Functional Specialization within the EXO70 Gene Family in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. EXO70A1 Can Be Functionally Substituted Only by EXO70A2 in Arabidopsis
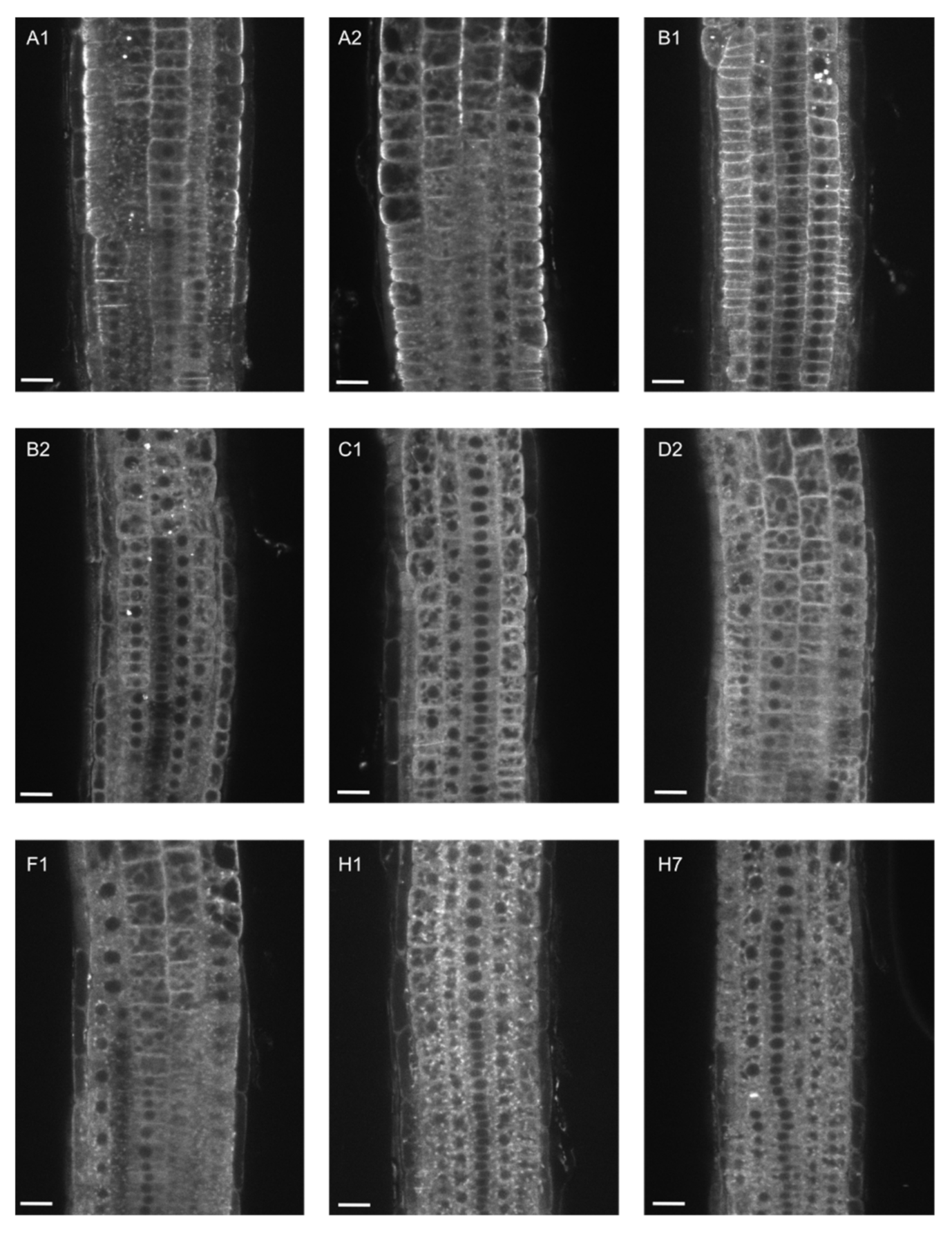
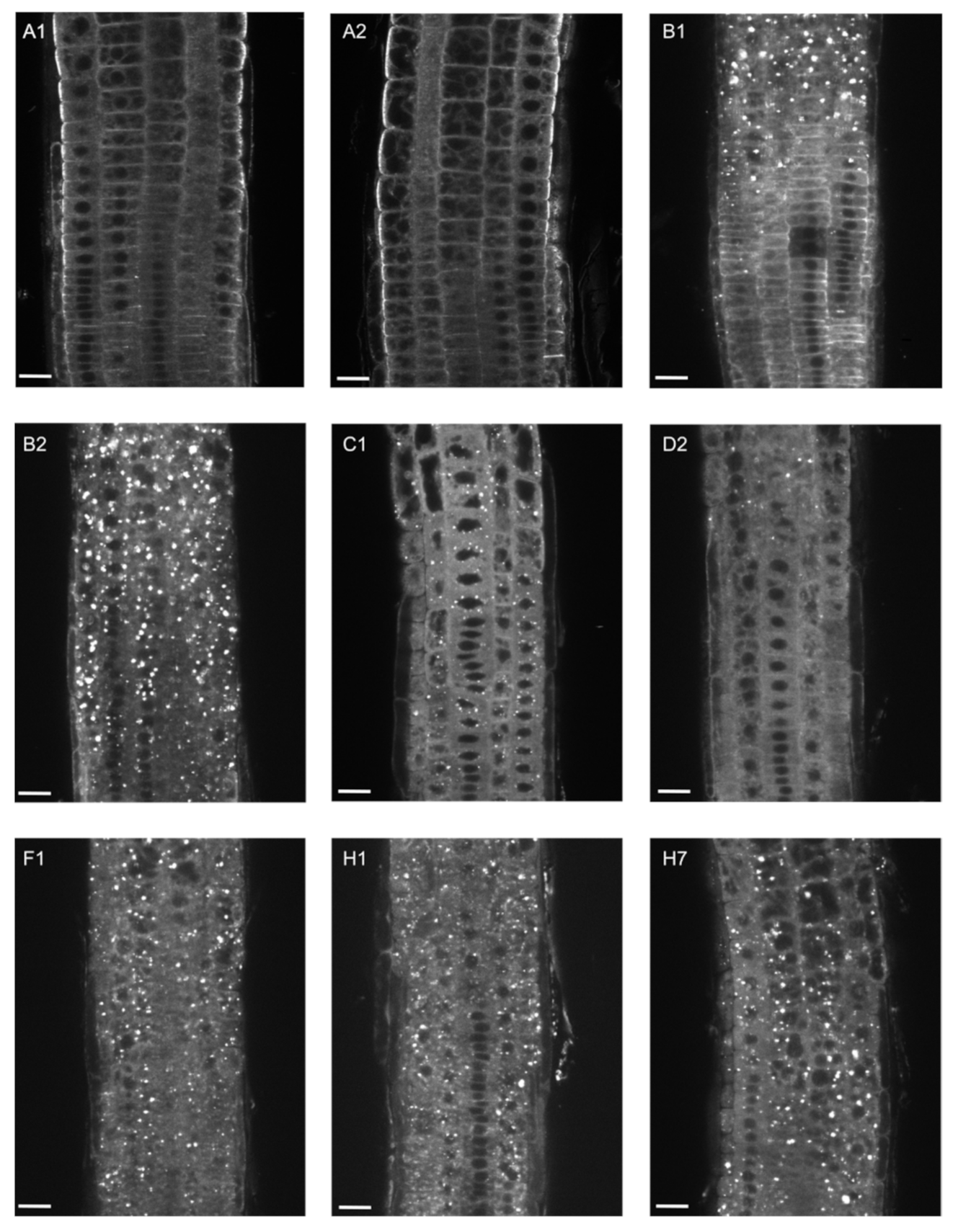
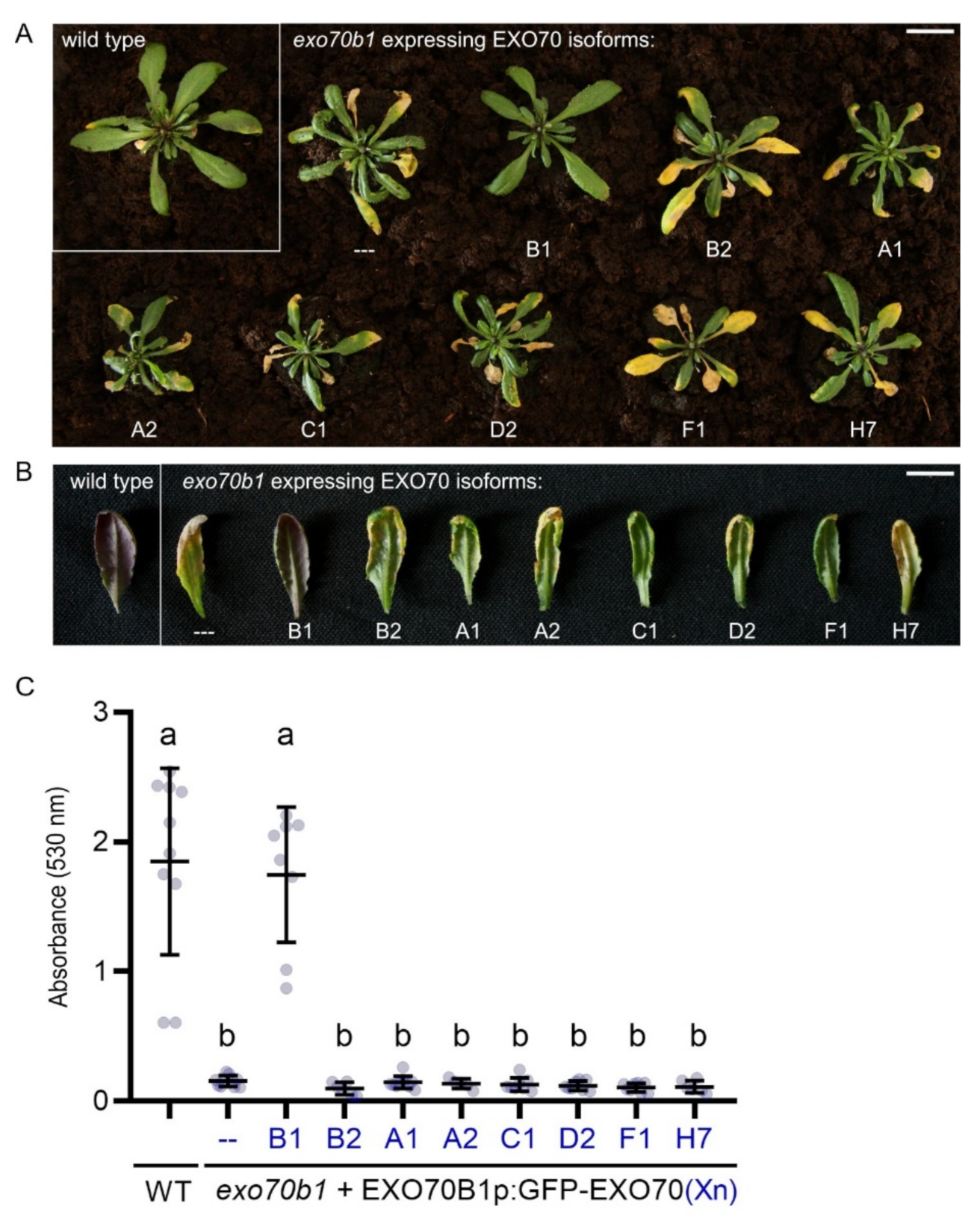
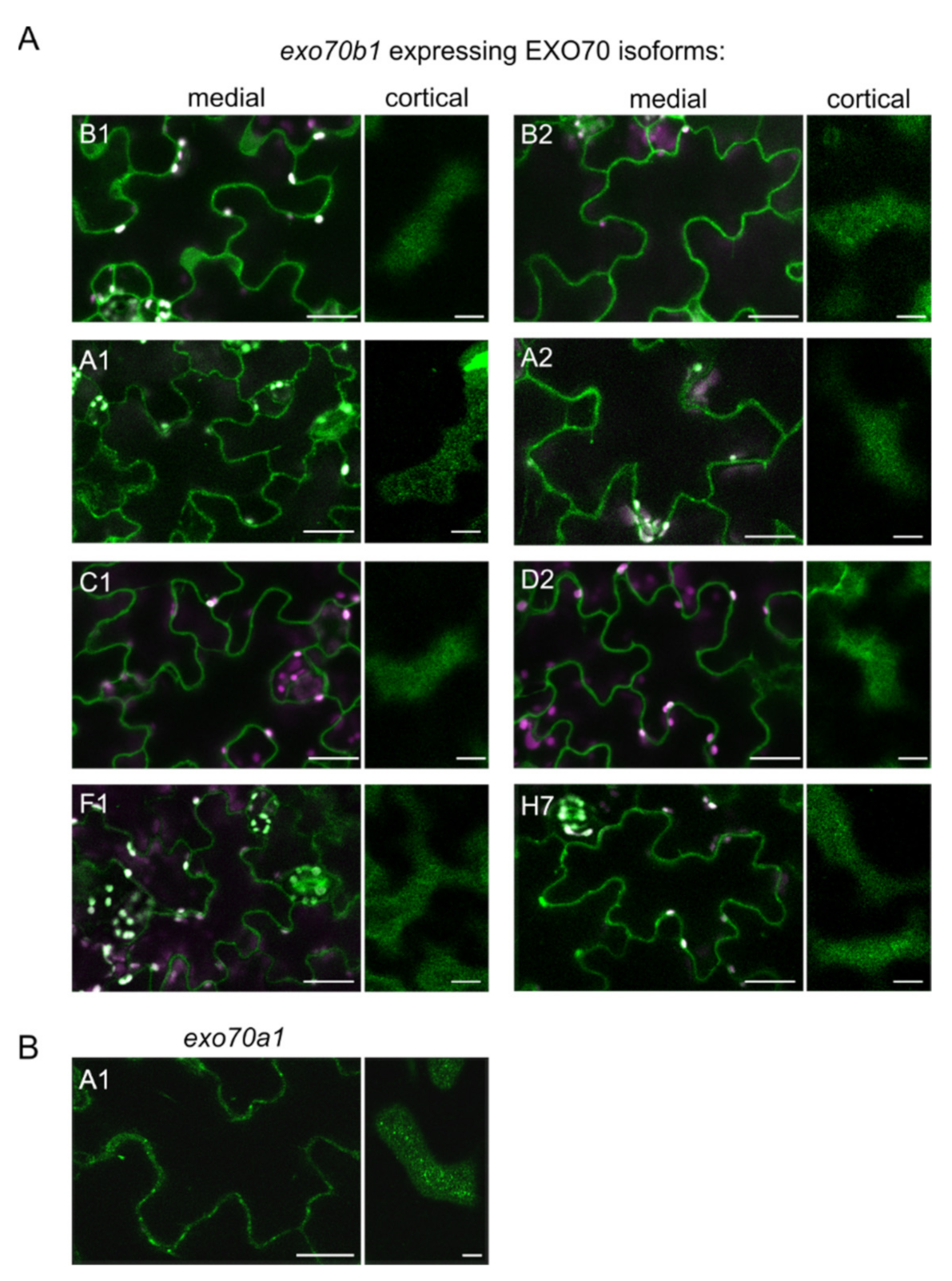
2.2. EXO70B1 Is a Highly Specialized EXO70 Isoform with No Functionally Redundant Paralogs
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Cloning of EXO70 Genes
4.3. Complementation Assays
4.4. Anthocyanin Induction and Assessment
4.5. Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GFP | green fluorescent protein |
| PM | plasma membrane |
| WT | wild type |
| PIP2 | phosphatidylinositol 4,5-bisphosphate |
| CDS | coding sequence |
References
- Novick, P.; Field, C.; Schekman, R. Identification of 23 Complementation Groups Required for Post-Translational Events in the Yeast Secretory Pathway. Cell 1980, 21, 205–215. [Google Scholar] [CrossRef]
- Guo, W.; Grant, A.; Novick, P. Exo84p Is an Exocyst Protein Essential for Secretion. J. Biol. Chem. 1999, 274, 23558–23564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- TerBush, D.R.; Maurice, T.; Roth, D.; Novick, P. The Exocyst Is a Multiprotein Complex Required for Exocytosis in Saccharomyces Cerevisiae. EMBO J. 1996, 15, 6483–6494. [Google Scholar] [CrossRef] [PubMed]
