Heat, and Other Abiotic Stresses in Capsicum annuum L. Front. Plant Sci. 2016, 7, 131. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy Is Rapidly Induced by Salt Stress and Is Required for Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef]
- Xiong, Y.; Contento, A.L.; Nguyen, P.Q.; Bassham, D.C. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007, 143, 291–299. [Google Scholar] [CrossRef]
- Liu, Y.; Xiong, Y.; Bassham, D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963. [Google Scholar] [CrossRef]
- Sun, X.; Wang, P.; Jia, X.; Huo, L.; Che, R.; Ma, F. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 2018, 16, 545–557. [Google Scholar] [CrossRef]
- Wang, P.; Sun, X.; Yue, Z.Y.; Liang, D.; Wang, N.; Ma, F.W. Isolation and characterization of MdATG18 alpha, a WD40-repeat AuTophaGy-related gene responsive to leaf senescence and abiotic stress in Malus. Sci. Hortic. 2014, 165, 51–61. [Google Scholar] [CrossRef]
- Slavikova, S.; Ufaz, S.; Avin-Wittenberg, T.; Levanony, H.; Galili, G. An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 2008, 59, 4029–4043. [Google Scholar] [CrossRef] [PubMed]
- Olenieva, V.; Lytvyn, D.; Yemets, A.; Bergounioux, C.; Blume, Y. Tubulin acetylation accompanies autophagy development induced by different abiotic stimuli in Arabidopsis thaliana. Cell Biol. Int. 2019, 43, 1056–1064. [Google Scholar] [CrossRef]
- Kuzuoglu-Ozturk, D.; Cebeci Yalcinkaya, O.; Akpinar, B.A.; Mitou, G.; Korkmaz, G.; Gozuacik, D.; Budak, H. Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta 2012, 236, 1081–1092. [Google Scholar] [CrossRef]
- Pei, D.; Zhang, W.; Sun, H.; Wei, X.; Yue, J.; Wang, H. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 2014, 33, 1697–1710. [Google Scholar] [CrossRef]
- Minina, E.A.; Moschou, P.N.; Vetukuri, R.R.; Sanchez-Vera, V.; Cardoso, C.; Liu, Q.; Elander, P.H.; Dalman, K.; Beganovic, M.; Lindberg Yilmaz, J.; et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 2018, 69, 1415–1432. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Yu, J.Q.; Chen, Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 2014, 5, 174. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Yoshimoto, K.; Ohsumi, Y.; Jeon, J.S.; An, G. OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells. 2009, 27, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Guo, Z.; Jia, X.; Sun, X.; Wang, P.; Gong, X.; Ma, F. Increased autophagic activity in roots caused by overexpression of the autophagy-related gene MdATG10 in apple enhances salt tolerance. Plant Sci. 2020, 294, 110444. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, X.; Jia, X.; Ma, F. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 2017, 256, 53–64. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011, 7, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Y.; Qi, J.; Chi, Y.; Fan, B.; Yu, J.Q.; Chen, Z. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014, 10, e1004116. [Google Scholar] [CrossRef] [PubMed]
- Johansen, T.; Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 2020, 432, 80–103. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Z. Broad and Complex Roles of NBR1-Mediated Selective Autophagy in Plant Stress Responses. Cells 2020, 9, 2562. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.J.; Fan, B.; Yu, J.Q.; Chen, Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013, 9, e1003196. [Google Scholar] [CrossRef]
- Jung, H.; Lee, H.N.; Marshall, R.S.; Lomax, A.W.; Yoon, M.J.; Kim, J.; Kim, J.H.; Vierstra, R.D.; Chung, T. Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J. Exp. Bot. 2020, 71, 73–89. [Google Scholar] [CrossRef]
- Thirumalaikumar, V.P.; Gorka, M.; Schulz, K.; Masclaux-Daubresse, C.; Sampathkumar, A.; Skirycz, A.; Vierstra, R.D.; Balazadeh, S. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90 and ROF1. Autophagy 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.; Li, X.; Fang, P.; Xia, X.; Shi, K.; Zhou, Y.; Zhou, J.; Yu, J. Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J. Exp. Bot. 2020, 71, 1092–1106. [Google Scholar] [CrossRef]
- Su, W.; Bao, Y.; Lu, Y.; He, F.; Wang, S.; Wang, D.; Yu, X.; Yin, W.; Xia, X.; Liu, C. Poplar Autophagy Receptor NBR1 Enhances Salt Stress Tolerance by Regulating Selective Autophagy and Antioxidant System. Front. Plant Sci. 2021, 11, 568411. [Google Scholar] [CrossRef]
- Honig, A.; Avin-Wittenberg, T.; Ufaz, S.; Galili, G. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 2012, 24, 288–303. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T.; Michaeli, S.; Honig, A.; Galili, G. ATI1, a newly identified Atg8-interacting protein, binds two different Atg8 homologs. Plant Signal. Behav. 2012, 7, 685–687. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, S.; Honig, A.; Levanony, H.; Peled-Zehavi, H.; Galili, G. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 2014, 26, 4084–4101. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, Z.; Wang, X.; Li, X.; Zhang, Z.; Fan, B.; Zhu, C.; Chen, Z. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy 2018, 14, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective Autophagy of BES1 Mediated by DSK2 Balances Plant Growth and Survival. Dev. Cell. 2017, 41, 33–46.e37. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin, Y.; Li, X.; Guo, S.; Huang, Y.; Xie, Q. Autophagy Dances with Phytohormones upon Multiple Stresses. Plants 2020, 9, 1038. [Google Scholar] [CrossRef]
- Vanhee, C.; Zapotoczny, G.; Masquelier, D.; Ghislain, M.; Batoko, H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 2011, 23, 785–805. [Google Scholar] [CrossRef] [PubMed]
- Balsemão-Pires, E.; Jaillais, Y.; Olson, B.J.; Andrade, L.R.; Umen, J.G.; Chory, J.; Sachetto-Martins, G. The Arabidopsis translocator protein (AtTSPO) is regulated at multiple levels in response to salt stress and perturbations in tetrapyrrole metabolism. BMC Plant Biol. 2011, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Guillaumot, D.; Guillon, S.; Déplanque, T.; Vanhee, C.; Gumy, C.; Masquelier, D.; Morsomme, P.; Batoko, H. The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum-Golgi-localized membrane protein. Plant J. 2009, 60, 242–256. [Google Scholar] [CrossRef]
- Hachez, C.; Veljanovski, V.; Reinhardt, H.; Guillaumot, D.; Vanhee, C.; Chaumont, F.; Batoko, H. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell 2014, 26, 4974–4990. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Q.; Feng, H.; Deng, J.; Zhang, R.; Wen, J.; Dong, J.; Wang, T. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 2020, 16, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Song, W.M.; Wang, P.; Yu, X.; Li, B.; Jiang, C.; Shiu, S.H.; Zhang, H.; Bassham, D.C. COST1 regulates autophagy to control plant drought tolerance. Proc. Natl. Acad. Sci. USA 2020, 117, 7482–7493. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Bassham, D.C. COST1 balances plant growth and stress tolerance via attenuation of autophagy. Autophagy 2020, 16, 1157–1158. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y. Links between drought stress and autophagy in plants. Plant Signal. Behav. 2020, 15, 1779487. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).