- Elias, M. The Exocyst Complex in Plants. Cell Biol. Int. 2003, 27, 199–201. [Google Scholar] [CrossRef]
- Cvrčková, F.; Grunt, M.; Bezvoda, R.; Hála, M.; Kulich, I.; Rawat, A.; Zárský, V. Evolution of the Land Plant Exocyst Complexes. Front. Plant Sci. 2012, 3, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Žárský, V.; Sekereš, J.; Kubátová, Z.; Pečenková, T.; Cvrčková, F. Three Subfamilies of Exocyst EXO70 Family Subunits in Land Plants: Early Divergence and Ongoing Functional Specialization. J. Exp. Bot. 2020, 71, 49–62. [Google Scholar] [CrossRef]
- Fendrych, M.; Synek, L.; Pecenková, T.; Drdová, E.J.; Sekeres, J.; de Rycke, R.; Nowack, M.K.; Zársky, V. Visualization of the Exocyst Complex Dynamics at the Plasma Membrane of Arabidopsis Thaliana. Mol. Biol. Cell 2013, 24, 510–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fendrych, M.; Synek, L.; Pecenková, T.; Toupalová, H.; Cole, R.; Drdová, E.; Nebesárová, J.; Sedinová, M.; Hála, M.; Fowler, J.E.; et al. The Arabidopsis Exocyst Complex Is Involved in Cytokinesis and Cell Plate Maturation. Plant Cell 2010, 22, 3053–3065. [Google Scholar] [CrossRef] [Green Version]
- Synek, L.; Schlager, N.; Eliás, M.; Quentin, M.; Hauser, M.-T.; Zárský, V. AtEXO70A1, a Member of a Family of Putative Exocyst Subunits Specifically Expanded in Land Plants, Is Important for Polar Growth and Plant Development. Plant J. 2006, 48, 54–72. [Google Scholar] [CrossRef] [Green Version]
- Drdová, E.J.; Synek, L.; Pečenková, T.; Hála, M.; Kulich, I.; Fowler, J.E.; Murphy, A.S.; Zárský, V. The Exocyst Complex Contributes to PIN Auxin Efflux Carrier Recycling and Polar Auxin Transport in Arabidopsis. Plant J. 2013, 73, 709–719. [Google Scholar] [CrossRef]
- Tan, X.; Feng, Y.; Liu, Y.; Bao, Y. Mutations in Exocyst Complex Subunit SEC6 Gene Impaired Polar Auxin Transport and PIN Protein Recycling in Arabidopsis Primary Root. Plant Sci. 2016, 250, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Cole, R.A.; McInally, S.A.; Fowler, J.E. Developmentally Distinct Activities of the Exocyst Enable Rapid Cell Elongation and Determine Meristem Size during Primary Root Growth in Arabidopsis. BMC Plant Biol. 2014, 14, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, T.-J.; Hochholdinger, F.; Sauer, M.; Bruce, W.; Schnable, P.S. The roothairless1 Gene of Maize Encodes a Homolog of sec3, Which Is Involved in Polar Exocytosis. Plant Physiol. 2005, 138, 1637–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalmbach, L.; Hématy, K.; De Bellis, D.; Barberon, M.; Fujita, S.; Ursache, R.; Daraspe, J.; Geldner, N. Transient Cell-Specific EXO70A1 Activity in the CASP Domain and Casparian Strip Localization. Nat. Plants 2017, 3, 17058. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, M.; Yu, D.; Ren, S.; Sun, S.; Liu, L.; Ketelaar, T.; Emons, A.-M.C.; Liu, C.-M. EXO70A1-Mediated Vesicle Trafficking Is Critical for Tracheary Element Development in Arabidopsis. Plant Cell 2013, 25, 1774–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vukašinović, N.; Oda, Y.; Pejchar, P.; Synek, L.; Pečenková, T.; Rawat, A.; Sekereš, J.; Potocký, M.; Žárský, V. Microtubule-Dependent Targeting of the Exocyst Complex Is Necessary for Xylem Development in Arabidopsis. New Phytol. 2017, 213, 1052–1067. [Google Scholar] [CrossRef]
- Marković, V.; Cvrčková, F.; Potocký, M.; Kulich, I.; Pejchar, P.; Kollárová, E.; Synek, L.; Žárský, V. EXO70A2 Is Critical for Exocyst Complex Function in Pollen Development. Plant Physiol. 2020, 184, 1823–1839. [Google Scholar] [CrossRef]
- Beuder, S.; Dorchak, A.; Bhide, A.; Moeller, S.R.; Petersen, B.L.; MacAlister, C.A. Exocyst Mutants Suppress Pollen Tube Growth and Cell Wall Structural Defects of Hydroxyproline O-Arabinosyltransferase Mutants. Plant J. 2020, 103, 1399–1419. [Google Scholar] [CrossRef]
- Ogura, T.; Goeschl, C.; Filiault, D.; Mirea, M.; Slovak, R.; Wolhrab, B.; Satbhai, S.B.; Busch, W. Root System Depth in Arabidopsis Is Shaped by EXOCYST70A3 via the Dynamic Modulation of Auxin Transport. Cell 2019, 178, 400–412.e16. [Google Scholar] [CrossRef]
- Kulich, I.; Pečenková, T.; Sekereš, J.; Smetana, O.; Fendrych, M.; Foissner, I.; Höftberger, M.; Zárský, V. Arabidopsis Exocyst Subcomplex Containing Subunit EXO70B1 Is Involved in Autophagy-Related Transport to the Vacuole. Traffic 2013, 14, 1155–1165. [Google Scholar] [CrossRef] [Green Version]
- Hong, D.; Jeon, B.W.; Kim, S.Y.; Hwang, J.-U.; Lee, Y. The ROP2-RIC7 Pathway Negatively Regulates Light-Induced Stomatal Opening by Inhibiting Exocyst Subunit Exo70B1 in Arabidopsis. New Phytol. 2016, 209, 624–635. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T.; Rui, L.; Li, J.; Nishimura, M.T.; Vogel, J.P.; Liu, N.; Liu, S.; Zhao, Y.; Dangl, J.L.; Tang, D. A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the exo70B1 Mutant. PLoS Genet. 2015, 11, e1004945. [Google Scholar] [CrossRef] [PubMed]
- Pecenková, T.; Hála, M.; Kulich, I.; Kocourková, D.; Drdová, E.; Fendrych, M.; Toupalová, H.; Zársky, V. The Role for the Exocyst Complex Subunits Exo70B2 and Exo70H1 in the Plant-Pathogen Interaction. J. Exp. Bot. 2011, 62, 2107–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stegmann, M.; Anderson, R.G.; Ichimura, K.; Pecenkova, T.; Reuter, P.; Žársky, V.; McDowell, J.M.; Shirasu, K.; Trujillo, M. The Ubiquitin Ligase PUB22 Targets a Subunit of the Exocyst Complex Required for PAMP-Triggered Responses in Arabidopsis. Plant Cell 2012, 24, 4703–4716. [Google Scholar] [CrossRef] [Green Version]
- Synek, L.; Vukašinović, N.; Kulich, I.; Hála, M.; Aldorfová, K.; Fendrych, M.; Žárský, V. EXO70C2 Is a Key Regulatory Factor for Optimal Tip Growth of Pollen. Plant Physiol. 2017, 174, 223–240. [Google Scholar] [CrossRef] [Green Version]
- Acheampong, A.K.; Shanks, C.; Cheng, C.-Y.; Schaller, G.E.; Dagdas, Y.; Kieber, J.J. EXO70D Isoforms Mediate Selective Autophagic Degradation of Type-A ARR Proteins to Regulate Cytokinin Sensitivity. Proc. Natl. Acad. Sci. USA 2020, 117, 27034–27043. [Google Scholar] [CrossRef]
- Wang, J.; Ding, Y.; Wang, J.; Hillmer, S.; Miao, Y.; Lo, S.W.; Wang, X.; Robinson, D.G.; Jiang, L. EXPO, an Exocyst-Positive Organelle Distinct from Multivesicular Endosomes and Autophagosomes, Mediates Cytosol to Cell Wall Exocytosis in Arabidopsis and Tobacco Cells. Plant Cell 2010, 22, 4009–4030. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Wang, J.; Chun Lai, J.H.; Ling Chan, V.H.; Wang, X.; Cai, Y.; Tan, X.; Bao, Y.; Xia, J.; Robinson, D.G.; et al. Exo70E2 Is Essential for Exocyst Subunit Recruitment and EXPO Formation in Both Plants and Animals. Mol. Biol. Cell 2014, 25, 412–426. [Google Scholar] [CrossRef]
- Ostertag, M.; Stammler, J.; Douchkov, D.; Eichmann, R.; Hückelhoven, R. The Conserved Oligomeric Golgi Complex Is Involved in Penetration Resistance of Barley to the Barley Powdery Mildew Fungus. Mol. Plant Pathol. 2013, 14, 230–240. [Google Scholar] [CrossRef]
- Fujisaki, K.; Abe, Y.; Ito, A.; Saitoh, H.; Yoshida, K.; Kanzaki, H.; Kanzaki, E.; Utsushi, H.; Yamashita, T.; Kamoun, S.; et al. Rice Exo70 Interacts with a Fungal Effector, AVR-Pii, and Is Required for AVR-Pii-Triggered Immunity. Plant J. 2015, 83, 875–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulich, I.; Vojtíková, Z.; Glanc, M.; Ortmannová, J.; Rasmann, S.; Žárský, V. Cell Wall Maturation of Arabidopsis Trichomes Is Dependent on Exocyst Subunit EXO70H4 and Involves Callose Deposition. Plant Physiol. 2015, 168, 120–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulich, I.; Vojtíková, Z.; Sabol, P.; Ortmannová, J.; Neděla, V.; Tihlaříková, E.; Žárský, V. Exocyst Subunit EXO70H4 Has a Specific Role in Callose Synthase Secretion and Silica Accumulation. Plant Physiol. 2018, 176, 2040–2051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Pumplin, N.; Ivanov, S.; Harrison, M.J. EXO70I Is Required for Development of a Sub-Domain of the Periarbuscular Membrane during Arbuscular Mycorrhizal Symbiosis. Curr. Biol. 2015, 25, 2189–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Synek, L.; Pleskot, R.; Sekereš, J.; Serrano, N.; Vukašinović, N.; Ortmannová, J.; Klejchová, M.; Pejchar, P.; Batystová, K.; Gutkowska, M.; et al. Plasma membrane electrostatic signature targets the plant exocyst complex via the EXO70 subunit. Proc. Natl. Acad. Sci. USA. under review.
- Wu, C.; Tan, L.; van Hooren, M.; Tan, X.; Liu, F.; Li, Y.; Zhao, Y.; Li, B.; Rui, Q.; Munnik, T.; et al. Arabidopsis EXO70A1 Recruits Patellin3 to the Cell Membrane Independent of Its Role as an Exocyst Subunit. J. Integr. Plant Biol. 2017, 59, 851–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Liu, N.; Gao, C.; Cai, H.; Romeis, T.; Tang, D. The Arabidopsis Exocyst Subunits EXO70B1 and EXO70B2 Regulate FLS2 Homeostasis at the Plasma Membrane. New Phytol. 2020, 227, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Irani, N.G.; Lu, Y.; Riedl, K.; Schwartz, S.; Grotewold, E. The formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol. Plant 2010, 3, 78–90. [Google Scholar] [CrossRef]
- Pečenková, T.; Potocká, A.; Potocký, M.; Ortmannová, J.; Drs, M.; Drdová, E.J.; Pejchar, P.; Synek, L.; Soukupová, H.; Žárský, V.; et al. Redundant and Diversified Roles Among Selected Arabidopsis Thaliana EXO70 Paralogs During Biotic Stress Responses. Front. Plant Sci. 2020, 11, 960. [Google Scholar] [CrossRef]
- Sabol, P.; Kulich, I.; Žárský, V. RIN4 Recruits the Exocyst Subunit EXO70B1 to the Plasma Membrane. J. Exp. Bot. 2017, 68, 3253–3265. [Google Scholar] [CrossRef]
- Vukašinović, N.; Cvrčková, F.; Eliáš, M.; Cole, R.; Fowler, J.E.; Žárský, V.; Synek, L. Dissecting a Hidden Gene Duplication: The Arabidopsis Thaliana SEC10 Locus. PLoS ONE 2014, 9, e94077. [Google Scholar]
- Hála, M.; Cole, R.; Synek, L.; Drdová, E.; Pecenková, T.; Nordheim, A.; Lamkemeyer, T.; Madlung, J.; Hochholdinger, F.; Fowler, J.E.; et al. An Exocyst Complex Functions in Plant Cell Growth in Arabidopsis and Tobacco. Plant Cell 2008, 20, 1330–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi, M.; Bleys, A.; Vanderhaeghen, R.; Hilson, P. Building Blocks for Plant Gene Assembly. Plant Physiol. 2007, 145, 1183–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method forAgrobacterium-Mediated Transformation ofArabidopsis Thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marković, V.; Kulich, I.; Žárský, V. Functional Specialization within the EXO70 Gene Family in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7595. https://doi.org/10.3390/ijms22147595
Marković V, Kulich I, Žárský V. Functional Specialization within the EXO70 Gene Family in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(14):7595. https://doi.org/10.3390/ijms22147595
Chicago/Turabian StyleMarković, Vedrana, Ivan Kulich, and Viktor Žárský. 2021. "Functional Specialization within the EXO70 Gene Family in Arabidopsis" International Journal of Molecular Sciences 22, no. 14: 7595. https://doi.org/10.3390/ijms22147595
APA StyleMarković, V., Kulich, I., & Žárský, V. (2021). Functional Specialization within the EXO70 Gene Family in Arabidopsis. International Journal of Molecular Sciences, 22(14), 7595. https://doi.org/10.3390/ijms22147595





